Abstract
The Chinese mitten crab (CMC) also known as Eriocheir sinensis has great significance in the aquaculture industry. The bacterial communities inhabiting the CMC’s gut may differ depending on the host habitat and can aid in their normal biological functioning. These microbes are also known to have certain effects on their flavor. In this study, we utilized MiSeq high-throughput sequencing technology to explore the diversity of bacterial communities in the gut of CMCs from three different geographical locations in Korea: the Geum (GD), Han (HD), and Tamjin (TD) rivers. Although most of the environmental parameters were similar at the three sites, significant differences in conductivity (CDS), dissolved oxygen (DO), and salinity were observed. The results show that CMCs sampled from these locations exhibited distinct microbial composition and abundance. For example, the genus Candidatus Hepatoplasma displayed significantly higher abundance in CMCs from HD than those in the other locations, suggesting nutritional stress. Similarly, the crabs collected from TD showed a higher abundance of pathogenic Helicobacter than those from HD and GD sites. We also observed differences in the amino acid, nucleotide, and lactic acid concentrations between different tissues such as the muscle, hepatopancreas, and testis of CMCs. However, only small differences were observed when these characteristics were compared in CMCs from different locations. Our results offer important insights into the intestinal bacterial composition in CMCs which in turn may help in designing better culturing strategies for these important species of crabs.
Key Contribution:
The present study describes the differences in the gut microbiome of Chinese mitten crabs collected from three distinct rivers in Korea. The findings of this study might be useful for future studies on improving CMC aquaculture.
1. Introduction
Eriocheir sinensis, commonly known as the Chinese mitten crab (CMC), is a freshwater crustacean of huge economic significance [1]. The adult CMC has the unique characteristics of spending its entire life in freshwater and then migrating to saline (brackish) water to undergo reproduction [2]. It is native to China and Korea, and in Korea, it is distributed along the west coast, including the Geum and Han rivers. The CMC is also found as an invasive crab in the United States of America (USA) and Europe [3,4]. CMCs rank among one of the most important aquaculture organisms in East Asia and are highly sought after by consumers for their unique flavor, characterized by umami and distinct sweetness [5,6]. Based on the recent report of The State of World Fisheries and Aquaculture (SOFIA), the annual worldwide production of CMCs comes close to 800 thousand tons and is among one of the top three crustaceans after whiteleg shrimp and red swamp crawfish (https://www.fao.org/home/en/, accessed on 6 April 2024). In China, the yield of CMCs has swiftly increased in the last three decades and reached around 800,000 tons in 2020, making it the top ranking aquacultural crab species [7]. According to 2009 records from Korea, about 2000 tons of mitten crabs are consumed by Korean people each year, of which 1500 tons are imported from China [8]. However, with the recent expansion of CMC farming, it was reported that the Korean government purchased 12,057,751 domestically farmed CMCs for KRW 2,177,873,000 (approximately USD 1,611,730) and released them in nature [9]. These trends have further expanded the economic scale of the aquacultured crab market in Korea and increased the focus on the quantity and quality of aquacultured CMCs.
In the aquaculture of CMCs, factors that have been shown to influence taste and quality include the geographical environment of the habitat, gut microbiota, and diet [10]. Among them, the environmental parameters of the habitat play crucial roles in the aquatic ecosystem, and any unfavorable environmental condition may create a stressful situation for CMCs and their capability for homeostasis may be disrupted [11]. These environmental parameters can regulate the survivability of CMCs by influencing different physiological processes in animals such as osmoregulation, growth, development, and reproduction [12,13,14]. Besides their impact on growth, development, and other physico-chemical processes, environmental parameters such as salinity have an impact on the flavor and meat quality as well. It has been reported that the environment of CMCs has a significant impact on their flavor as well as market price [10].
The digestive tract harbors trillions of microbial cells known as gut microbiota, which plays essential roles in maintaining the host’s health, by performing many beneficial functions for the host such as balancing the immune response, absorbing nutrients, and maintaining homeostasis [14,15]. The host environment is also one of the most crucial factors that can also regulate the composition of gut microbial communities [16,17]. Environmental factors such as salinity can lead to stress (hyposaline or hypersaline) to CMCs, which can influence the growth condition of the host [18] and can also affect the host microbiota. For example, depending on the salinity tolerance, strict freshwater bacteria may be destroyed while saline tolerant bacteria may live, and marine bacteria could immigrate [19] in the gastrointestinal trails of a gastropod living in freshwaters and mesohaline waters. In spite of these facts, a lot of information is available regarding the mechanisms of the intestinal immune regulation of the host and its relationship with the symbiotic microorganisms on the gut mucosa surface [20]. However, limited information regarding the gut immune mechanisms of invertebrates at the barrier epithelia [21] is available especially for CMCs. Additionally, the relationship between the salinity, intestinal microbiota, and growth condition of the CMCs remains unclear. Therefore, maintaining a functional and stable gut microbiota is essential not only for the health of CMCs but it can also have a significant impact on their flavor and market price [10].
To the best of our knowledge, no studies have been conducted to identify any potential relationship between the geographical environment of the habitat and the flavor characteristics of cultivated CMCs in Korea. To date, research on the flavor properties of CMCs has mainly focused on analyzing taste-related characteristics such as fatty acids, amino acids, and nucleotides and their association with diet [6]. Recently, although some studies have attempted to identify the differences in the intestinal microbiome and flavor substances in CMCs from freshwater lakes and rivers [10], these studies represent geographically distant locations from three main rivers (Geum, Han, and Tamjin rivers) in Korea that represent the CMC habitat. In this work, an attempt was made to determine the influence of different geographical locations on the gut microbiota of CMCs and its potential implications on crab physiology. To investigate the differences in the gut bacterial communities of CMCs inhabiting different environments, we exploited MiSeq technology to sequence the bacterial 16S rRNA sequences. Additionally, to understand the potential associations between different geographical environments of the CMCs’ habitat and flavor characteristics, we also analyzed the taste-determining metabolites through HPLC analysis. Therefore, this is the first study to investigate the influence of geographical location on the gut microbiome of CMCs in Korea, which in turn may affect their culturing and flavor. Our results may suggest what environmental conditions may be a good solution for culturing CMCs within Korea and may have consequences for their flavor and quality, cultivation, and adaptation.
2. Materials and Methods
2.1. Sample Collection
A total of 54 healthy adult male crabs were collected from three different locations in Korea, namely 18 specimens from the Geum River (GD), Chungcheongnam-do, 18 specimens from the Han River (HD), Seoul, and 18 specimens from the Tamjin River (TD), Jeollanam-do (Figure 1, Table S1). The environmental factors, including water temperature (WT), salinity, pH, and dissolved oxygen (DO), were measured using a portable meter (YK-2001PHA, LUTRON Co., Taipei City, Taiwan) during sample collection (Table S2). The crabs were transported to the laboratory while alive and stabilized for an hour. To extract DNA, the carapace of twenty-seven adult male crabs was irrigated thoroughly using sterile water and disinfected with 70% ethanol for three minutes in the laboratory. The crabs were dissected immediately after washing. For the DNA extraction and bacterial 16S rRNA gene sequencing of the gut microbiome, the digestive tracts of CMCs were aseptically removed and stored at −80 °C for subsequent analysis. For HPLC analysis, the muscle, hepatopancreas, and testis were extracted from twenty-seven healthy adult male crabs and immediately stored at −80 °C for subsequent analysis. The remaining tissues were stored in 99% ethanol at room temperature as voucher specimens.
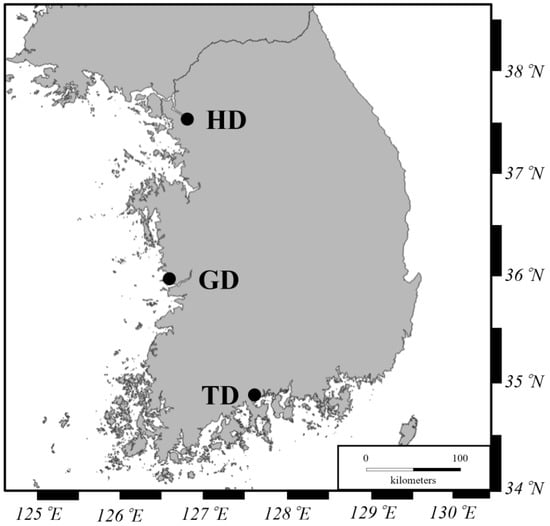
Figure 1.
Three sampling locations for Chinese mitten crabs in Korea; HD = Han River, GD = Geum River, and TD = Tamjin River.
