Gray Level Co-Occurrence Matrix, Fractal and Wavelet Analyses of Discrete Changes in Cell Nuclear Structure following Osmotic Stress: Focus on Machine Learning Methods
Abstract
:1. Introduction
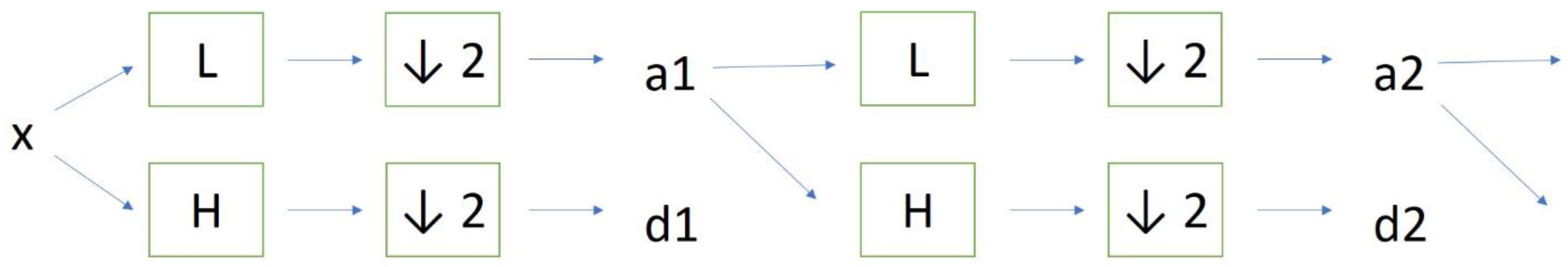
2. Materials and Methods
3. Results
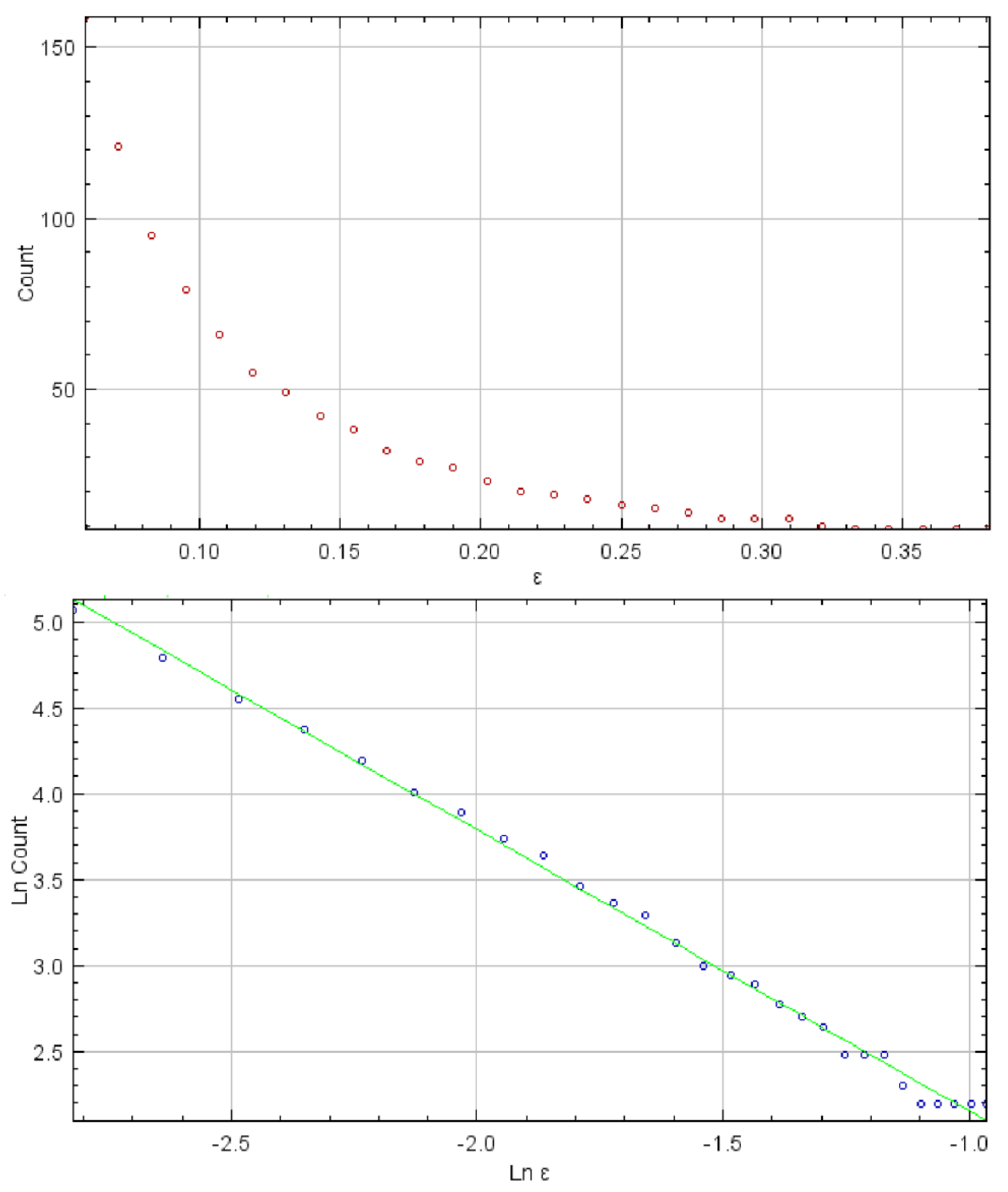
3.1. Results of GLCM, Fractal, and DWT Analyses
3.2. Machine Learning Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- AlKubeyyer, A.; Ben Ismail, M.M.; Bchir, O.; Alkubeyyer, M. Automatic detection of the meningioma tumor firmness in MRI images. J. X-Ray Sci. Technol. 2020, 28, 659–682. [Google Scholar] [CrossRef] [PubMed]
- Althubiti, S.A.; Paul, S.; Mohanty, R.; Mohanty, S.N.; Alenezi, F.; Polat, K. Ensemble Learning Framework with GLCM Texture Extraction for Early Detection of Lung Cancer on CT Images. Comput. Math. Methods Med. 2022, 2022, 2733965. [Google Scholar] [CrossRef]
- Alyami, J.; Sadad, T.; Rehman, A.; Almutairi, F.; Saba, T.; Bahaj, S.A.; Alkhurim, A. Cloud Computing-Based Framework for Breast Tumor Image Classification Using Fusion of AlexNet and GLCM Texture Features with Ensemble Multi-Kernel Support Vector Machine (MK-SVM). Comput. Intell. Neurosci. 2022, 2022, 7403302. [Google Scholar] [CrossRef]
- Anand, L.; Mewada, S.; Shamsi, W.; Ritonga, M.; Aflisia, N.; KumarSarangi, P.; NdoleArthur, M. Diagnosis of Prostate Cancer Using GLCM Enabled KNN Technique by Analyzing MRI Images. BioMed. Res. Int. 2023, 2023, 3913351. [Google Scholar] [CrossRef] [PubMed]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical Image Analysis using Convolutional Neural Networks: A Review. J. Med. Syst. 2018, 42, 226. [Google Scholar] [CrossRef] [Green Version]
- Yu, K.H.; Beam, A.L.; Kohane, I.S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2018, 2, 719–731. [Google Scholar] [CrossRef]
- Pantic, I.; Cumic, J.; Dugalic, S.; Petroianu, G.; Corridon, P. Gray level co-occurrence matrix and wavelet analyses reveal discrete changes in proximal tubule cell nuclei after mild acute kidney injury. Sci. Rep. 2022, 13, 4025. [Google Scholar] [CrossRef]
