A Neutrosophic Cubic Hesitant Fuzzy Decision Support System, Application in the Diagnosis and Grading of Prostate Cancer
Abstract
:1. Introduction
2. Preliminaries
3. Neutrosophic Cubic Hesitant Fuzzy Sets (NCHFSs)
- Truth-internal neutrosophic cubic hesitant fuzzy number (T-internal) if the following inequality is satisfied ;
- Indeterminacy-internal neutrosophic cubic hesitant fuzzy number (I-internal) if the following inequality is satisfied
- Falsity-internal neutrosophic cubic hesitant fuzzy number (F-internal) if the following inequality is satisfied
- A neutrosophic cubic hesitant fuzzy number (NCHFNs) is called internal neutrosophic cubic hesitant fuzzy number if it is T-internal, I-internal, and F-internal.
- Truth-external neutrosophic cubic hesitant fuzzy number (T-external) if the following inequality is satisfied
- Indeterminacy-external neutrosophic cubic hesitant fuzzy number (I-external) if the following inequality is satisfied
- Falsity-external neutrosophic cubic hesitant fuzzy number (F-external) if the following inequality is satisfied
- A neutrosophic cubic hesitant fuzzy number (NCHFN) is called external neutrosophic cubic hesitant fuzzy number if it is T-external, I-external, and F-external.
4. Similarity Measures of NCHFSs
- If we consider each NCHFN and by the weight where with and , then generalized NCHFN weighted distance is defined asThe following proposition is related to weighted distance measure .
- The relation between the similarity measure and the distance measure is called weighted-similarity measure, which is defined asThe following proposition is related to weighted similarity measure .
5. Risk Evaluation Method of Prostate Cancer (PC)
- Gleason’s score of 3 to 5 indicates low risk;
- Gleason’s score of 6 to 8 suggests medium-grade cancer;
- Gleason’s score of 8 to 10 indicates high-grade cancer.
- Gleason’s score of 0 to 2 indicates low risk;
- Gleason’s score of 3 to 5 indicates intermediate risk;
- Gleason’s score of 6 to 10 indicates high risk.
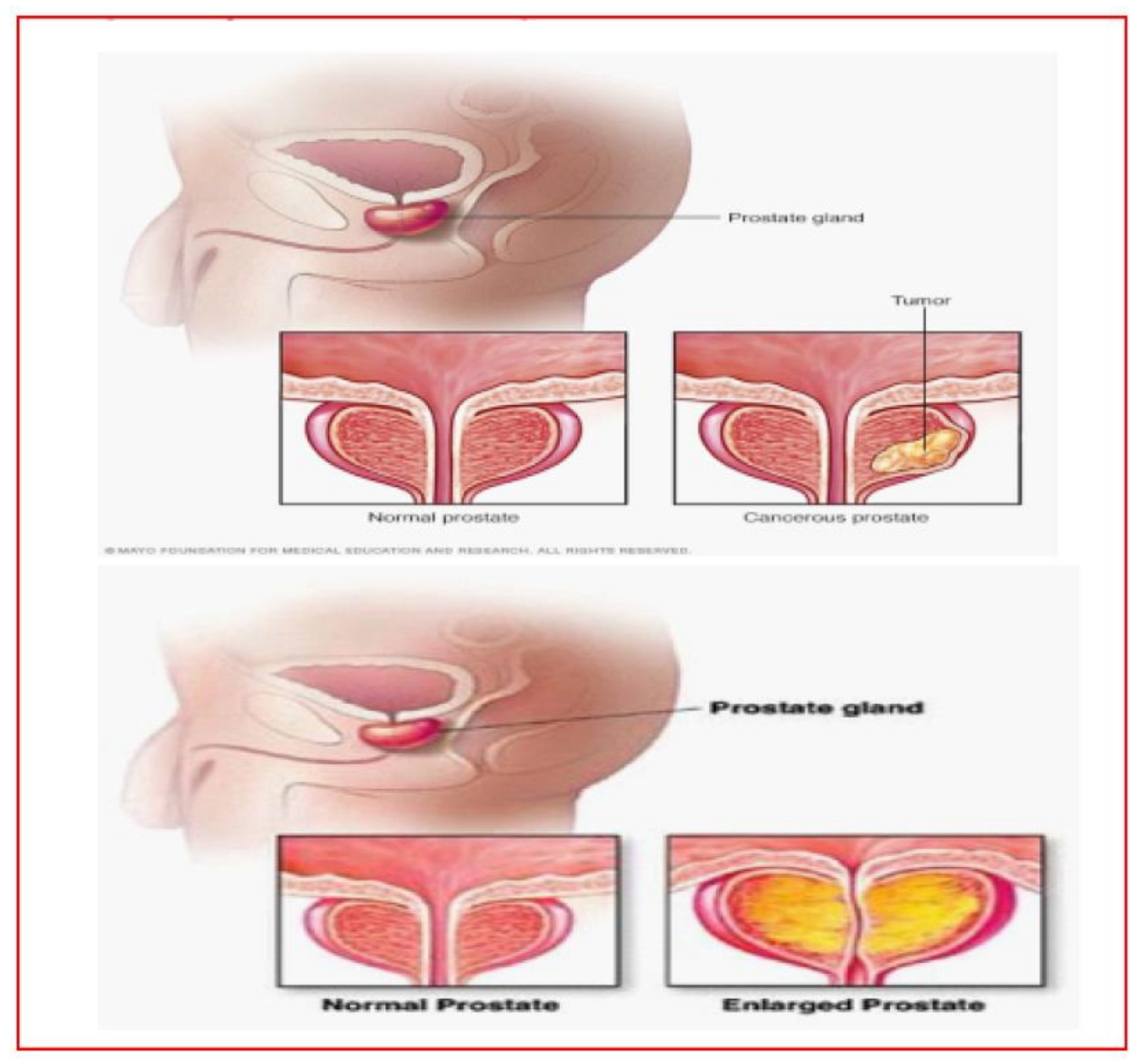
- Gleason’s Pattern scale is shown in Figure 1.
