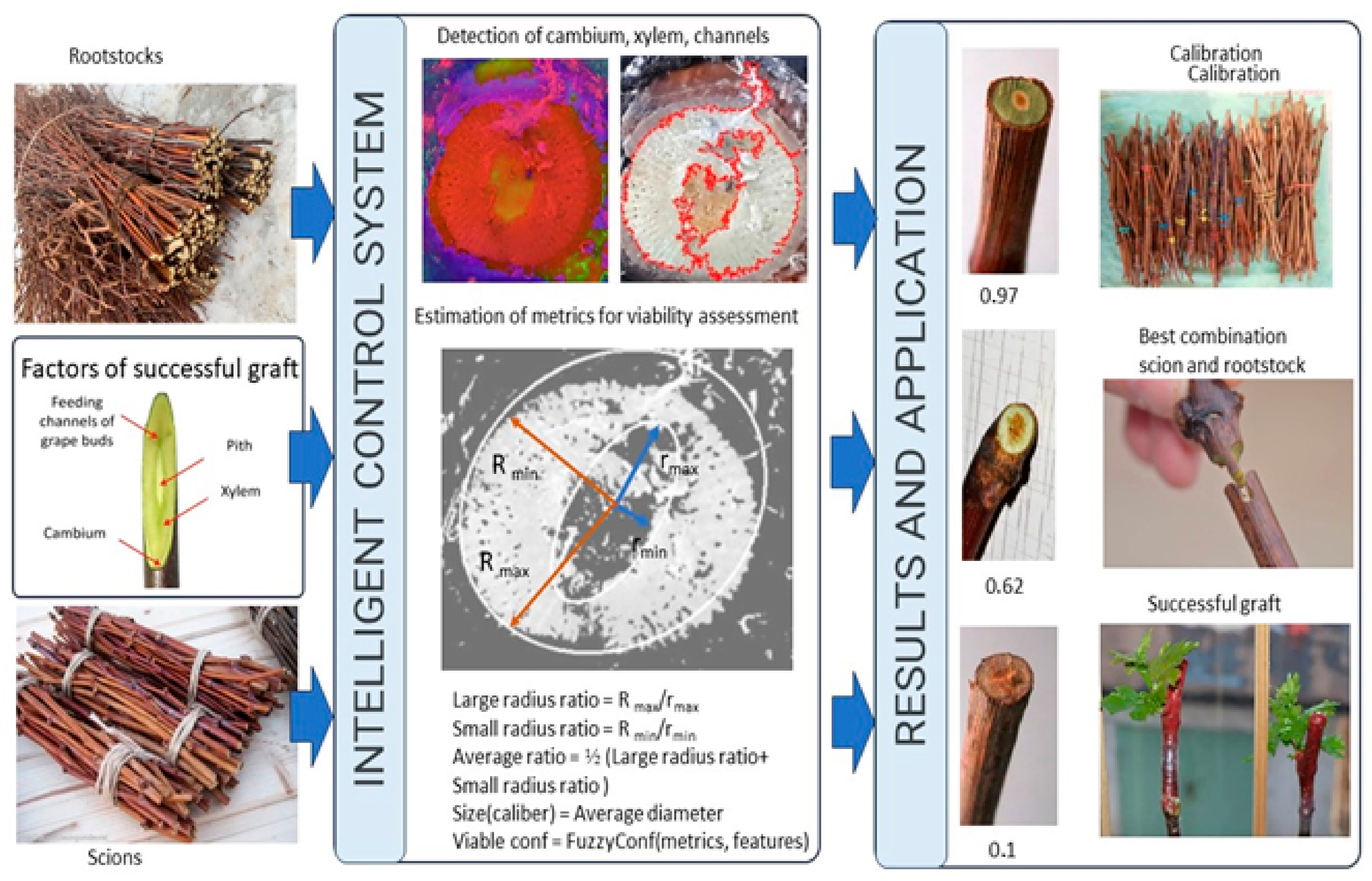
1. Introduction
Viticulture is a traditional branch of the agro-industrial complex in the south of Russia. The favorable soil and climatic conditions of this region allow for growing high yields of grapes during various ripening periods. However, the effectiveness of these conditions is not yet fully utilized, primarily due to miscalculations made during the establishment and operation of industrial plantations. One of the main reasons for the unsatisfactory state of perennial plantations lies in the quality of planting material. The quality of seedlings in the future will depend on the yield of plantations, the period of their productive operation, the volume of investments, and the payback period. In vineyards planted with certified planting material, the service life is increased 1.5 times, productivity is increased by 30–50%, and the volume of capital investments is significantly reduced, which is accompanied by an increase in the competitiveness of domestic grape and wine producers [
1,
2,
3,
4].