- Davidovic, L.M.; Cumic, J.; Dugalic, S.; Vicentic, S.; Sevarac, Z.; Petroianu, G.; Corridon, P.; Pantic, I. Gray-Level Co-occurrence Matrix Analysis for the Detection of Discrete, Ethanol-Induced, Structural Changes in Cell Nuclei: An Artificial Intelligence Approach. Microsc. Microanal. Off. J. Microsc. Soc. Am. Microbeam Anal. Soc. Microsc. Soc. Can. 2021, 28, 265–271. [Google Scholar] [CrossRef]
- Dimitriadis, I.; Zaninovic, N.; Badiola, A.C.; Bormann, C.L. Artificial intelligence in the embryology laboratory: A review. Reprod. Biomed. Online 2021, 44, 435–448. [Google Scholar] [CrossRef]
- Hudson, I.L. Data Integration Using Advances in Machine Learning in Drug Discovery and Molecular Biology. Methods Mol. Biol. 2021, 2190, 167–184. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.M.; Khan, R.A.; Arif, S.; Sajid, U. Artificial intelligence for breast cancer analysis: Trends & directions. Comput. Biol. Med. 2022, 142, 105221. [Google Scholar] [CrossRef]
- Pantic, I.V.; Shakeel, A.; Petroianu, G.A.; Corridon, P.R. Analysis of Vascular Architecture and Parenchymal Damage Generated by Reduced Blood Perfusion in Decellularized Porcine Kidneys Using a Gray Level Co-occurrence Matrix. Front. Cardiovasc. Med. 2022, 9, 797283. [Google Scholar] [CrossRef] [PubMed]
- Alkhodari, M.; Fraiwan, L. Convolutional and recurrent neural networks for the detection of valvular heart diseases in phonocardiogram recordings. Comput. Methods Programs Biomed. 2021, 200, 105940. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR Conference 2015, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Zhang, T.; Zeng, Y.; Zhang, Y.; Zhang, X.; Shi, M.; Tang, L.; Zhang, D.; Xu, B. Neuron type classification in rat brain based on integrative convolutional and tree-based recurrent neural networks. Sci. Rep. 2021, 11, 7291. [Google Scholar] [CrossRef]
- Tan, J.; Gao, Y.; Liang, Z.; Cao, W.; Pomeroy, M.J.; Huo, Y.; Li, L.; Barish, M.A.; Abbasi, A.F.; Pickhardt, P.J. 3D-GLCM CNN: A 3-Dimensional Gray-Level Co-Occurrence Matrix-Based CNN Model for Polyp Classification via CT Colonography. IEEE Trans. Med. Imaging 2020, 39, 2013–2024. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.C.; Ritter, L.J.; Whitty, A.; Fernandez, R.C.; Moran, L.J.; Robertson, S.A.; Thompson, J.G.; Brown, H.M. Gray level Co-occurrence Matrices (GLCM) to assess microstructural and textural changes in pre-implantation embryos. Mol. Reprod. Dev. 2016, 83, 701–713. [Google Scholar] [CrossRef]