- T1a: The clinical staging score of 1 to 4 of tumor is 5% or less of the prostate tissue removed during surgery;
- T1b: The clinical staging score of 5 to 7 of tumors is in more than 5% of the prostate tissue removed during surgery;
- T1c: The clinical staging score of 8 to 10 of tumor is found during a needle biopsy, usually because the patient has an elevated PSA level;
- T2: The tumor is found only in the prostate, not other parts of the body;
- T2a: The clinical staging score of 4 to 6 of the tumor involves one-half of one side of the prostate;
- T2b: The clinical staging score of 6 to 7 of a tumor involves more than one-half of one side of the prostate but not both sides;
- T2c: The tumor’s clinical staging score of 7 to 10 has grown in both sides of the prostate;
- Different stages of the prostate are shown in Figure 2.
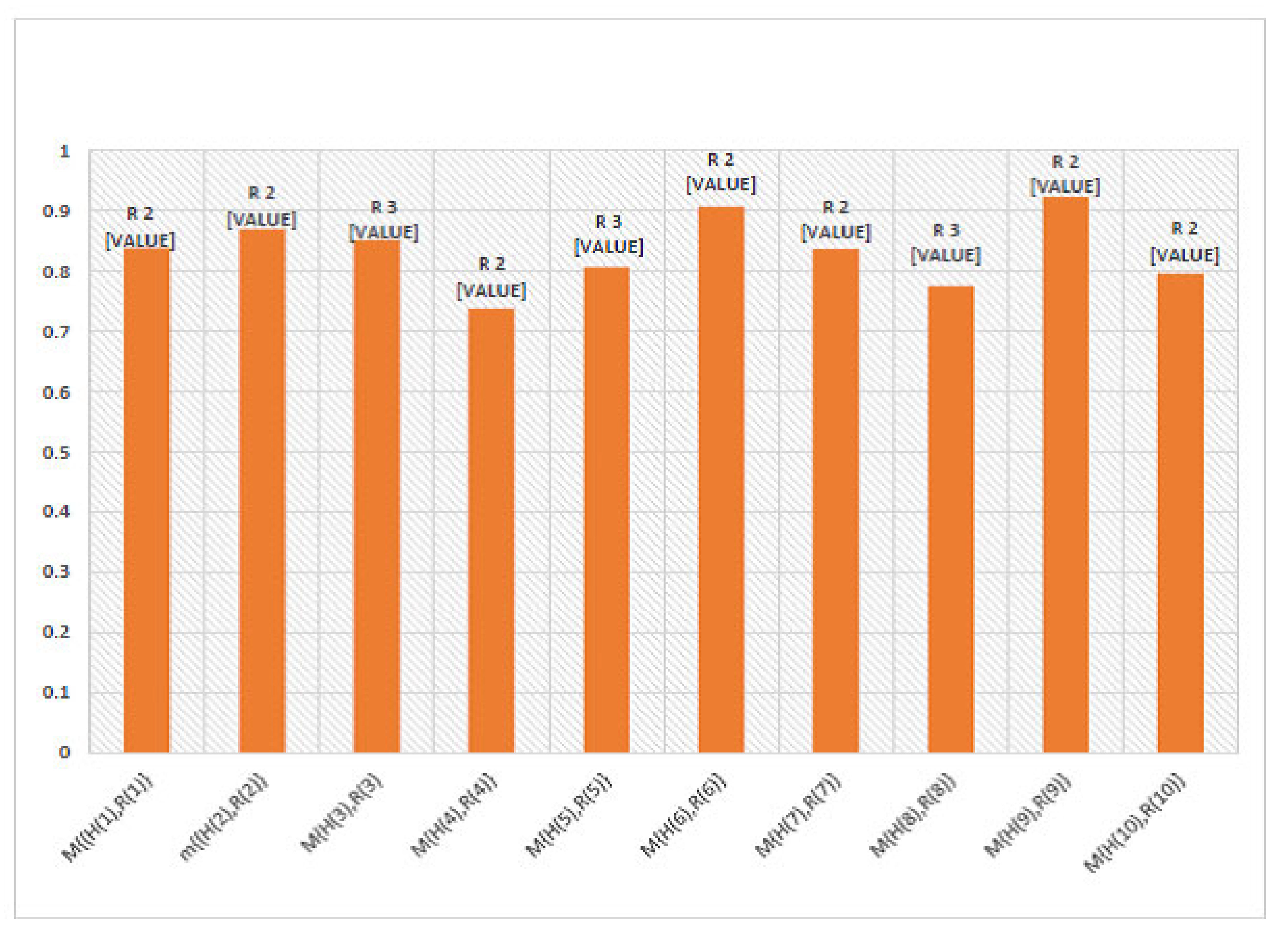
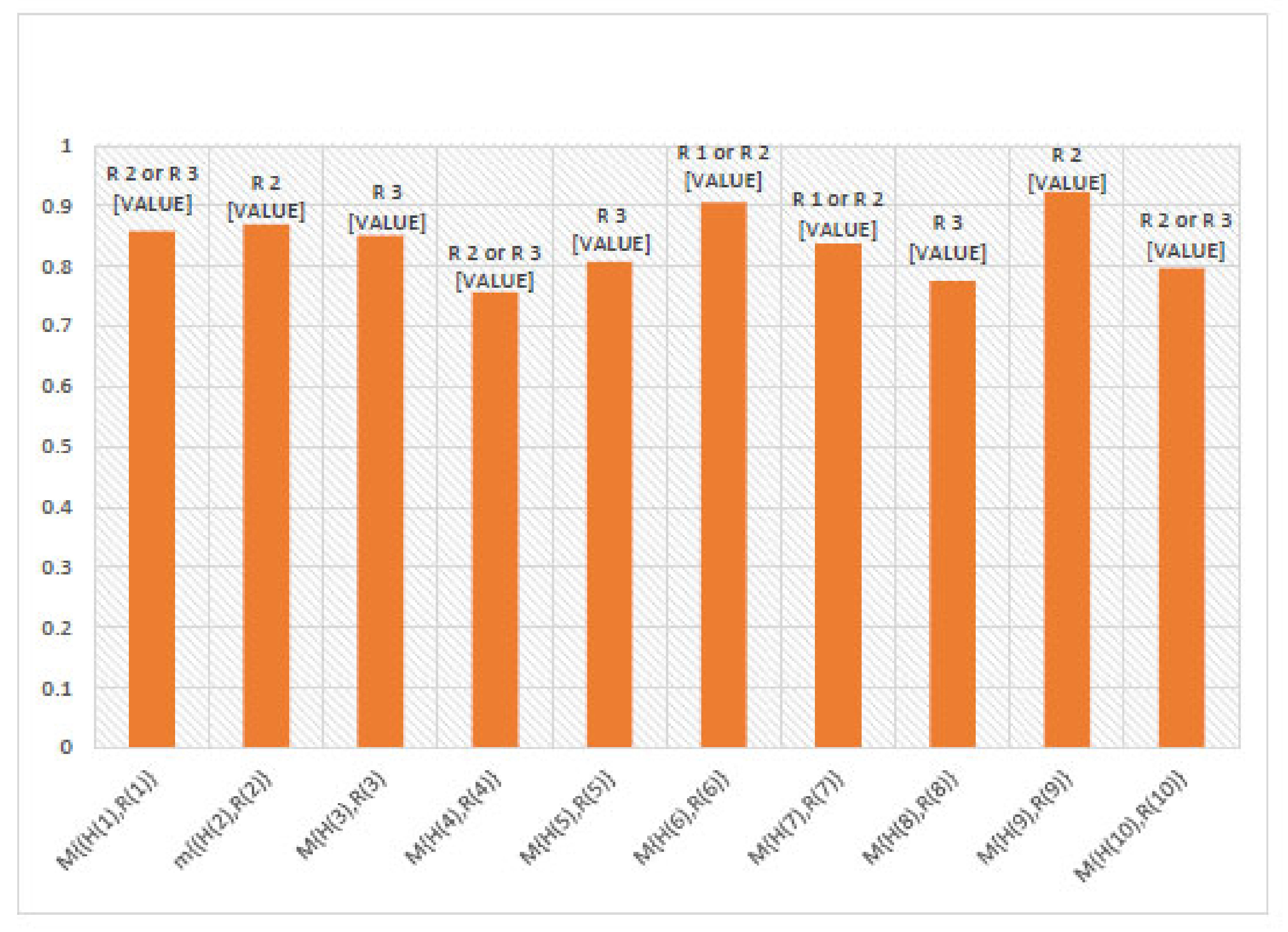
6. Numerical Example
7. Comparison Analysis
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lin, P.-H.; Liu, J.-M.; Hsu, R.-J.; Chuang, H.-C.; Chang, S.-W.; Pang, S.-T.; Chang, Y.-H.; Chuang, C.-K.; Lin, S.-K. Depression negatively impacts survival of patients with metastatic prostate cancer. Int. J. Environ. Res. Public Health 2018, 15, 2148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jemal, A.; Siegel, R.; Ward, E.; Murray, T.; Xu, J.; Smigal, C.; Thun, M.J. Cancer statistics, 2006. CA Cancer J. Clin. 2006, 56, 106–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, H.B.; Albertsen, P.C.; Barry, M.J.; Etzioni, R.; Freedland, S.J.; Greene, K.L.; Holmberg, L.; Kantoff, P.; Konety, B.R.; Murad, M.H. Early detection of prostate cancer: Aua guideline. J. Urol. 2013, 190, 419–426. [Google Scholar] [CrossRef] [Green Version]
- Cao, K.; Arthurs, C.; Atta-ul, A.; Millar, M.; Beltran, M.; Neuhaus, J.; Horn, L.-C.; Henrique, R.; Ahmed, A.; Thrasivoulou, C. Quantitative analysis of seven new prostate cancer biomarkers and the potential future of the ‘biomarker laboratory’. Diagnostics 2018, 8, 49. [Google Scholar] [CrossRef] [Green Version]