2.2. Species Identification
Species identification was performed using tissue from the pereopods in each specimen. DNA extraction was performed using a Qiagen DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. A Maestro Nano spectrophotometer (Maestrogen, Hsinchu, Taiwan) was used to evaluate the purity and concentration of the extracted DNA. To amplify the mitochondrial cytochrome c oxidase subunit I (COI) gene, polymerase chain reaction (PCR) was performed using the LCO1490/HCO2198 primer set [22]. The PCR conditions included an initial denaturation at 95 °C for 2 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 48 °C for 45 s, and extension at 72 °C for 1 min, with a final extension step at 72 °C for 5 min. After amplification, the PCR products were observed by electrophoresis on 1% agarose gels in tris-acetate buffer. All specimens were identified as E. sinensis (accession numbers: OR842741-OR842794) by both morphological characteristics and molecular phylogenetic analysis based on the COI sequence. The preserved E. sinensis specimens, stored in 99% absolute ethanol, were deposited in the Department of Biotechnology at Sangmyung University, assigned voucher numbers from SMU00218 to SMU00271.
2.3. DNA Extraction and Bacterial 16S rDNA Gene Sequencing of CMC Gut
About 0.5 g of the intestinal tissue samples was placed in 1.5 mL microcentrifuge tubes, and the DNA was isolated using Qiagen DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. The bacterial 16S rDNA V3-V4 regions were utilized to analyze and compare the gut microbiota among CMCs originating from various habitats [23]. The 16S rDNA was sequenced using high-throughput technology with the MiSeq platform (Illumina, San Diego, CA, USA). The paired-end sequencing data of CMCs from three different locations were deposited in NCBI’s Sequence Read Archive (SRA) SRR26926093~SRR26926095 under BioProject PRJNA1044169.
2.4. Bioinformatic Analyses
The paired-end sequencing data of CMCs from three different locations were first imported into the QIIME2 ver 2022.2 [24] pipeline. Next, the DADA2 plugin within the QIIME2 pipeline was used to improve the joining of forward and reverse reads by filtering the poorest quality ends of the reads and keeping the read length enough for what is required for overlap. The output of this step comprises the amplicon sequencing variant (ASV) table and representative sequences. The sequencing depth was estimated using rarefaction analysis. To estimate the genera richness and α-diversity, the Simpson, Chao 1, and Shannon index and Good’s coverage in each sample were calculated by exporting the QIIME2 output to the R package Phyloseq ver 1.32.0 [25]. In general, microbiome datasets are sparse; therefore, it is indispensable to filter the dataset by excluding the low quality or superfluous variables (ASVs) for better downstream analyses [26]. In this work, we retained only those ASVs which have at least two counts in at least 11% of the samples and normalized the data by the rarefying method. To assign a species-level taxonomy to the unclassified species, we performed a BLAST-based search against the NCBI non-redundant nucleotide database. The R vegan (https://github.com/vegandevs/vegan. accessed on 14 February 2024) package was used to perform the statistical analyses.
2.5. Determination of Free Amino Acid Content
Analyses of taste-determining metabolites through HPLC analysis were performed to understand the potential associations between different geographical environments of the CMCs’ habitat and flavor characteristics. To determine the free amino acid content in hepatopancreas, muscle, and testis tissues, equal amounts of tissue samples were taken from nine crabs and mixed to a total of 1 g, and the mixture was quickly ground using a mortar and pestle. Then samples were pretreated according to Tao et al. [10]. The concentration of free amino acid was determined using the high-performance liquid chromatography–fluorescence detector (HPLC-FLD) system (Shimadzu HPLC 40 Series-FLD; Kyoto, Japan) by Korea Quality Testing Institute (KQT, Suwon, Korea). Pre-column derivatization with FMOC-Cl (9-fluorenylmethyl chloroformate) and OPA (o-Phthalaldehyde) was carried out. The column used was a C18 Column (250 × 4.6 mm, 5 μm). The gradient elution for the HPLC analysis was conducted using 20 mM KH2PO4 solution with 225 μL TEA (triethylamine) and 5 mL THF (tetrahydrofuran) per liter (mobile phase A) and the mixture of deionized water, acetonitrile, and methanol in a volume ratio of 15:45:40 (mobile phase B). The linear gradient conditions are shown in Table S3. The operating conditions were as follows: a flow rate of 0.8 mL/minute; column temperature, 35 °C; injection volume, 2 μL; excitation wavelength, 350 nm and 266 nm; and emission wavelength, 450 nm and 305 nm.
2.6. Determination of Free Nucleotide Content
To determine the free nucleotide content in hepatopancreas, muscle, and testis tissues, samples were pretreated according to modified methods of Tao et al. [10]. Equal amounts of tissue samples were taken from nine crabs and mixed to a total of 1 g, and the mixture was quickly ground using a mortar and pestle. Then, 5 mL of 0.6 M perchloric acid was added, homogenized for 2 min, and shaken for 20 min. After that, the mixture was centrifuged at 13,500× g for 10 min, and the pH of the supernatant was adjusted to 7.0 using 1.0 M potassium hydroxide and subsequently centrifuged at 13,500× g for 10 min. The supernatant was collected, adjusted to a total volume of 10 mL, filtered through a 0.45 μm membrane filter, and finally analyzed by the HPLC–diode array detector system (HPLC-DAD, Hitachi, Tokyo, Japan). The analysis was carried out under the following conditions: column, INNO column C18 (250 × 4.6 mm, 5 μm); detector, DAD at 260 nm; and mobile phase, 13.6 g of KH2PO4 dissolved in 1 L water (pH 5.6) (A) and 13.6 g of KH2PO4 dissolved in 750 mL water and 150 mL methanol (pH 5.6) (B). The gradient analysis was set as follows: start, 100% A; 30 min, 100% A; 65 min, 100% B; 90 min, 100% B; 95 min, 100% A; and 130 min, 100% A at a flow rate of 0.5 mL/minute.
2.7. Determination of Lactic Acid Content
To determine the lactic acid content in hepatopancreas, muscle, and testis tissues, equal amounts of tissue samples were taken from nine crabs and mixed to a total of 1 g, and the mixture was quickly ground using a mortar and pestle. Then, samples were pretreated according to Tao et al. [10]. The concentration of lactic acid was determined using the HPLC-RID (refractive index detector) system (Shimadzu HPLC-RID-10A; Kyoto, Japan). The column used was a Shodex SUGAR SH1011 H+ ion exclusion column (8 mm × 300 mm). The isocratic elution was performed with 0.005 N H2SO4 at a flow rate of 0.8 mL/minute. The temperature of the column oven was set to 50 °C, and the sample injection volume was 20 μL.
3. Results
3.1. Differences in the Environmental Parameters
The environmental parameters for each sampling site are provided in Table S2. Since Korea is a country with a narrow range of latitudes, the climate is generally similar across the region; the WT in the geographical locations was similar. Overall, the WT ranged between 16.3 °C (HD) and 19.2 °C (TD). The WT in GD was found to be 17.3 °C. However, we observed significant differences in a few parameters such as CD (conductivity), DO, and salinity. Specifically, DO levels were the highest (21.6 mg/L) in HD, followed by TD (12.7 mg/L) and GD (10.2 mg/L), respectively. In contrast, GD showed the highest concentration of CD and the lowest salinity.