The duration of the productive period of rootstock combinations will largely depend on the affinity, which is determined by the proximity of the components in terms of morphological and anatomical features, the type of metabolism, and the agricultural technology used. Affinity can also change depending on the environmental conditions. A grafted plant consists of two parts that are different in their biological properties, each of which reacts in its own way to certain environmental conditions by changing the metabolic rate. With the same type of reaction of both parts, i.e., while maintaining a similar nature of their metabolism, the grafted plant retains high viability and productivity [
5,
6,
7,
8,
9].
An analysis of numerous studies has shown that the splicing of rootstock combinations depends on many factors that can be conditionally divided into mechanical and physiological compatibility according to the signs of a clear manifestation. Mechanical compatibility is determined by the high level of splicing of the graft components. Studies have shown that the incompatibility of the mechanical type can be interconnected with differences in the anatomical structure of the grafting components. Therefore, in grapes with equal rootstock and scion diameters, there can be significant differences in the structural ratio of the core area to wood. This may affect the mechanical strength of the rootstock combination, and the fusion will not be circular. With mechanical action on the seedling, breaks are observed at the grafting site.
The probability of the coincidence of the tissues of the core and wood in rootstock varieties with the graft, according to many authors, is considered one of the main factors in the high level of fusion of grafted cuttings after stratification, as well as the release of standard planting material from grape schools [
2,
10,
11,
12,
13,
14].
This study aims to use computer vision to improve the selection of a suitable graft to rootstock based on the analysis of a section of a grape seedling.
Computer vision (CV) is receiving increasing attention in agriculture, as it represents a set of methods that allow for the automatic collection of images and precise and efficient extraction of valuable information [
15]. CV-based systems typically include an image acquisition phase and image analysis methods that can distinguish areas of interest for detection and classification such as plants and leaves [
16,
17]. There are many image analysis methods, but deep learning (DL) [
18] is considered the most effective for detecting objects in agriculture such as diseases [
19], crops and weeds [
20], and fruits [
21]. Deep learning is based on machine learning and is capable of automatically extracting features from unstructured data [
22]. Convolutional neural networks (CNN) are being tested for various tasks to support precision agriculture production systems, including phenological monitoring [
23,
24]. Thus, high-performance plant phenotyping methods [
25] based on CV in combination with DL (CV and DL) can assess the spatiotemporal dynamics of crop features related to their phenophases in the context of precision agriculture [
26,
27,
28].
However, to the best of our knowledge, computer vision technologies have not been applied to improve the affinity of rootstock-scion combinations of vine seedlings. This is a differential and innovative aspect of the present work, the purpose of which is to show a new method for the analysis of seedlings, which will reduce the need for laborious and time-consuming manual operations and checks.
Computer vision algorithms can automatically determine the main characteristics of the seedling, such as the diameter of the stem, the number and location of the vessels, the presence of damage, etc. To achieve this, we need to take a photo of a cut of a seedling and process it using special computer vision programs. Based on these data, it is possible to determine which scion will be most suitable for a given seedling and rootstock. This approach can significantly speed up and simplify the process of selecting a scion for a rootstock, which will save time and money on growing grapes.
Figure 1 shows a schematic representation of the use of computer vision to improve the affinity of rootstock-graft combinations.
In addition to the benefits mentioned above, computer vision can also assist in identifying and diagnosing diseases or pests affecting vine seedlings. By analyzing images of the seedlings, computer vision algorithms can detect visual cues or abnormalities that indicate the presence of diseases or pests. This early detection can help farmers take timely actions to prevent the spread of diseases and minimize crop losses.