- Vidya, K.S.; Ng, E.Y.; Acharya, U.R.; Chou, S.M.; Tan, R.S.; Ghista, D.N. Computer-aided diagnosis of Myocardial Infarction using ultrasound images with DWT, GLCM and HOS methods: A comparative study. Comput. Biol. Med. 2015, 62, 86–93. [Google Scholar] [CrossRef]
- Gupta, S.; Savala, R.; Gupta, N.; Dey, P. Fractal dimension and chromatin textural analysis to differentiate follicular carcinoma and adenoma on fine needle aspiration cytology. Cytopathol. Off. J. Br. Soc. Clin. Cytol. 2020, 31, 491–493. [Google Scholar] [CrossRef]
- Mattos, A.C.; Florindo, J.B.; Adam, R.L.; Lorand-Metze, I.; Metze, K. The Fractal Dimension Suggests Two Chromatin Configurations in Small Cell Neuroendocrine Lung Cancer and Is an Independent Unfavorable Prognostic Factor for Overall Survival. Microsc. Microanal. Off. J. Microsc. Soc. Am. Microbeam Anal. Soc. Microsc. Soc. Can. 2022, 28, 522–526. [Google Scholar] [CrossRef]
- Metze, K.; Adam, R.; Florindo, J.B. The fractal dimension of chromatin—A potential molecular marker for carcinogenesis, tumor progression and prognosis. Expert Rev. Mol. Diagn. 2019, 19, 299–312. [Google Scholar] [CrossRef]
- Brocker, C.; Thompson, D.C.; Vasiliou, V. The role of hyperosmotic stress in inflammation and disease. Biomol. Concepts 2012, 3, 345–364. [Google Scholar] [CrossRef] [PubMed]
- Colin, L.; Ruhnow, F.; Zhu, J.K.; Zhao, C.; Zhao, Y.; Persson, S. The cell biology of primary cell walls during salt stress. Plant Cell 2023, 35, 201–217. [Google Scholar] [CrossRef]
- Reiling, J.H.; Sabatini, D.M. Stress and mTORture signaling. Oncogene 2006, 25, 6373–6383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadowska, A.; Kameda, T.; Krupkova, O.; Wuertz-Kozak, K. Osmosensing, osmosignalling and inflammation: How intervertebral disc cells respond to altered osmolarity. Eur. Cells Mater. 