- Kelly, W.K.; Scher, H.I.; Mazumdar, M.; Vlamis, V.; Schwartz, M.; Fossa, S.D. Prostate-specific antigen as a measure of disease outcome in metastatic hormone-refractory prostate cancer. J. Clin. Oncol. 1993, 11, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.Y.; Partin, A.W.; Walsh, C.P.; Epstein, J.I. Prognostic significance of Gleason score 3+4 versus Gleason score 4+3 tumor at radical prostatectomy. Urology 2000, 56, 823–827. [Google Scholar] [CrossRef]
- Edge, S.B.; Byrd, D.R.; Compton, C.C.; Fritz, A.G.; Greene, F.L.; Trotti, A., III. AJCC Cancer Staging Manual, 7th ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 457–468. [Google Scholar]
- Matzkin, H.; Eber, P.; Todd, B.; van der Zwaag, R.; Soloway, M.S. Prognostic significance of changes in prostate-specific markers after endocrine treatment of stage D2 prostatic cancer. Cancer 1992, 70, 2302–2309. [Google Scholar] [CrossRef]
- Partin, W.; Kattan, M.W.; Subong, E.N.; Walsh, P.C.; Wojno, K.J.; Oesterling, J.E.; Scardino, P.T.; Pearson, J.D. Combination of prostate-specific antigen, clinical stage, and Gleason score to predict pathological stage of localized prostate cancer. A multi-institutional update. J. Am. Med Assoc. 1997, 277, 1445–1451. [Google Scholar] [CrossRef]
- Pisansky, T.M.; Cha, S.S.; Earle, J.D.; Durr, E.D.; Kozelsky, T.F.; Wieand, H.S.; Oesterling, J.E. Prostate-specific antigen as a pretherapy prognostic factor in patients treated with radiation therapy for clinically localized prostate cancer. J. Clin. Oncol. 1993, 11, 2158–2166. [Google Scholar] [CrossRef]
- Ronco, A.L.; Fernandez, R. Improving Ultrasonographic Diagnosis of Prostate Cancer with Neural Networks. Ultrasound Med. Biol. 1999, 25, 729–733. [Google Scholar] [CrossRef]
- Metlin, C.; Lee, F.; Drago, J.; Murphy, G.P.; Investigators of the American Cancer Society National Prostate Cancer Detection Project. The American Cancer Society National Prostate Cancer Detection, Project: Findings on the detection of Early Prostate Cancer in 2425 Men. Cancer 1991, 67, 2949–2958. [Google Scholar] [CrossRef]
- Cosma, G.; McArdle, S.E.; Reeder, S.; Foulds, G.A.; Hood, S.; Khan, M.; Pockley, A.G. Identifying the presence of prostate cancer in individuals with psa levels <20ngml-1 using computational data extraction analysis of high dimensional peripheral blood flow cytometric phenotyping data. Front. Immunol. 2017, 8, 1771. [Google Scholar] [PubMed] [Green Version]