Furthermore, computer vision can provide valuable insights into the growth and development of vine seedlings. By analyzing images captured at different stages of growth, computer vision algorithms can track the progress of key phenological traits such as leaf emergence, bud break, and fruit development. This information can aid in optimizing irrigation, fertilization, and other agronomic practices to ensure optimal growth conditions for the vines.
The integration of computer vision with deep learning techniques such as convolutional neural networks (CNNs) enables a more accurate and robust analysis of vine seedlings. CNNs are capable of learning complex patterns and features from large datasets, allowing them to effectively classify and identify various characteristics of the seedlings. This includes distinguishing between different scion varieties, assessing the health and quality of the seedlings, and predicting their future growth potential.
The application of computer vision in the context of rootstock-scion combinations for vine seedlings offers a novel and efficient approach to streamlining the selection process. By automating the analysis of seedling characteristics, farmers can save time and resources, leading to more precise and successful grafting decisions.
2. Materials and Methods
2.1. Plant Materials
The studies were carried out at the V.I. Vernadsky Crimean Federal University’s grape grafting complex. The objects of research were vines of rootstock and graft varieties, from which stratified cuttings of various variety-rootstock combinations were subsequently produced, represented by technical zoned varieties. For the graft group, the native varieties Dzhevatkara, Sarypandas, Ekim Kara, Kefessia, and Kokur Beliy grafted onto the rootstocks of the varieties Berlandieri x Riparia Kober 5BB, Riparia x Rupestris 101–14, and Berlandieri x Riparia CO4 were studied. Most of the rootstock varieties have an interspecific hybridization of American species with European varieties of the species Vitisvinifera, which has certain morphological and anatomical distinctive features from varieties of European and Asian origin of the species Vitisvinifera. These differences lead to varying degrees of incompatibility since the development of wood and, as a result, the conducting system, may be different depending on the variety-rootstock combination. The output of stratified cuttings, along with other factors, depends on the safety of the buds in the eyes of the graft vine.
In 2022, manually conducted grafting operations at the complex numbered approximately 260,000. With the implementation of the semi-automated system in 2023, this number increased to half a million grafts. The goal for 2024 is to reach maximum capacity, aiming for one million grafts per year.
Regarding the training dataset, a size of 1000 pairs of photographs was used to train the computer vision system.
The detailed economic calculations and analysis are presented in a separate publication, providing a comprehensive understanding of the economic implications and benefits of implementing a semi-automated computer vision system in grape grafting processes.
Before grafting, as a part of the process of connecting the rootstock and graft parts of the cutting, the quality of the graft and rootstock material was assessed with three repetitions of 30 pieces each. In the period from the second half of November to the first ten days of December, the harvesting of rootstock and graft vines was carried out and their biometric indicators were assessed. The vines, after soaking in a 0.5% solution of chinosol, were kept in a refrigerator until the vaccination campaign was carried out. The storage temperature was +2…+4 °C. In March, the preserved vines of the rootstock and graft varieties were prepared for the production of grafts. The rootstock vine was cut to a length of at least 40 cm (lower cut under the eye) with blinding eyes on the vine using a knife. The graft vine was cut into one-eyed cuttings, leaving part of the internodes (below the peephole without a length limitation, but not less than 5 cm, above the peephole within 1.5 cm). The prepared inoculations, in accordance with the scheme of the experiment, were bundled, labeled, and placed in boxes, which were transferred to the stratification chamber. Stratification was carried out using an open method on water in a humid atmosphere at an air temperature of 25–27 °C. The relative air humidity was maintained within 80…90%.
The biometric assessment of their quality is important for studying vines of graft and rootstock varieties. The biometric indicators include the total length of the vines, m; average length of the vine, m; average internode length, cm; average diameter of the vine, mm; average core diameter, mm; average ratio of the total diameter of the vine to the diameter of the core; average cross-sectional area of the vine, mm
2; average cross-sectional area of the core, mm
2; average cross-sectional area of wood, mm
2; conditional coefficient of shoot ripening. It was found that in the graft and rootstock vines, on average, for varieties and years, the lengths of internodes differed. This corresponds to the fact that in the methods for determining grape varieties for varietal compliance, the biometric indicators of the vine can change depending on the conditions of the place and year of cultivation [
2,
4,
5,
6,
7,
8,
9,
10,
11].