2018, 36, 231–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itto-Nakama, K.; Watanabe, S.; Ohnuki, S.; Kondo, N.; Kikuchi, R.; Nakamura, T.; Ogasawara, W.; Kasahara, K.; Ohya, Y. Prediction of ethanol fermentation under stressed conditions using yeast morphological data. J. Biosci. Bioeng. 2023, 135, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Pereira, D.R.; Papa, J.P.; Saraiva, G.F.R.; Souza, G.M. Automatic classification of plant electrophysiological responses to environmental stimuli using machine learning and interval arithmetic. Comput. Electron. Agric. 2018, 145, 35–42. [Google Scholar] [CrossRef] [Green Version]
- Santos, T.A.; Maistro, C.E.; Silva, C.B.; Oliveira, M.S.; Franca, M.C., Jr.; Castellano, G. MRI Texture Analysis Reveals Bulbar Abnormalities in Friedreich Ataxia. AJNR Am. J. Neuroradiol. 2015, 36, 2214–2218. [Google Scholar] [CrossRef] [Green Version]
- Kociołek, M.; Materka, A.; Strzelecki, M.; Szczypinski, P. Discrete wavelet transform—Derived features for digital image texture analysis. In Proceedings of the International Conference on Signals and Electronic Systems, Lodz, Poland, 18–21 September 2001; pp. 163–168. [Google Scholar]
- Strzelecki, M.; Szczypinski, P.; Materka, A.; Klepaczko, A. A software tool for automatic classification and segmentation of 2D/3D medical images. Nucl. Instrum. Methods Phys. Res. A 2013, 702, 137–140. [Google Scholar] [CrossRef]
- Szczypinski, P.; Strzelecki, M.; Materka, A. MaZda—A Software for Texture Analysis. In Proceedings of the 2007 International Symposium on Information Technology Convergence, ISITC 2007, Jeonju, Republic of Korea, 23–24 November 2007; pp. 245–249. [Google Scholar]
- Szczypinski, P.; Strzelecki, M.; Materka, A.; Klepaczko, A. MaZda-A software package for image texture analysis. Comput. Methods Programs Biomed. 2009, 94, 66–76. [Google Scholar] [CrossRef]
- Mallat, S. A Wavelet Tour of Signal Processing; Academic Press: San Diego, CA, USA, 1998. [Google Scholar]
- Karperien, A. FracLac for ImageJ. Available online: http://rsb.info.nih.gov/ij/plugins/fraclac/FLHelp/Introduction.htm (accessed on 28 January 2023).