- Ren, S.; Wang, F.; Shen, J.; Sun, Y.; Xu, W.; Lu, J.; Wei, M.; Xu, C.; Wu, C.; Zhang, Z.; et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived mini RNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur. J. Cancer 2013, 49, 2949–2959. [Google Scholar] [CrossRef]
- Stamey, T.A.; Yang, N.; Hay, A.R.; McNeal, J.E.; Freiha, F.S.; Redwine, E. Prostate-specific antigen as a serum marker for adenocarcinoma of the prostate. N. Engl. J. Med. 1987, 317, 909–916. [Google Scholar] [CrossRef]
- Saritas, I.; Allahverdi, N.; Sert, I.U. A Fuzzy Expert System Design for Diagnosis of Prostate Cancer. In Proceedings of the International Conference on Computer Systems and Technologies—CompSysTech’03, Sofia, Bulgaria, 19–20 June 2003; pp. 345–351. [Google Scholar]
- Saritas, I.; Allahverdi, N.; Sert, U. A fuzzy approach for determination of prostate cancer. Int. J. Intell. Syst. Appl. Eng. 2013, 1, 1–7. [Google Scholar]
- Benecchi, L. Neuro-fuzzy system for prostate cancer diagnosis. Urology 2006, 68, 357–361. [Google Scholar] [CrossRef]
- Yuksel, S.; Dizman, T.; Yildizdan, G.; Sert, U. Application of soft sets to diagnose the prostate cancer risk. J. Inequal. Appl. 2013, 2013, 229. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Ye, J.; Cui, W. An evaluation method of risk grades for prostate cancer using similarity measure of cubic hesitant fuzzy sets. J. Biomed. Inform. 2018, 87, 131–137. [Google Scholar] [CrossRef]
- Smarandache, F. Neutrosophic Logic. Neutrosophy, Neutrosophic Set, Neutrosophic Probability; Amrican Reserch Press: Rehoboth, NM, USA, 1999. [Google Scholar]
- Ye, J. Multiple-attribute decision-making method using similarity measures of single-valued neutrosophic hesitant fuzzy sets based on least common multiple cardinality. J. Intell. Fuzzy Syst. 2018, 34, 4203–4211. [Google Scholar] [CrossRef]
- Aslam, M.; Albassam, M. Application of Neutrosophic Logic to Evaluate Correlation between Prostate Cancer Mortality and Dietary Fat Assumption. Symmetry 2019, 11, 330. [Google Scholar] [CrossRef]