2.2. Fuzzy Classification Model for Characteristics and Damage of Vine Seedlings
The vector of the results of the characteristics and damage of vine seedlings should be supported by metrics that characterize the significance and proximity of the state of vine seedlings to a set of traits of a particular class. In practice, as one of the methods of confirmation, a comparison with similar conclusions made previously by specialists can be performed. To achieve this, a database of “reference” classes of characteristics and lesions of grape seedlings, characteristic of a particular localization of agricultural production, can be formed, containing the characteristics and descriptions of objects in the image. Analyzing on the basis of comparison, a specialist or artificial intelligence can draw reasonable conclusions based on their proximity to the standards according to certain rules. The formulation of the problem of fuzzy classification based on image analysis can be reduced to the classical problem of classifying the elements of a set of characteristics and damage of grape seedlings using a tuple of fuzzy variables, together with an additional neural network for fuzzy classification.
The fuzzy classifier software modules were developed and distributed under the MIT license. However, we decided to build our own fuzzy classifier, which allowed us to enter and take into account information and expert assessments from specialists using given membership functions. The proposed mechanism made it possible to more accurately and flexibly tune the classifier, taking into account the complexity of formalizing knowledge and expert opinions, and retrain the fuzzy model. Under these conditions, the method of convoluting tuples of image objects using classes with the formation of a fuzzy estimate of their form factors, such as the fill factor (the share of the area occupied by class objects), presence factor (the share of class objects in the tuple), and degree of severity of the class structure (assessment of the degree of assigning an object to a class), was used. Also, the estimates of the SNA of the entire image, the estimate of informativeness (the share of the area occupied by all objects), and the uniformity of the distribution of objects were also converted to fuzzy variables.
This study uses fuzzy classification rules, each of which describes one of the types of damage to grapes in the data set. The a priori rule is a fuzzy description in the n-dimensional property space, and the sequence of rules is a fuzzy class label from the given set (1):
where
n denotes the number of properties,
is the input vector, and
Aij are the fuzzy sets, represented by the fuzzy ratios of the output of the
i-th rule and the input vector or the previous fuzzy rule. The degree of activation of the
i-th rule from Set (2),
M, is calculated as:
The classifier output is determined by Rule (3), which has the highest degree of activation,
ai:
At this stage, we have implemented only 21 rules for drawing a conclusion regarding the type of lesion and its degree. The degree of confidence in the solution is given by the normalized degree of rule triggering (4):
However, a feature of grape damage is its combination of several classes, which requires information about all reliable lesions found in the image in accordance with a fuzzy assessment of confidence. Therefore, the result is determined by Vector (5)
where
,
is the threshold value for a given class of damage to grapes. It also allows for combining estimates and conclusions obtained after the detection of several images from the same source location. The union is a convolution using the maximum and average values of the metrics to activate classes. The output of the classifier activates the score gain among the set
L of images acquired at the selected location (6).
The complex lesion vector collapses the estimates using the maximum weighted one from the set
L relative to the threshold
Dm (7).
In the future, the rules will be reinforced by tracking the dynamics of the development of the process when using certain factors in the processing and protection of grapes. This a posteriori information forms a set of meta-rules or reflex patterns that are evaluated and ranked by disease, proven potency, and grape variety. Given the variety of drugs and technologies for combating grape diseases, we have chosen the path of accumulating the proposed templates and technologies as rules that have their own fuzzy assessment of effectiveness µ.
3. Results
3.1. Search Subject by Bloom
Color is a property of bodies that reflects or emits visible radiation of a certain spectral composition and intensity. Everywhere we are surrounded by color indicators, including traffic lights, white and yellow road markings, corporate products, road signs, and various other indicators. For example, for visually impaired people, yellow circles are glued to the doors of shops (yellow is the only color they see) so as not to confuse, for example, a glass showcase with a glass door. There are also yellow stripes on pedestrian crossings. This color is one of the brightest and makes it easier for people with poor eyesight to navigate the city. Searching by color is influenced by many factors, for example, illumination. We must also not forget that the visible color is the result of an interaction between the spectrum of the emitted light and the surface. If a white sheet is illuminated by the light of a red bulb, then the sheet will also appear red [
29].