- Dincic, M.; Todorovic, J.; Nesovic Ostojic, J.; Kovacevic, S.; Dunderovic, D.; Lopicic, S.; Spasic, S.; Radojevic-Skodric, S.; Stanisavljevic, D.; Ilic, A.Z. The Fractal and GLCM Textural Parameters of Chromatin May Be Potential Biomarkers of Papillary Thyroid Carcinoma in Hashimoto’s Thyroiditis Specimens. Microsc. Microanal. Off. J. Microsc. Soc. Am. Microbeam Anal. Soc. Microsc. Soc. Can. 2020, 26, 717–730. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Kim, J.; Kwon, N.; Chang, S.; Kim, K.T.; Lee, D.; Kim, S.; Yun, S.J.; Hwang, D.; Kim, J.W.; Hwu, Y.; et al. Altered branching patterns of Purkinje cells in mouse model for cortical development disorder. Sci. Rep. 2011, 1, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cozzini, T.; Piona, C.; Marchini, G.; Merz, T.; Brighenti, T.; Bonetto, J.; Marigliano, M.; Olivieri, F.; Maffeis, C.; Pedrotti, E. In vivo confocal microscopy study of corneal nerve alterations in children and youths with Type 1 diabetes. Pediatr. Diabetes 2021, 22, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, X.; Wang, Z.H.; Lin, Z.; Yang, J.; Chen, J.; Wang, R.; Ye, W.; Li, Y.; Wu, Y.; et al. Changes in dendritic complexity and spine morphology following BCG immunization in APP/PS1 mice. Hum. Vaccines Immunother. 2022, 18, 2121568. [Google Scholar] [CrossRef] [PubMed]
- Blomberg, A. Yeast osmoregulation—Glycerol still in pole position. FEMS Yeast Res. 2022, 22, foac035. [Google Scholar] [CrossRef]
- Saxena, A.; Sitaraman, R. Osmoregulation in Saccharomyces cerevisiae via mechanisms other than the high-osmolarity glycerol pathway. Microbiology 2016, 162, 1511–1526. [Google Scholar] [CrossRef]
- de Nadal, E.; Posas, F. The HOG pathway and the regulation of osmoadaptive responses in yeast. FEMS Yeast Res. 2022, 22, foac013. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Pantic, I.; Paunovic, J.; Cumic, J.; Valjarevic, S.; Petroianu, G.A.; Corridon, P.R. Artificial neural networks in contemporary toxicology research. Chem. Biol. Interact. 2023, 369, 110269. [Google Scholar] [CrossRef]




| Osmotic Stress | Controls | |
|---|---|---|
| Angular second moment | 0.00076 ± 0.00092 ** | 0.0015 ± 0.0011 |
| Inverse difference moment | 0.154 ± 0.012 ** | 0.167 ± 0.013 |
| Contrast | 59.87 ± 8.78 ** | 51.03 ± 9.17 |
| Correlation | 0.0019 ± 0.0023 ** | 0.0041 ± 0.0037 |
| Variance | 799.39 ± 363.96 ** | 507.17 ± 434.37 |
| Fractal dimension | 1.454 ± 0.155 ** | 1.538 ± 0.142 |
| DWT coefficient energy | 0.369 ± 0.089 * | 0.250 ± 0.084 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pantic, I.; Valjarevic, S.; Cumic, J.; Paunkovic, I.; Terzic, T.; Corridon, P.R. Gray Level Co-Occurrence Matrix, Fractal and Wavelet Analyses of Discrete Changes in Cell Nuclear Structure following Osmotic Stress: Focus on Machine Learning Methods. Fractal Fract. 2023, 7, 272. https://doi.org/10.3390/fractalfract7030272
Pantic I, Valjarevic S, Cumic J, Paunkovic I, Terzic T, Corridon PR. Gray Level Co-Occurrence Matrix, Fractal and Wavelet Analyses of Discrete Changes in Cell Nuclear Structure following Osmotic Stress: Focus on Machine Learning Methods. Fractal and Fractional. 2023; 7(3):272. https://doi.org/10.3390/fractalfract7030272
Chicago/Turabian StylePantic, Igor, Svetlana Valjarevic, Jelena Cumic, Ivana Paunkovic, Tatjana Terzic, and Peter R. Corridon. 2023. "Gray Level Co-Occurrence Matrix, Fractal and Wavelet Analyses of Discrete Changes in Cell Nuclear Structure following Osmotic Stress: Focus on Machine Learning Methods" Fractal and Fractional 7, no. 3: 272. https://doi.org/10.3390/fractalfract7030272
APA StylePantic, I., Valjarevic, S., Cumic, J., Paunkovic, I., Terzic, T., & Corridon, P. R. (2023). Gray Level Co-Occurrence Matrix, Fractal and Wavelet Analyses of Discrete Changes in Cell Nuclear Structure following Osmotic Stress: Focus on Machine Learning Methods. Fractal and Fractional, 7(3), 272. https://doi.org/10.3390/fractalfract7030272