3.2. A 16S rDNA Metabarcoding Analysis of the CMC Gut Microbiome
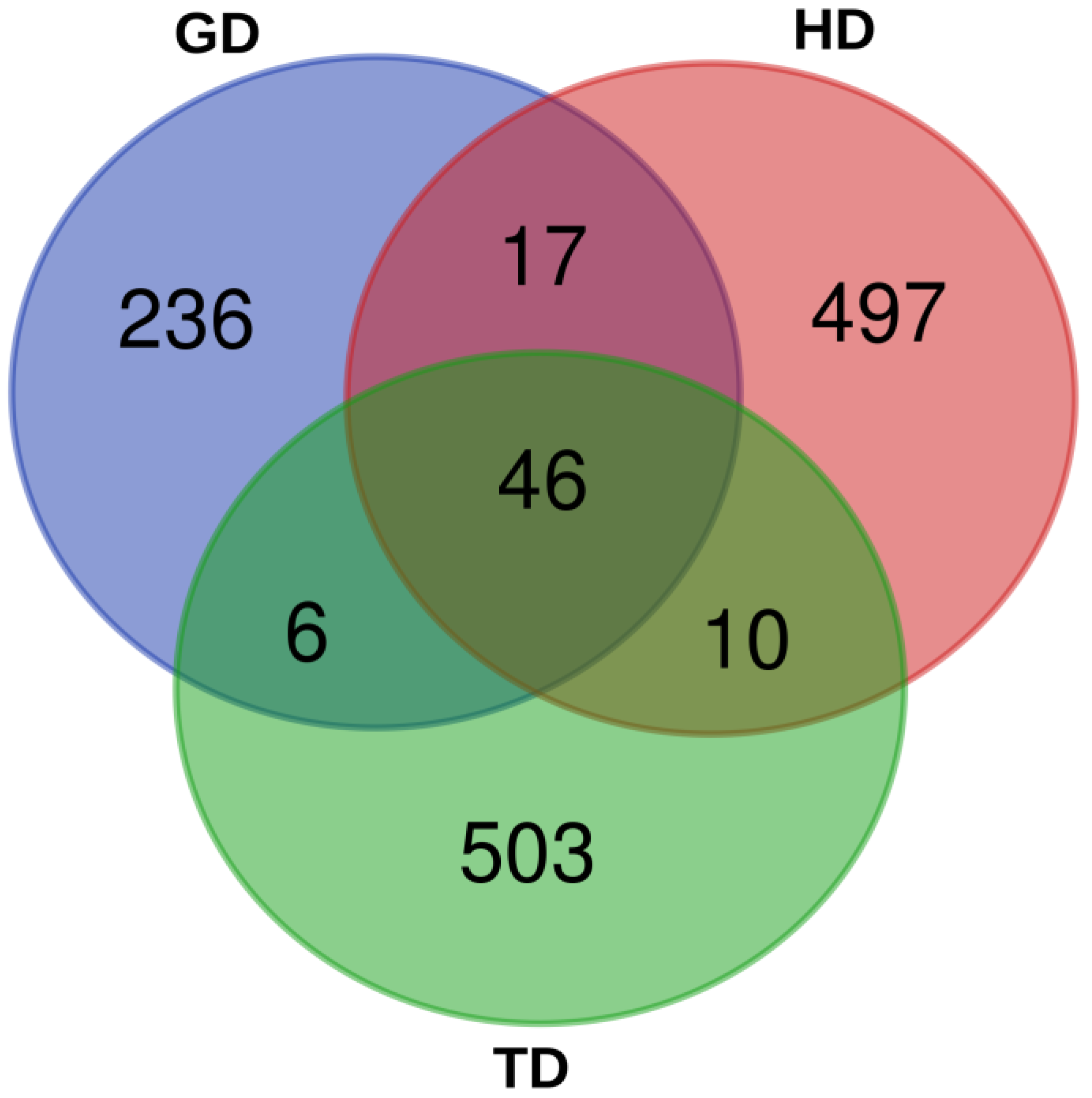
The 16S rDNA of the gut microbiome of the CMCs from three different habitats was sequenced. A total of 1,977,372 reads were generated from HD, whereas 1,778,436 and 1,645,806 reads were produced from TD and GD, respectively. The total read bases range between 495.4 Mbp and 595.2 Mbp. Table 1 shows the raw data statistics for each sampling site. The read length from all the three habitats was 301 bp, and a total of about 76% of the reads passed the input filter. These data indicate that the sequencing results were reliable and that all three datasets could be compared and analyzed. Subsequently, a total of 305, 570, and 565 ASVs were identified from the gut microbiota of GD, HD, and TD habitats, respectively. A total of 46 ASVs were shared by all samples, whereas the number of unique ASVs for GD, HD, and TD were 236, 497, and 503, respectively (Figure 2). Rarefaction analysis suggested that an adequate sampling depth was achieved for each sampling location (Figure S1). Additionally, in order to assess and compare the bacterial diversity in each location, bacterial richness and diversity indices were calculated from the proportion of ASVs. The microbial complexities in the guts of CMCs were estimated based on alpha-diversity indices (Chao1 and Shannon indices). The results indicate that the bacterial community in crabs from HD and TD showed higher alpha-diversity indices than those from GD (Table S4).

Table 1.
Raw data statistics of CMCs from three different locations. Here, Q20 and Q30 represent the ratio of bases that have Phred quality scores of over 20 and 30, respectively.
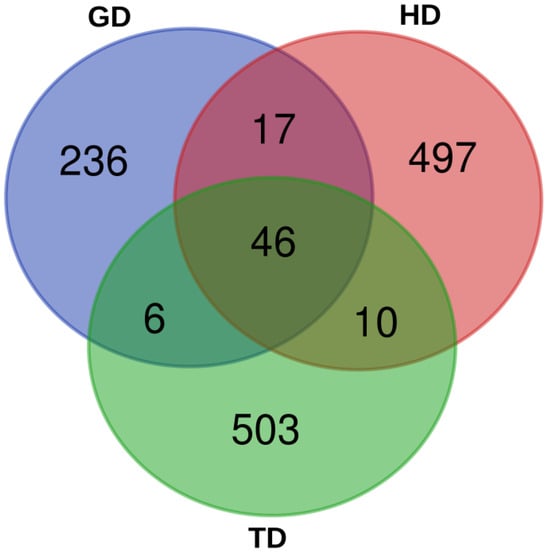
Figure 2.
A Venn diagram showing the number of shared and unique bacterial amplicon sequence variants (ASVs) between the guts of Chinese mitten crabs (CMCs) collected from different environmental locations.
3.3. Microbial Community Composition and Structure
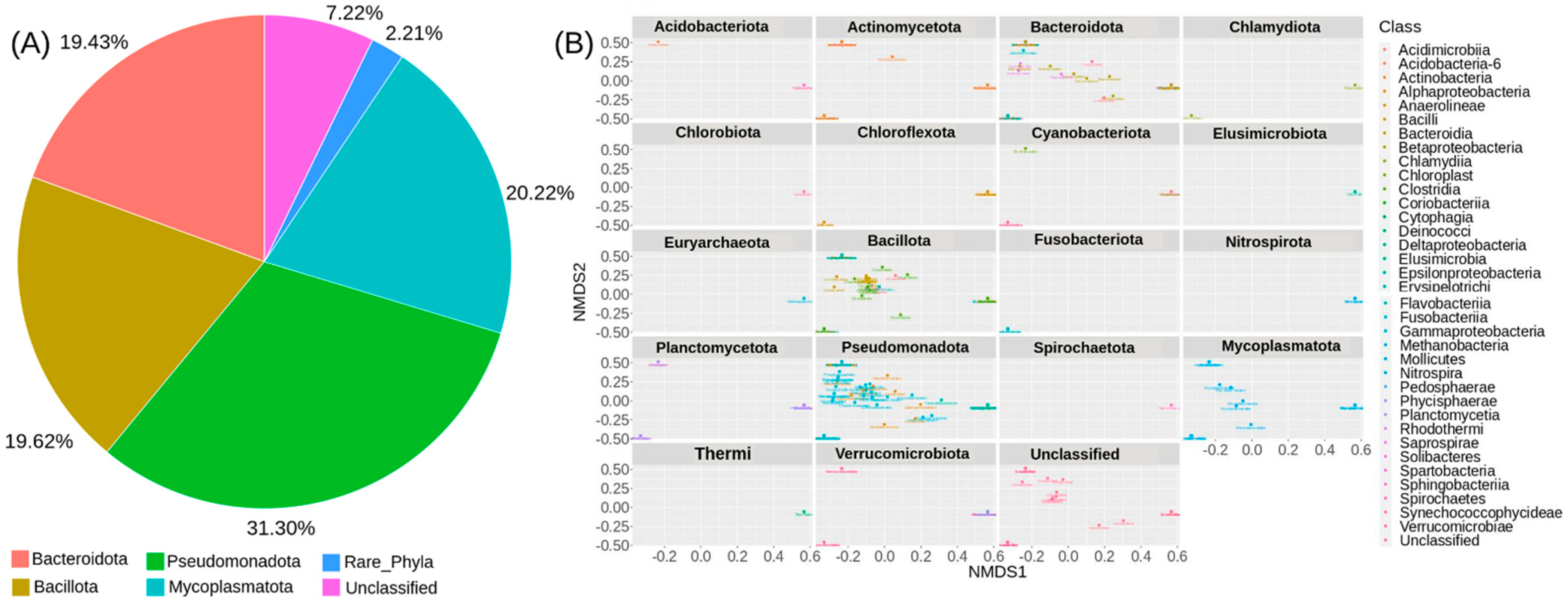
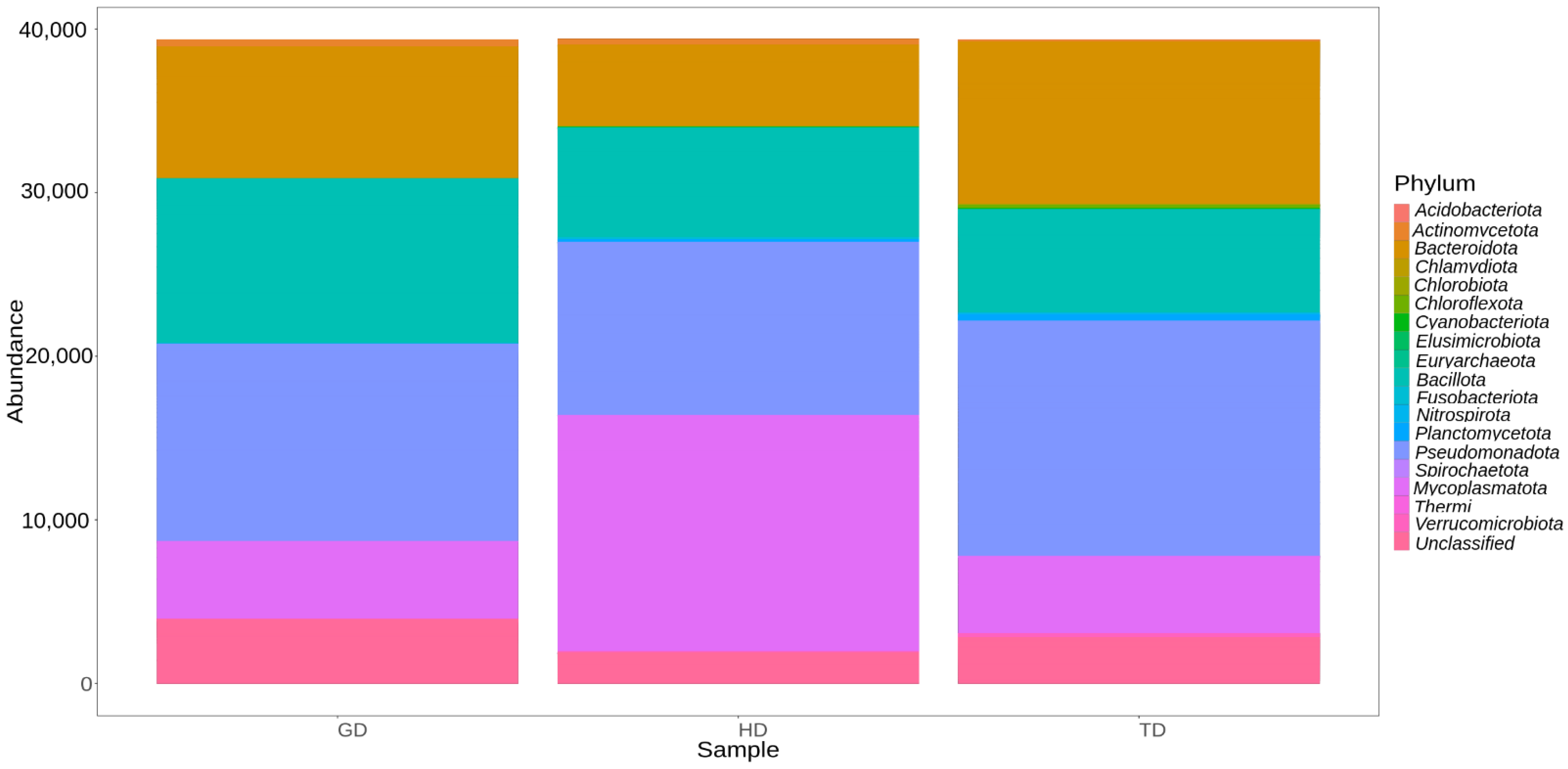