The main difficulty of object localization is the most difficult aspect of object detection. There are many ways to search for objects in an image, and one of these methods is using a sliding window of different sizes to search for objects in an image. This method is called “exhaustive search”. The “exhaustive search” method consumes enormous computing resources since it is necessary to detect an object in thousands of windows, given the small size of the image. To improve the operation of the method and increase the speed of data processing, we optimized the processing algorithm, namely we changed the window sizes in different ratios (instead of 23 magnifications by several pixels). The result of this optimization was an increase in the speed of the algorithm, but the efficiency leaves much to be desired.
This paper discusses a selective search algorithm that uses both an exhaustive method and segmentation (a method of separating objects of different shapes in an image by assigning different colors to them). To determine the color characteristics, a program was written that helps highlight the contours in the image. To determine these parameters, a program was written for the RGB color of an image captured by video camera no. 1. The results of this program are shown in
Figure 2. The resulting images were obtained by coloring the original image in red, green, and blue and defining the mask [
30].
By applying the output information, it is possible to find the pixels of the required color using a function that selects the image pixels that fall within the specified range of colors. This function examines the array elements (pixels) element by element and checks whether the colors are in the list of values of two different matrices.
3.2. Algorithm for Contour Analysis of the Image of a Cut of a Grape Seedling for the Selection of the Optimal Graft to the Rootstock
The contour analysis algorithm is one of the most important and useful methods for describing, recognizing, comparing, and searching for graphic images (objects). A contour is the outer outline of an object.
The contour analysis is performed under the following conditions:
- −
The contour contains sufficient information about the shape of the object;
- −
Due to having the same brightness as the background, the object may not have a clear border or it may be noisy with interference, making it impossible to select the contour (successful use only with a clearly defined object against a contrasting background and no interference);
- −
The overlapping of objects or their grouping leads to the contour being incorrectly selected and not corresponding to the border of the object;
- −
The internal points of the object are not taken into account. Therefore, a number of restrictions are imposed on the scope of the contour analysis, which are mainly related to the problems of contour detection in images.
Among the methods for obtaining a binary image, one can single out, for example, thresholding or selecting an object by color. The OpenCV library contains convenient methods for detecting and manipulating image edges. The find contours function is used to find contours in a binary image. This function can find the outer and nested contours and determine their nesting hierarchy. There are also other functions that are often used, such as minAreaRect2(), which returns the smallest possible rectangle that can wrap around the path but can be rotated relative to the image coordinate system at a certain angle. An example of how contour analysis works is shown in
Figure 3a. When the algorithm is running with the cv. CHAIN_APPROX_SIMPLE parameter (
Figure 3b), all boundary points are preserved. It removes all unnecessary points, compresses the contour, and also saves memory [
31].
3.3. Development of a Software Module for the Analysis of Grape Seedlings
To find the radii of the selected contours with the support of two cameras, the following method was developed. The investigated object moved along the assembly line, and when it entered the frame of video camera no. 2, its contour was located by subtracting the snow-white background. As soon as the middle of the plant reached the center of the frame, the assembly line stopped and focused on the prevailing paint of the resulting contour. Subsequently, it started processing the image from video camera no. 1. With a given color, a specific color palette was used to determine the contour of the ellipse and find its center.
Before use, the camera was calibrated. Calibration allows us to take into account the distortion introduced by the optical system. Camera calibration comes down to obtaining the internal and external parameters of the camera from the available photos or videos captured by it. Camera calibration is usually performed at the initial stage of many computer vision problems. In addition, this procedure allows us to correct distortion in photos and videos.
Before the sensor was embedded, the image was converted to shades of gray to reduce computational costs. First, the image was smoothed to remove useless noise, and then a Gaussian filter was applied. To achieve this, we used a filter that can be approximated by 1 Gaussian derivative. Then came the calculation of the gradients. Object contours were marked where the image gradient contained the largest values. After that, non-maximums were removed, and only local maxima were marked as boundaries. This was followed by double-threshold filtering. The output contained clear contours on a black-and-white binary image. The results of the analysis of a current image of grape cuttings are shown in
Figure 4.