- Wang, H.; Smarandache, F.; Zhang, Y.Q.; Sunderaman, R. Interval Neutrosophic Sets and Logic, Theory and Applications in Computing; Hexis: Phoenix, AZ, USA, 2005. [Google Scholar]
- Thai, H.D.; Huh, J.H. Optimizing patient transportation by applying cloud computing and big data analysis. J. Supercomput. 2022, 1–30. [Google Scholar] [CrossRef]
- Fu, J.; Ye, J.; Cui, W. The Dice measure of cubic hesitant fuzzy sets and its initial evaluation method of benign prostatic hyperplasia symptoms. Sci. Rep. 2019, 9, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, J.; Ye, J. Similarity measure with indeterminate parameters regarding cubic hesitant neutrosophic numbers and its risk grade assessment approach for prostate cancer patients. Appl. Intell. 2020, 50, 2120–2131. [Google Scholar] [CrossRef]
- Choi, W.H.; Huh, J.H. A Survey to Reduce STDs Infection in Mongolia and Big Data Virtualization Propagation. Electronics 2021, 10, 3101. [Google Scholar] [CrossRef]
- Ho, T.; Thanh, T.D. Discovering community Interests approach to topic model with time factor andclustering methods. J. Inf. Process. Syst. 2021, 17, 163–177. [Google Scholar]
- Kadian, R.; Kumar, S. A novel intuitionistic Renyi’s–Tsallis discriminant information measure and itsapplications in decision-making. Granul. Comput. 2021, 6, 901–913. [Google Scholar] [CrossRef]
- Torra, V. Hesitant fuzzy sets. Int. J. Intell. Syst. 2010, 25, 529–539. [Google Scholar] [CrossRef]
- Jun, Y.B.; Kim, C.S.; Yang, K.O. Cubic sets. Ann. Fuzzy Math. Inform. 2012, 4, 83–98. [Google Scholar]
- Applegate, C.; Rowles, J.; Ranard, K.; Jeon, S.; Erdman, J. Soy consumption and the risk of prostate cancer: An updated systematic review and meta-analysis. Nutrients 2018, 10, 40. [Google Scholar] [CrossRef] [Green Version]
- Seker, H.; Odeyato, M.; Petrovic, D.; Naguib, R.N.G. A Fuzzy Logic Based Method for Prognostic Decision Making in Breast and Prostate Cancers. IEEE Trans. Inf. Technol. Biomed. 2003, 7, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Torra, V.; Narukawa, Y. On hesitant fuzzy sets and decision. In Proceedings of the 18th IEEE International Conference on Fuzzy Systems, Jeju Island, Korea, 20–24 August 2009; pp. 1378–1382. [Google Scholar]