The gut microbiota of CMCs from three different sites mainly comprised bacteria belonging to four phyla, Pseudomonadota (31.3%), Mycoplasmatota (20.2%), Bacillota (19.6%), and Bacteroidota (19.4%). About 7% remained unclassified, and taxa with abundance <1% such as Acidobacteriota, Planctomycetota, and Chloroflexota were grouped as rare phyla (Figure 3). The relative distribution of four dominant phyla was similar at each sampling site; however, different abundance and variation trends show according to the locations. Specifically, we observe that Bacteroidota were more dominant in GD and TD compared to HD. In contrast, site HD showed more dominance of Mycoplasmatota than GD and TD sampling sites (Figure 4).
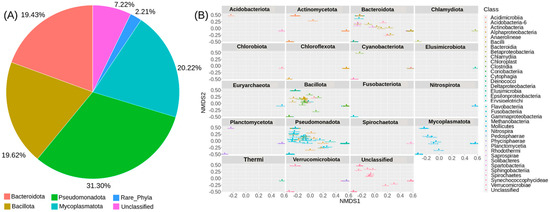
Figure 3.
The overall microbial diversity identified by 16S rDNA metabarcoding. (A) The relative abundance of detected bacterial ASVs among the CMCs collected from 3 different sampling sites. (B) A multivariate analysis plot based on the Bray–Curtis distance and NMDS ordination.
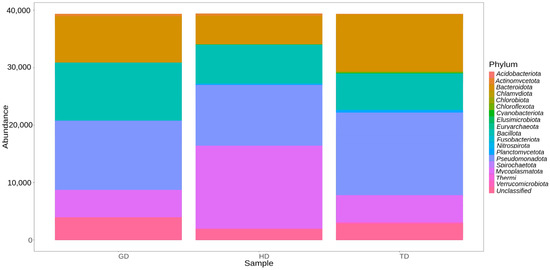
Figure 4.
Abundance of different bacterial phyla in crabs collected from three different locations.
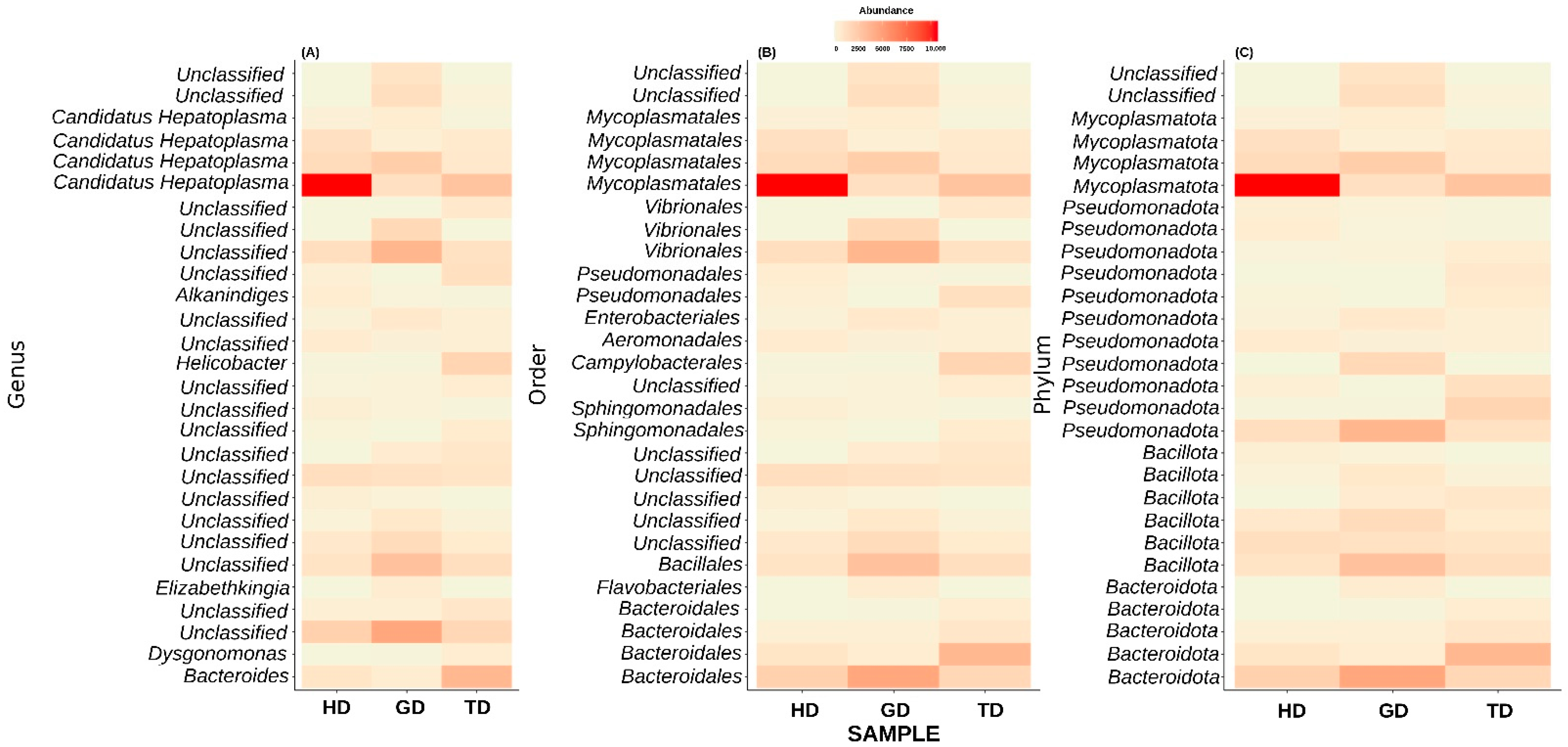
Next, we explored only those taxa that represent at least 1% of reads in at least one sample. This step resulted in the top 28 most abundant taxonomic groups (Figure 5). At the genus level, most of these ASVs could not be assigned to a well-defined taxonomic group (Figure 5A). Nevertheless, the genus-level abundance heatmap exhibited that Candidatus Hepatoplasma had significantly high abundance in CMCs retrieved from the HD sampling location (Figure 5A). Similarly, CMCs obtained from the GD sampling location showed a higher abundance of certain unclassified bacteria at the genus level. These unclassified bacteria mainly belonged to the order Bacteroidales, Bacillales, or Vibrionales (Figure 5B). Interestingly, the crabs collected from the TD sampling location displayed a higher abundance of Helicobacter than HD and GD sites. At the phylum level, the top 28 most abundant groups again represented Pseudomonadota, Mycoplasmatota, Bacillota, and Bacteroidota, although with varying abundance levels (Figure 5C). Additionally, two unclassified phyla showed dominance in the guts of crabs from GD sites. These results are in line with the previous studies which have identified these four phyla as the major groups in the CMC gut microbiome [10].