3.4. Analysis of the Results
Here, we consider the results obtained during the testing of the plant contour analysis algorithm and also present the comparative results of the algorithm for five different seedlings. Each sapling has two multi-colored circles in the center (
Figure 5a), which are needed to assess the quality of the sapling and together form a two-dimensional representation of the internal structure of the sapling. The selection of the contour on the seedling is displayed as a red connecting line (
Figure 5b), but the program also highlighted unnecessary parts due to excess material in the images (such as the film that is visible in the picture), which is why the algorithm did not perfectly determine the contour.
The next step was to convert the image to a black-and-white format (
Figure 5c). After that, we used a mask to select the contours by cutting off the extra points (
Figure 5d) [
32,
33]. As we can see in the view of the contour from above (
Figure 5d), the algorithm selected an extra part of the seedling due to an incorrect source code (it captured a part of the film in which the seedling was wrapped).
Next, we calculated the radii of the ellipses and determined how much space the inner ellipse occupied relative to the outer one (
Figure 6a). If it occupied more than half of the seedling, then it was considered unsuitable. However, the visual processing of the program in this case was incorrect and a first-order error occurred since we used incorrect image data. On this account, it is possible to increase the error in the calculation of the ellipses (
Figure 6b), but this is only suitable for unique cases such as when seedlings with external physical interference are used.
Let us move on to the second test. This seedling turned was illuminated by a flash (
Figure 7a), which is why the white background strongly predominated on the left side, which led to the incorrect operation of the algorithm. We used image processing based on information about the color of the object. First, the algorithm colored the original picture in red, green, and blue (
Figure 7b). Next, came the calculation of the mask for all three colors: red, green, and blue (
Figure 7c). Then, came the testing for the set of HSV colors (
Figure 7d).
The next step was to calculate the mask for all three colors: red, green, and blue (
Figure 8a). As we can see in the images, too much noise was obtained during the determination, which did not allow us to conduct a qualitative assessment of the seedling. The next step was to use the contour analysis algorithm (
Figure 8b), but due to the image being overexposed by the flash, the algorithm could not highlight the center. Next, we converted the image to a black-and-white format (
Figure 8c). After that, we used a mask to select the contours by cutting off the extra points (
Figure 8d). It turned out that there was a very small part in the center that the algorithm did not highlight. However, visually, since the center was quite light, indicating that the dead part occupied a small space, the seedling was considered to be of high quality.
The next photo of the seedling used for testing is shown in
Figure 9a. The image of the seedling turned out well, without unnecessary defects and noise. The next step used the contour analysis algorithm (
Figure 9b). After that, we used a mask to select the contours by cutting off the extra points (
Figure 9c). Further, the use of the algorithm for selecting the contours is shown in
Figure 9d. The algorithm highlighted the contours of the seedling clearly along the edges, the center in this experiment, which means that the dead part occupied a small space and the seedling was high quality.
The fourth seedling had damage in the very center in the form of a fault (
Figure 10a). The next step used the contour analysis algorithm (
Figure 10b). After that, we used a mask to select the contours by cutting off the extra points (
Figure 10c). Further, the use of the algorithm for selecting contours is shown in
Figure 10d. It was not possible to qualitatively highlight the contours of the seedling, as there were too many changes in this picture and the algorithm could not cope with the task of evaluating the dead part relative to the center of the healthy part. Due to the pronounced defects, it was considered to be of poor quality.
We conducted one last test. This seedling (
Figure 11a) was without visible damage but the image was slightly blurry. The next step used the contour analysis algorithm (
Figure 11b). Then, we used a mask to select the contours by cutting off the extra points (
Figure 11c,d). This qualitatively highlighted the contours of the seedling. In this picture, it can be seen that there was a small black area that was not selected and it was not clear how critical this was—perhaps the problem was due to the fuzzy image as a whole. The algorithm coped with its task and determined the seedling to be a high-quality one.