| Risk Grades | PSA (ng/mL) | Gleason Score | Clinical Staging |
|---|---|---|---|
| Small-risk | <10, 4.0, 3.5 | ≤6 | ≤ |
| Fair-risk | 10–20, 4.0–10, 3.5–4.5 | 7 | |
| Large-risk | >20, 10, 4.5 | ≥8 | ≥ |
| Staging | PSA (ng/mL) | Description |
|---|---|---|
| Stag 1 | Low PSA level | |
| Stag 2 | Moderate PSA level | |
| Stag 3 | High PSA level |
| Gleason Grade | Gleason Score | Histological Description |
|---|---|---|
| Grade cannot be evaluate | Not exist | |
| Well differentiated | ||
| Moderately differentiated | ||
| Poorly differentiated |
| Clinical Staging | Description | Score |
|---|---|---|
| Tumor gains ≤50% of one lobe | ||
| Tumor gains >50% of not both lobes but one lobe | ||
| Tumor gains both of the lobes |
| Risk Grades | : PSA (ng/mL) | : Gleason Score | : Clinical Staging |
|---|---|---|---|
| Risk Grades | |||
|---|---|---|---|
| (Small-risk) | |||
| (Fair-risk) | |||
| (Large-risk) |
| Patiens | : PSA(ng/mL) | : Gleason Score | : Clinical Staging |
|---|---|---|---|
| Patients | Risk Grade Based on Table 1 | |
|---|---|---|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madasi, J.D.; Al-Shbeil, I.; Cătaş, A.; Aloraini, N.; Gulistan, M.; Azhar, M. A Neutrosophic Cubic Hesitant Fuzzy Decision Support System, Application in the Diagnosis and Grading of Prostate Cancer. Fractal Fract. 2022, 6, 648. https://doi.org/10.3390/fractalfract6110648
Madasi JD, Al-Shbeil I, Cătaş A, Aloraini N, Gulistan M, Azhar M. A Neutrosophic Cubic Hesitant Fuzzy Decision Support System, Application in the Diagnosis and Grading of Prostate Cancer. Fractal and Fractional. 2022; 6(11):648. https://doi.org/10.3390/fractalfract6110648
Chicago/Turabian StyleMadasi, Joseph David, Isra Al-Shbeil, Adriana Cătaş, Najla Aloraini, Muhammad Gulistan, and Muhammad Azhar. 2022. "A Neutrosophic Cubic Hesitant Fuzzy Decision Support System, Application in the Diagnosis and Grading of Prostate Cancer" Fractal and Fractional 6, no. 11: 648. https://doi.org/10.3390/fractalfract6110648