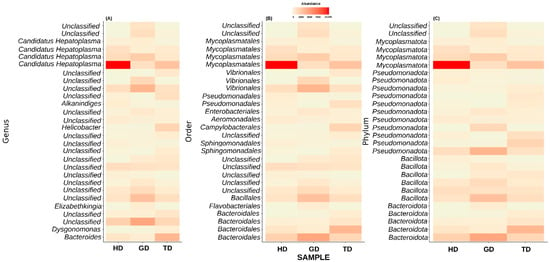
Figure 5.
Composition of top 28 bacterial communities in crabs from different locations at genus (A), order (B), and phylum (C) level. Majority of bacterial sequences remained unclassified at genus level.
At the genus or species level, most of the top 28 taxa remain unclassified, except Alkanindiges illinoisensis and Dysgonomonas gadei. Next, in anticipation of assigning a genus-level taxonomy to the unclassified top 28 most abundant genera, we performed a BLAST-based search against the NCBI non-redundant nucleotide database. Interestingly, the BLAST analysis results also identified most of these taxa as uncultured bacteria (Table 2). Only two phyla could be assigned at the genus level. Among these, one exhibited the best percent identity of 98.49% with Pseudomonas paralcaligenes MRCP1333. The BLAST-based percent identity for other taxa ranged between 96.55 and 99.78%. Additionally, a conflicting taxonomic assignment was also observed for the other taxonomic group by the QIIME2 pipeline and BLAST analysis. For example, QIIME2 identified one of the species as Alkanindiges illinoisensis, whereas the BLAST-based analysis suggested the best match to be Acinetobacter soli S-X6A (sequence identity = 99.14%).

Table 2.
An overview of the BLAST analysis for the 28 most abundant taxa. It should be noted that both the QIIME2 and BLAST-based approaches were unable to assign a genus-level taxonomy to most of these ASVs.
3.4. Microbial Community Similarity among Three Sampling Sites
A principal coordinate analysis (PCoA) was performed to compare the similarity in the bacterial compositions of CMCs from GD, HD, and TD sampling sites. Based on this analysis, bacteria from the guts of CMCs collected at the three locations separated from each other suggesting that we can differentiate between the samples harvested in GD, HD, and TD (Figure S2). The PCoA plot represents every sample as a dot, which is colored based on the sampling site (GD, HD, and TD). However, at this stage, it is difficult to comment on the intra-variability between the samples collected from the three different sites because of limited sampling. Therefore, it will require a large amount of sample collection over a long period of time followed by DNA sequencing.
3.5. Comparison of Free Amino Acids, Nucleotides, and Lactic Acid Content
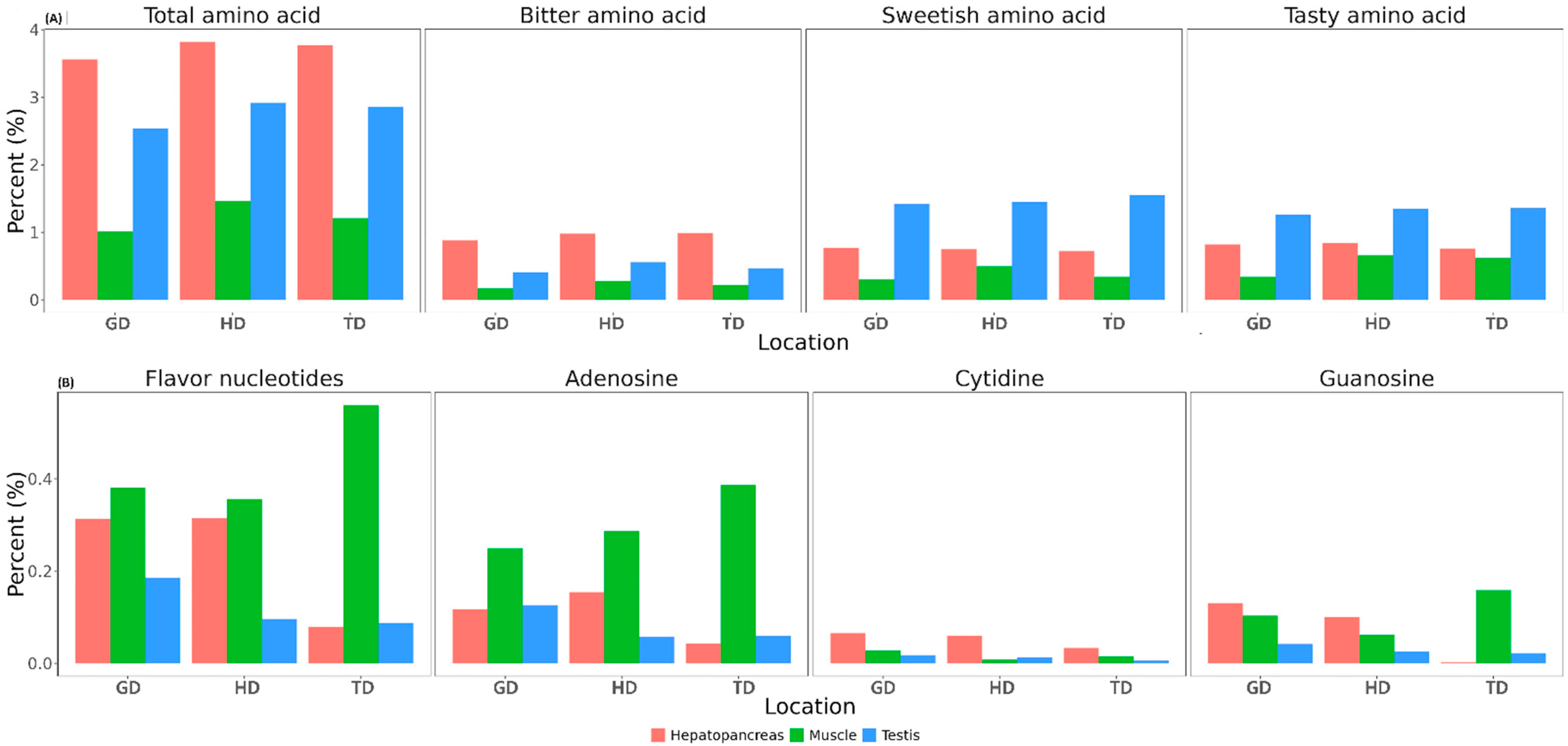
The contents of free amino acids, nucleotides, and lactic acid, which are known to affect the flavor characteristics of CMCs [10], were determined in the hepatopancreas, testis, and muscle of the edible portion of the crabs. The content of free amino acids in the three tissues of the CMCs is presented in Figure 6A. For the total free amino acids, the highest amount was detected in the hepatopancreas, while the muscle exhibited the lowest quantity. All tissues from HD had the highest amount of free amino acids, while GD had the lowest amount of free amino acids in all tissues (Figure 6A). Tasty amino acids and sweetish amino acids presented a similar distribution, with the highest amounts detected in the testis, followed by the hepatopancreas and muscle (Figure 6A). Meanwhile, bitter amino acids were enriched in the hepatopancreas compared to the testis and muscle tissues (Figure 6A). As shown in Figure 6A, free amino acid contents showed considerable differences between tissues, and differences were also observed between habitats. However, these differences were not statistically significant.
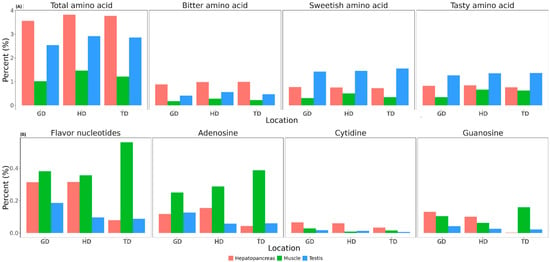
Figure 6.
Content of free amino acids (A) and nucleotides (B) in three different tissues of CMCs collected from three different locations. Bitter amino acids = valine, leucine, isoleucine, and L-arginine; sweetish amino acids = serine, glycine, alanine, and proline; and tasty amino acids = aspartic acid, glutamic acid, glycine, and alanine.