3.5. Pre-Training of the Neural Network for Detecting Specified Object Classes (Grape Diseases) in Images
This section presents images demonstrating the results of pre-training the neural network for detecting grape diseases in images. Thanks to this process, the authors obtained accurate and reliable data on the condition of grapevines, which allows for improving crop quality and preserving plant health. The dataset labeling information for the initial training is shown in
Figure 12. The dataset labeling information for the final training is shown in
Figure 13.
The confusion matrix is a table that visualizes the effectiveness of the classification algorithm by comparing the predicted value of the target variable with its actual value. The performance graph for the algorithm used in the initial models is presented in
Figure 14, and it can be seen that the performance was at a low level. The performance graph for the algorithm used in the final model training is presented in
Figure 15, and it can be seen that the performance was at a sufficiently high level.
To evaluate the quality of the algorithm’s performance on each class separately, we used precision and recall metrics. Precision can be interpreted as the proportion of objects classified as positive by the classifier that are truly positive, whereas recall shows the proportion of positive class objects found by the algorithm out of all positive class objects.
Figure 16 shows the plots for the final model training, which indicate that the accuracy increased and the number of undetected affected areas decreased with each epoch, indicating the correctness of the training and high key performance indicators, as well as the absence of model overfitting.
The accuracy and recall increased with each epoch, which may indicate that the model was improving and becoming more accurate in detecting affected areas. If there was no overfitting, this is an even more positive result.
4. Discussion
As noted above, the monitoring of phenology and seedling growth using computer vision has received considerable attention in the literature [
20,
24,
25,
34,
35,
36,
37,
38,
39,
40,
41]. It is therefore important to relate our proposal to these related works. Although each of these papers is devoted to quantifying some aspect of the early stages of plant development, they include a wide variety of approaches that are related to the word “seedling”.
A number of studies are devoted to the use of computer vision and robotics for the winter pruning of vines, reducing the need for human intervention. At the heart of this approach is an architecture that allows a robot to easily move through vineyards, identify vines with high accuracy, and approach them for pruning [
42,
43].
However, we did not find studies and publications on the use of computer vision and the automation of the processes of grafting grape cuttings.
The appearance of a seedling is determined visually. On examination, a person may not notice the damage to the seedling, so some authors have proposed solutions based on computer vision.
Some of the techniques used in computer vision include image segmentation, object recognition, feature extraction, and image classification. These techniques are used to analyze and interpret visual data to provide insights, detect anomalies, and make predictions. In addition, computer vision can help determine the quality of grape seedlings, which is also important when choosing a suitable scion for a rootstock. Analysis of a cutting section of a seedling can reveal the presence of diseases, damage, or other defects that can affect the growth and development of the plant. Thus, computer vision can help growers increase the efficiency of growing grapes and obtain a better harvest.
Also, computer vision can help improve the study of grape seedlings and grape diseases by providing accurate and fast data on plant health. For example, computer vision can automatically detect and classify grape diseases, allowing us to quickly and accurately determine which plants need treatment. We can also use computer vision to analyze the growth and development of grape seedlings, which will help determine the optimal conditions for growing them. In addition, computer vision can help collect data on grape varieties, which will improve the selection and cultivation of grapes.
There are grape diseases that can only be identified by cutting grape seedlings. For example, bacterial cancer of grapes, which manifests itself in the form of dark spots on the wood and can lead to the death of the plant. Also, when cut, root rot caused by a fungus can be detected. This disease can lead to a weakening of the plant and a decrease in yields. Therefore, when buying grape seedlings, it is recommended to pay attention to the condition of the roots and wood, as well as take preventive measures to prevent the development of diseases.
Computer vision can be used to examine vine seedlings at the scion to determine the presence of disease or other problems. To achieve this, it is necessary to cut a vine seedling and obtain an image using a microscope or other imaging device. The image can then be processed using computer vision algorithms that can automatically detect the presence of diseases or other problems on the grape seedling. For example, algorithms can look for signs, such as changes in color or texture, which may indicate the presence of a disease.
Unlike scenarios where UAV images integrated with an artificial neural network (ANN) model are analyzed, we use fixed-scale macro photography to obtain accurate sizes. We also apply filters to the image to highlight areas and evaluate geometric indicators. These are different approaches.