The total content of flavor nucleotides, including adenylic acid (AMP), guanylic acid (GMP), and cytidylic acid (CMP), was the highest in the muscle and the lowest in the testis (Figure 6B). The TD population showed the most flavor nucleotides in the muscle, while the lowest flavor nucleotides were found in the hepatopancreas. AMP was the most abundant in the muscle (Figure 6B), while CMP and GMP exhibited variations between different samples; their quantities were minute.
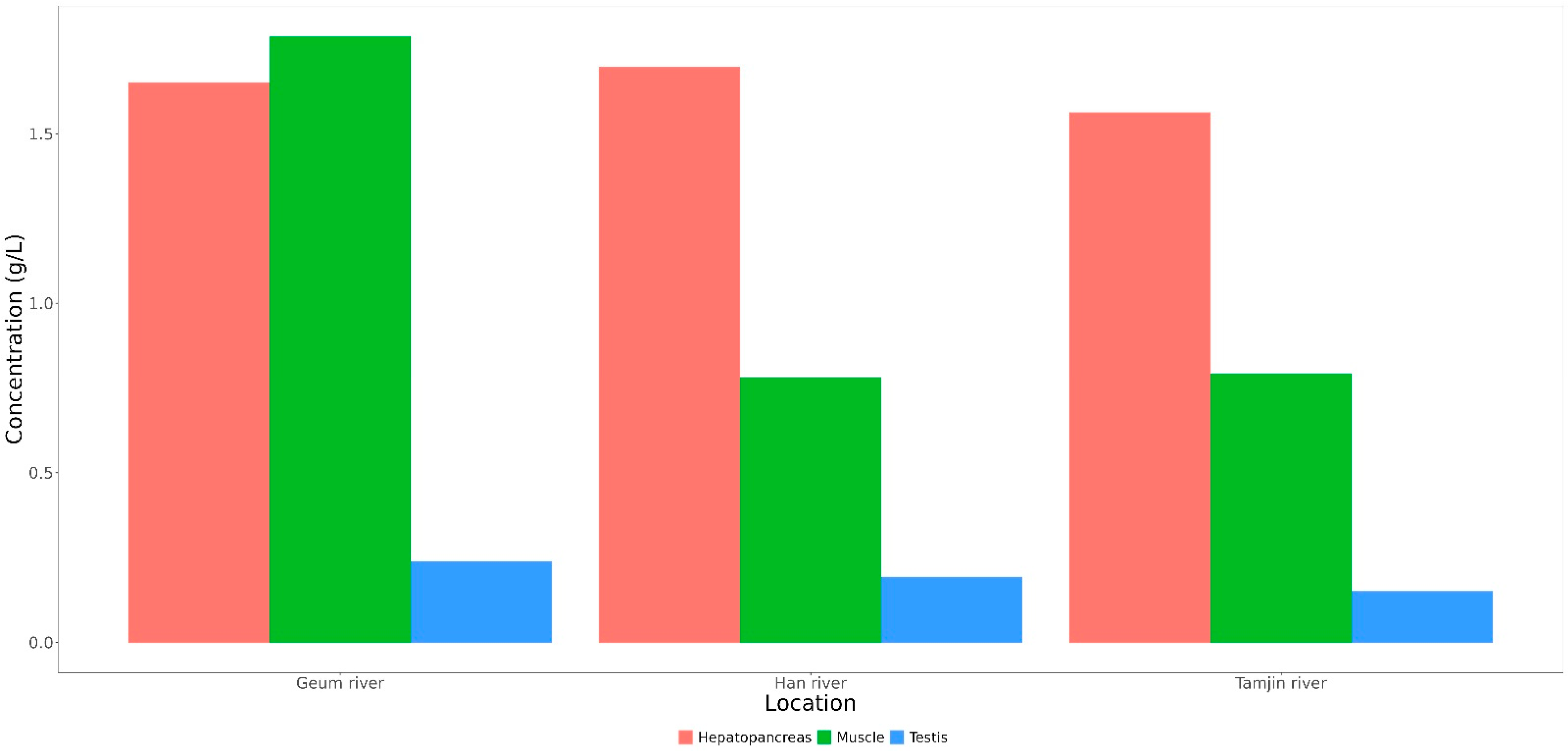
The lactic acid content in various tissues from the three locations was also compared in this study. The results revealed a difference in the lactic acid content among hepatopancreas, testis, and muscle tissues (Figure 7). Specifically, the lactic acid content was more enriched in the hepatopancreas and muscle tissues than in the testis of CMCs sampled from the three sites.
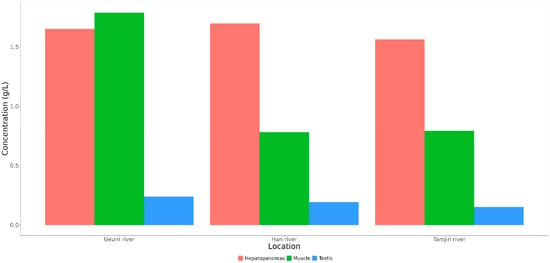
Figure 7.
Content of lactic acids in three tissues of CMCs collected from three different sampling sites.
4. Discussion
Eriocheir sinensis (Chinese mitten crab), also known as the hairy or river crab, is a crustacean of great economic significance [27]. The gut microbiome [10,28], bait [29], and growth environment [30] are considered as some of the most important factors that can regulate the flavor of crabs. Among these, salinity is one of the key factors which can regulate the development of their sexual organs, migration, and adaptation to the changing osmotic pressure [31,32]. In addition, to having a direct effect on the CMC life cycle [33], salinity is one of the essential environmental parameters that influences the composition of microbial communities associated with the host [16,17]. The differences in the host bacterial communities may also affect the flavor substances found in the CMCs harboring disparate habitats [10]. Despite the variations in their habitats, the major taxonomic phyla that are associated with CMCs include Pseudomonadota, Bacillota, Mycoplasmatota, and Bacteroidota. In this work, we also found these groups as the major taxa linked to the CMCs sampled from three distinct locations in South Korea. However, the variations in their abundances do exist. To the best of our knowledge, this study is the first to investigate the differences in the gut microbiota and flavor in CMCs from three separate locations in Korea.
The uncultured bacteria identified from the guts of CMCs (Table 2) have also been reported in previous studies on CMCs [34,35], blue crabs [36], black tiger shrimp [37], and yellow catfish [38]. Moreover, bacterial organisms such as Candidatus Hepatoplasma, a hepatopancreatic colonizing Mycoplasma-like symbiont [39], was observed to be highly abundant in CMCs retrieved from the HD sampling location (Figure 5A). Candidatus Hepatoplasma is a well-known bacterial symbiont found in the midgut glands of terrestrial isopods [40]. Studies have shown that isopods associated with Candidatus Hepatoplasma have developed a capability to prevail in nutritional stress conditions [41]. Studies have also shown a higher abundance of Candidatus Hepatopancreas in the hepatopancreas of CMCs with hepatopancreatic necrosis disease (HPND) suggesting abnormalities in the absorption of nutrients [42]. Additionally, according to recent reports, sick crabs are continuously being caught in nets in HD. It has been argued that the impact of the effluent discharged into HD from two sewage and waste treatment plants might have perturbed the underwater ecosystem of HD [43]. Therefore, the higher abundance of Candidatus Hepatopancreas suggests this may be related to the diseased state of the crabs. However, there are no direct studies linking HD pollution and the diseased crabs. Therefore, additional studies are required to better understand the effect of the changing underwater ecosystem of HD, especially on organisms with significance in aquaculture. Another interesting bacterial species that we identified belongs to the genus Helicobacter in TD. The members of the genus Helicobacter are Gram-negative bacteria with a characteristic helical shape [44]. They inhabit the gastrointestinal tract and liver of mammals and birds [45]. Among various Helicobacter species, H. pylori is the most common pathogenic species known to infect humans [46]. In addition to mammals and birds, H. pylori has been detected in fishes such as tilapia [47]. Members of the Helicobacter group can survive the acidic environment of the host stomach by producing substantial amounts of the urease enzyme [46]. They move fast with the help of their flagella, are microaerophilic canophiles, and are also known to be susceptible to antibiotics [48,49]. With an exponential increase in the aquaculture industry of CMCs, several diseases have evolved in recent times, hence rendering substantial economic losses [50]. Therefore, additional studies exploring the roles of pathogenic bacteria such as Helicobacter are necessary for the better management of culturing CMCs.
For consumers, the hepatopancreas and gonads are some of the most cherished tissues [10]. This can be explained by the fact that when these crabs attain the age of sexual maturity (the best time for their sale in the markets), their gonads grow quickly and accumulate nutrients. In contrast, the hepatopancreas witnesses a rapid reduction in its nutrient content [51,52]. One of the major components of flavor substances in products of aquatic origin is organic acids. The metabolic activities of organisms are closely associated with the contents and composition of these organic acids [10]. Metabolites such as lactic acid are found in fishes [53] and are known to enhance their flavor [54]. In crustaceans such as CMCs, an elevated quantity of lactic acid is observed because of the stress induced by early developmental stages. However, they could keep producing lactic acid even after they have adapted to stress conditions, primarily due to the carbohydrate metabolism pathway [55].
Although most of the environmental parameters such as temperature and pH were more or less similar at the three sampling sites, salinity and DO seem to be the possible reasons for the differences in the gut microbiota of CMCs and their flavor substances. Studies have shown that such differences in the environmental parameters can not only influence the flavor but also their microbiota [16,17,18], fatty acid composition, texture of the flesh, and taste [27,30,56,57].
5. Conclusions
This study represents a preliminary analysis showing the diversity of the gut microbiota of CMCs from three different locations in Korea. Specifically, we observed that the genus Candidatus Hepatoplasma representing the phylum Mycoplasmatota more completely dominated the gut of CMCs from the Han River (HD) than crabs from the Geum (GD) and Tamjin rivers (TD). Although the environment did not seem to influence the flavor characteristics of the crabs, differences in the flavor substances were observed between different tissues. In the future, to better understand the relationship between the gut microbiome, environmental parameters, and flavor characteristics, it is essential to include multiple replicates, samples from more locations, and an equal representation of both genders should be included in the analysis. Nevertheless, our findings can facilitate further research on better management and develop optimal strategies for cultivating this economically important crab.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/fishes9040144/s1, Figure S1: Rarefaction curve showing the sequencing data in this study were reasonable; Figure S2: A principal coordinate analysis plot to compare the similarity in the bacterial compositions of CMCs from GD, HD, and TD sampling sites; Table S1: Information of general biological indicators for crab samples; Table S2: Summary of environmental parameters for each sampling site; Table S3: Gradient elution of HPLC in determination of free amino acids; Table S4: Summary of alpha-diversity metrics at three sampling sites.
Author Contributions
Conceptualization, A.M. and C.-B.K.; methodology, H.-E.A.; validation, H.-E.A. and K.H.L.; formal analysis, H.-E.A., A.M., J.L., M.-H.M. and K.H.L.; investigation, H.-E.A., J.L. and M.-H.M.; resources, H.Y.Y. and C.-B.K.; writing—original draft, H.-E.A., A.M. and J.L.; writing—review and editing, A.M., H.Y.Y. and C.-B.K.; visualization, H.-E.A.; supervision, A.M. and C.-B.K. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Research Foundation of Korea (NRF), funded by the Ministry of Science and ICT (2021R1I1A1A01056363).
Institutional Review Board Statement
Not applicable. Since the crabs are invertebrates, therefore no “Ethics Committee” or “Institutional Review Board” approval is required in Korea.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors have no conflicts of interest to report.
References
- Shen, H.; Hu, Y.; Zhou, X. Sex-lethal gene of the Chinese mitten crab Eriocheir sinensis: cDNA cloning, induction by eyestalk ablation, and expression of two splice variants in males and females. Dev. Genes Evol. 2014, 224, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Qi, T.; Liu, J.; Liu, Q.; Jiang, S.; Zhang, H.; Wang, Z.; Ding, G.; Tang, B. Adaptively differential expression analysis in gill of Chinese mitten crabs (Eriocheir japonica sinensis) associated with salinity changes. Int. J. Biol. Macromol. 2018, 120, 2242–2246. [Google Scholar] [CrossRef] [PubMed]
- Anger, K. Effects of temperature and salinity on the larval development of the Chinese mitten crab Eriocheir sinensis (Decapoda: Grapsidae). Mar. Ecol. Prog. Ser. 1991, 72, 103–110. [Google Scholar] [CrossRef]
- Jeong, H.; Jeon, D. An Integrated Ecological-Economic System Dynamics Model Analysis on the Ecosystem Restoration Policy (II): Extensions and Relaxations of the Model of King Crabs in the Imjin River, Korea. Korean Syst. Dyn. Rev. 2006, 7, 97–120. [Google Scholar]
- Wang, H.Z.; Wang, H.J.; Liang, X.M.; Cui, Y.D. Stocking models of Chinese mitten crab (Eriocheir japonica sinensis) in Yangtze lakes. Aquaculture 2006, 255, 456–465. [Google Scholar] [CrossRef]
- Qiu, J.; Luo, C.; Ren, L.; Li, W.; Dai, T.; Wang, G.; Sun, X.; Moua, K.C.; Sima, Y.; Xu, S. Black soldier fly larvae replace traditional iced trash fish diet to enhance the delicious flavor of Chinese mitten crab (Eriocheir sinensis). Front. Mar. Sci. 2023, 9, 1089421. [Google Scholar] [CrossRef]
- Jiang, H.; Bao, J.; Xing, Y.; Cao, G.; Li, X.; Chen, Q. Metabolomic and metagenomic analyses of the Chinese mitten crab Eriocheir sinensis after challenge with Metschnikowia bicuspidata. Front. Microbiol. 2022, 13, 990737. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.; Myung, N. Eco-Friendly Chinese Mitten Crab Production Process Using Microbiota; Ministry of SMEs and Startups: Sejong-si, Republic of Korea, 2012.
- FIRA Seed Release Management System. Available online: https://seed.fira.or.kr/index.jsp (accessed on 19 December 2023).
- Tao, H.; Du, B.; Wang, H.; Dong, H.; Yu, D.; Ren, L.; Sima, Y.; Xu, S. Intestinal microbiome affects the distinctive flavor of Chinese mitten crabs in commercial farms. Aquaculture 2018, 483, 38–45. [Google Scholar] [CrossRef]
- An, H.; Choi, T.; Kim, C. Comparative Transcriptome Analysis of Eriocheir sinensis from Wild Habitats in Han River, Korea. Life 2022, 12, 2027. [Google Scholar] [CrossRef] [PubMed]
- Long, X.; Wu, X.; Zhao, L.; Ye, H.; Cheng, Y.; Zeng, C. Effects of salinity on gonadal development, osmoregulation and metabolism of adult male Chinese mitten crab, Eriocheir sinensis. PLoS ONE 2017, 12, e0179036. [Google Scholar] [CrossRef]
- Yuan, Q.; Wang, Q.; Zhang, T.; Li, Z.; Liu, J. Effects of water temperature on growth, feeding and molting of juvenile Chinese mitten crab Eriocheir sinensis. Aquaculture 2017, 468, 169–174. [Google Scholar] [CrossRef]
- Hooper, L.V.; Midtvedt, T.; Gordon, J.I. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 2002, 22, 283–307. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Macpherson, A.J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 2010, 10, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Crump, B.C.; Hopkinson, C.S.; Sogin, M.L.; Hobbie, J.E. Microbial biogeography along an estuarine salinity gradient: Combined influences of bacterial growth and residence time. Appl. Environ. Microbiol. 2004, 70, 1494–1505. [Google Scholar] [CrossRef]
- Herlemann, D.P.R.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef]
- Zhang, M.; Sun, Y.; Liu, Y.; Qiao, F.; Chen, L.; Liu, W.-T.; Du, Z.; Li, E. Response of gut microbiota to salinity change in two euryhaline aquatic animals with reverse salinity preference. Aquaculture 2016, 454, 72–80. [Google Scholar] [CrossRef]
- Kivistik, C.; Knobloch, J.; Käiro, K.; Tammert, H.; Kisand, V.; Hildebrandt, J.-P.; Herlemann, D.P.R. Impact of salinity on the gastrointestinal bacterial community of Theodoxus fluviatilis. Front. Microbiol. 2020, 11, 683. [Google Scholar] [CrossRef] [PubMed]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Garcia-Garcia, E.; Galindo-Villegas, J.; Mulero, V. Mucosal immunity in the gut: The non-vertebrate perspective. Dev. Comp. Immunol. 2013, 40, 278–288. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, A.R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Kumar, P.S.; Brooker, M.R.; Dowd, S.E.; Camerlengo, T. Target region selection is a critical determinant of community fingerprints generated by 16S pyrosequencing. PLoS ONE 2011, 6, e20956. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Bliek, T.; Frans, V.D.K.; Marc, G. RNA-seq lesson. ScienceParkStudyGroup. Available online: https://github.com/ScienceParkStudyGroup/rnaseq-lesson (accessed on 7 November 2023).