Such approaches can be useful for the rapid and accurate detection of grape diseases, which can help vine growers and farmers take more effective disease control measures and increase yields. In the context of analyzing grape seedlings, computer vision techniques can be used to identify and classify different types of damage, such as diseases and frost infestations, based on the visual features present in the seedling images. This can then inform decisions regarding the treatment and management of the plants.
This study is expected to contribute to the field of viticulture by providing an automated and accurate method for analyzing seedlings, which will enable farmers to detect and treat diseases or frost infestations early enough to prevent devastating crop losses. The proposed solution is also expected to reduce the need for labor-intensive and time-consuming manual inspections while improving the accuracy and reliability of disease and frost detection.
This research relates to the field of viticulture, where there is a growing interest in the use of computer vision techniques for plant analysis and disease detection. The authors of this study used existing computer vision methods and techniques, such as image segmentation, object recognition, feature extraction, and image classification, to develop a software module for the analysis of grape seedlings. These methods are widely used in various fields, including medical imaging, robotics, and surveillance.
In addition, the authors proposed their original approach to the use of fuzzy logic models in the classification of the characteristics and damage of grape seedlings. Fuzzy logic is a mathematical approach that allows us to display uncertainty and inaccuracy in data. It has been used in a variety of applications, including control systems, decision making, and pattern recognition.
In general, the proposed approach is based on existing research in the field of viticulture and computer vision and also presents new methods and techniques for the analysis of grape seedlings. The software complex mentioned in this study implements the best approaches to vineyard control systems using computer vision technologies. The decision support system software can be adapted to solve other similar tasks. The commercialization plan of the software product is aimed at automating and robotizing agriculture, and it will serve as a basis for the development of the next type of similar software.
The results of this study show the efficiency of using artificial neural networks to classify grape lesions in images. The solution to these problems is impossible without the widespread use of convolutional neural networks. However, the most valuable conclusions are based on a comprehensive assessment of the results of recognizing both the entire image and individual objects using detection. The apparatus of fuzzy logic, with the help of linguistic and fuzzy variables, allows presenting the obtained conclusions in the form of a full-fledged conclusion.
The developed system for localization manages the delivered task but also has its disadvantages. The system is sensitive to background noise, which can lead to biased results, especially for images with relatively edgy contours. Computer vision algorithms do not always work correctly, especially when there are superfluous materials in the images like crumpled or cut film, when the data are strongly illuminated, or when the seedlings are strongly damaged.
5. Conclusions
This study discusses the use of computer vision to increase the affinity of rootstock-graft combinations and detect diseases in grape seedlings. A very important task has been set for domestic viticulture: to multiply the volume of production of its own planting material in a short time. Partial mechanization in viticulture has low efficiency and quality due to manual labor and a large amount of waste. The automation of the processes of grafting grape cuttings based on computer vision will help solve this problem.
CV analyzes the cut image of a grape seedling to find the optimal graft for the rootstock. This technology will allow evaluating the parameters and viability of seedlings; controlling the positioning of seedlings before grafting and the quality of the grafting result; and monitoring the development of seedlings to determine the time of planting. For this, image processing algorithms are used that automatically determine the characteristics of the seedling such as the diameter of the trunk, the presence of damage, etc.
This paper also presents a mathematical model based on fuzzy logic for classifying the features of grapevine cuts. The model is based on cluster analysis of the obtained images to optimize rootstock combinations and diagnose diseases or pests of grape seedlings. An algorithm for contour analysis of the image of a grape seedling is proposed. A software module for the analysis of grape seedlings is presented, and an analysis of the results obtained is carried out. This project can be used by agronomists and growers as a tool to quickly and accurately identify growth problems and take appropriate action to eliminate them. In addition, the project can be used for educational purposes to improve knowledge about computer vision and its applications in agriculture.
Furthermore, computer vision can assist in identifying diseases and defects in seedlings, which is crucial for determining their quality. The benefits of automating this process include increased efficiency, improved quality, and cost reduction through a decrease in manual labor and waste.
Based on these studies, it will be possible to develop an automated line for grafting grape cuttings. The line will be equipped with two conveyor belts, a delta robot, and a computer vision system, which will increase the productivity of the process.