- Qin, K.X.; Ruan, T.S.; Chen, Y.H.; Liang, G.L.; Wang, H.; Mu, C.K.; Wang, C.L. Effects of temporary rearing time under salinity 7 on the non-volatile flavorings and fatty acids of Eriocheir sinensis. J. Food Compos. Anal. 2022, 107, 104366. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, C.; Wang, H.; Ma, H.; Huang, Y.Q.; Lu, J.X.; Li, X.C.; Zhang, X.W. Involvement of a newly identified atypical type II crustin (SpCrus5) in the antibacterial immunity of mud crab Scylla paramamosain. Fish Shellfish Immunol. 2018, 75, 346–356. [Google Scholar] [CrossRef]
- Wu, X.; Chang, G.L.; Cheng, Y.; Zeng, C.; Southgate, P.C.; Lu, J. Effects of dietary phospholipid and highly unsaturated fatty acid on the gonadal development, tissue proximate composition, lipid class and fatty acid composition of precocious Chinese mitten crab, Eriocheir sinensis. Aquac. Nutr. 2010, 16, 25–36. [Google Scholar] [CrossRef]
- Tang, L.; Wang, H.; Wang, C.L.; Mu, C.K.; Wei, H.L.; Yao, H.Z.; Ye, C.Y.; Chen, L.Z.; Shi, C. Temperature potentially induced distinctive flavor of mud crab Scylla paramamosain mediated by gut microbiota. Sci. Rep. 2020, 10, 3720. [Google Scholar] [CrossRef] [PubMed]
- Kinne, O. Salinity: Animals-invertebrates. In Marine Ecology, Environmental Factors, Part 2; John Wiley and Sons: New York, NY, USA, 1971; Volume 1, pp. 821–996. [Google Scholar]
- Wang, H.; Tang, L.; Wei, H.; Mu, C.; Wang, C. “Butter Crab”: An environment-induced phenotypic variation of Scylla paramamosain with special nutrition and flavour. Aquac. Res. 2019, 50, 541–549. [Google Scholar] [CrossRef]
- Malik, A.; Kim, C.-B. Role of transportome in the gills of Chinese mitten crabs in response to salinity change: A meta-analysis of rna-seq datasets. Biology 2021, 10, 39. [Google Scholar] [CrossRef]
- Li, K.; Guan, W.; Wei, G.; Liu, B.; Xu, J.; Zhao, L.; Zhang, Y. Phylogenetic analysis of intestinal bacteria in the Chinese mitten crab (Eriocheir sinensis). J. Appl. Microbiol. 2007, 103, 675–682. [Google Scholar] [CrossRef]
- Chen, X.; Di, P.; Wang, H.; Li, B.; Pan, Y.; Yan, S.; Wang, Y. Bacterial community associated with the intestinal tract of Chinese mitten crab (Eriocheir sinensis) farmed in Lake Tai, China. PLoS ONE 2015, 10, e0123990. [Google Scholar] [CrossRef] [PubMed]
- Givens, C.E.; Burnett, K.G.; Burnett, L.E.; Hollibaugh, J.T. Microbial communities of the carapace, gut, and hemolymph of the Atlantic blue crab, Callinectes sapidus. Mar. Biol. 2013, 160, 2841–2851. [Google Scholar] [CrossRef]
- Rungrassamee, W.; Klanchui, A.; Chaiyapechara, S.; Maibunkaew, S.; Tangphatsornruang, S.; Jiravanichpaisal, P.; Karoonuthaisiri, N. Bacterial population in intestines of the black tiger shrimp (Penaeus monodon) under different growth stages. PLoS ONE 2013, 8, e60802. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Tian, J.; Wang, G.; Li, W.; Zou, H. Characterization of bacterial community in the stomach of yellow catfish (Pelteobagrus fulvidraco). World J. Microbiol. Biotechnol. 2012, 28, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Stingl, U.; Anton-Erxleben, F.; Geisler, S.; Brune, A.; Zimmer, M. “Candidatus Hepatoplasma crinochetorum,” a new, stalk-forming lineage of Mollicutes colonizing the midgut glands of a terrestrial isopod. Appl. Environ. Microbiol. 2004, 70, 6166–6172. [Google Scholar] [CrossRef] [PubMed]
- Bouchon, D.; Zimmer, M.; Dittmer, J. The terrestrial isopod microbiome: An all-in-one toolbox for animal–microbe interactions of ecological relevance. Front. Microbiol. 2016, 7, 1472. [Google Scholar] [CrossRef] [PubMed]
- Fraune, S.; Zimmer, M. Host-specificity of environmentally transmitted Mycoplasma-like isopod symbionts. Environ. Microbiol. 2008, 10, 2497–2504. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.S.; Zang, Y.N.; Song, K.; Ma, Y.C.; Dai, T.H.; Serwadda, A. A meta-transcriptomics survey reveals changes in the microbiota of the Chinese mitten crab Eriocheir sinensis infected with Hepatopancreatic necrosis disease. Front. Microbiol. 2017, 8, 732. [Google Scholar] [CrossRef] [PubMed]
- Jeon, I.J. Han River Chinese Mitten Crab with Shells Turned to Black...Fisherman “Never Seen Anything Like It”. The JoongAng. Available online: https://www.joongang.co.kr/article/25194822#home (accessed on 23 September 2023).
- Goodwin, C.S.; Armstrong, J.A.; Chilvers, T.; Peters, M.; Collins, M.D.; Sly, L.; McConnell, W.; Harper, W.E. Transfer of Campylobacter pylori and Campylobacter mustelae to Helicobacter gen. nov. as Helicobacter pylori comb. nov. and Helicobacter mustelae comb, nov., respectively. Int. J. Syst. Evol. Microbiol. 1989, 39, 397–405. [Google Scholar] [CrossRef]
- Ryan, K.J.; Ray, C.G. Sherris Medical Microbiology, 4th ed.; McGraw Hill: New York, NY, USA, 2004. [Google Scholar]
- Dunn, B.E.; Cohen, H.; Blaser, M.J. Helicobacter pylori. Clin. Microbiol. Rev. 1997, 10, 720–741. [Google Scholar] [CrossRef]
- Abdel-Moein, K.A.; Saeed, H.; Samir, A. Novel detection of Helicobacter pylori in fish: A possible public health concern. Acta Trop. 2015, 152, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Hua, J.-S.; Zheng, P.-Y.; Ho, B. Species differentiation and identification in the genus of Helicobacter. World J. Gastroenterol. 1999, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Rust, M.; Schweinitzer, T.; Josenhans, C. Helicobacter flagella, motility and chemotaxis. In Helicobacter pylori: Molecular Genetics and Cellular Biology; Yamaoka, Y., Ed.; Horizon Scientific Press: Norfolk, UK, 2008; pp. 61–86. [Google Scholar]
- Shen, H.; Ma, Y.; Hu, Y. Near-full-length genome sequence of a novel reovirus from the Chinese mitten crab, Eriocheir sinensis. Genome Announc. 2015, 3, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Spaargaren, D.H.; Haefner, P.A., Jr. Interactions of ovary and hepatopancreas during the reproductive cycle of Crangon crangon (L.). II. Biochemical relationships. J. Crustac. Biol. 1994, 14, 6–19. [Google Scholar] [CrossRef]
- Boucard, C.G.V.; Levy, P.; Ceccaldi, H.J.; Brogren, C.H. Developmental changes in concentrations of vitellin, vitellogenin, and lipids in hemolymph, hepatopancreas, and ovaries from different ovarian stages of Indian white prawn Fenneropenaeus indicus. J. Exp. Mar. Biol. Ecol. 2002, 281, 63–75. [Google Scholar] [CrossRef]
- Lalitha, K.V.; Sonaji, E.R.; Manju, S.; Jose, L.; Gopal, T.K.S.; Ravisankar, C.N. Microbiological and biochemical changes in pearl spot (Etroplus suratensis Bloch) stored under modified atmospheres. J. Appl. Microbiol. 2005, 99, 1222–1228. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Asakawa, A.; Yamaguchi, K.; Konosu, S. Studies on flavor components in boiled crabs. III. Sugars, organic acids, and minerals in the extracts. Nippon Suisan Gakkaishi 1979, 45, 1325–1329. [Google Scholar] [CrossRef][Green Version]
- Tao, H.; Liu, H.J.; Cheng, Y.Q.; Sima, Y.H.; Yin, W.M.; Xu, S.Q. Parental environmental exposure leads to glycometabolic disturbances that affect fertilization of eggs in the silkworm Bombyx mori: The parental transcript legacy. J. Comp. Physiol. B 2015, 185, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, X.; Huang, Y.; Bu, X.; Xiao, S.; Qin, C.; Qiao, F.; Qin, J.G.; Chen, L. Effects of dietary T-2 toxin on gut health and gut microbiota composition of the juvenile Chinese mitten crab (Eriocheir sinensis). Fish Shellfish Immunol. 2020, 106, 574–582. [Google Scholar] [CrossRef]
- Du, X.; Zhang, W.; He, J.; Zhao, M.; Wang, J.; Dong, X.; Fu, Y.; Xie, X.; Miao, S. The impact of rearing salinity on flesh texture, taste, and fatty acid composition in largemouth bass Micropterus salmoides. Foods 2022, 11, 3261. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).