Molecular Docking and ADME-TOX Profiling of Moringa oleifera Constituents against SARS-CoV-2
Abstract
:Highlights
- Bioactive compounds of Moringa oleifera exhibited activity against SARS-CoV-2.
- Computational approaches to studying the antiviral activity of natural compounds against SARS-CoV-2 might be a time- and money-saving option in the drug discovery and development process.
- The antiviral potential of Moringa oleifera against SARS-CoV-2 may contribute to an advanced level of pharmaceutical research.
- Advanced computational methods can be used to search for novel anti-SARS-CoV-2 agents from natural products.
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Binders
2.2. Molecular Docking
2.3. ADME-TOX Prediction
3. Results
3.1. Molecular Docking
3.2. ADME-TOX Prediction
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, D.; Wu, T.; Liu, Q.; Yang, Z. The SARS-CoV-2 outbreak: What we know. Int. J. Infect. Dis. 2020, 94, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Beg, A.Z. Antimicrobial and phytochemical studies on 45 Indian medicinal plants against multi-drug resistant human pathogens. J. Ethnopharmacol. 2001, 74, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Patra, B.; Schluttenhofer, C.; Wu, Y.; Pattanaik, S.; Yuan, L. Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2013, 1829, 1236–1247. [Google Scholar] [CrossRef] [PubMed]
- Anwar, F.; Latif, S.; Ashraf, M.; Gilani, A.H. Moringa oleifera: A food plant with multiple medicinal uses. Phytother. Res. 2007, 21, 17–25. [Google Scholar] [CrossRef]
- Gassenschmidt, U.; Jany, K.D.; Bernhard, T.; Niebergall, H. Isolation and characterization of a flocculating protein from Moringa oleifera Lam. Biochem. Biophys. Acta BBA Gen. Subj. 1995, 1243, 477–481. [Google Scholar] [CrossRef]
- Matos, F.J.A. Living Pharmacies: A System for Using Medicinal Plants Designed for Small Communities, 4th ed.; Editora Imprensa Universitária: Fortaleza, Brazil, 2002. [Google Scholar]
- Bezerra, A.M.E.; Momenté, V.G.; Medeiros Filho, S. Seed germination and seedling development of moringa (Moringa oleifera Lam.) as a function of seed weight and substrate type. Hortic Bras. 2004, 22, 295–299. [Google Scholar] [CrossRef]
- Makkar, H.; Becker, K. Nutrional value and antinutritional components of whole and ethanol extracted Moringa oleifera leaves. Anim. Feed. Sci. Technol. 1996, 63, 211–228. [Google Scholar] [CrossRef]
- Mehta, K.; Balaraman, R.; Amin, A.; Bafna, P.; Gulati, O. Effect of fruits of Moringa oleifera on the lipid profile of normal and hypercholesterolaemic rabbits. J. Ethnopharmacol. 2003, 86, 191–195. [Google Scholar] [CrossRef]
- Jaiswal, D.; Rai, P.K.; Kumar, A.; Mehta, S.; Watal, G. Effect of Moringa oleifera Lam. leaves aqueous extract therapy on hyperglycemic rats. J. Ethnopharmacol. 2009, 123, 392–396. [Google Scholar] [CrossRef]
- Waterman, C.; Cheng, D.M.; Rojas-Silva, P.; Poulev, A.; Dreifus, J.; Lila, M.A.; Raskin, I. Stable, water extractable isothiocyanates from Moringa oleifera leaves attenuate inflammation in vitro. Phytochemistry 2014, 103, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, C.; Peter, K.V.; Gopalakrishnan, P.K. Drumstick (Moringa oleifera): A multipurpose Indian vegetable. Econ. Bot. 1980, 34, 276–283. [Google Scholar] [CrossRef]
- Gasteiger, J. The central role of chemoinformatics. Chemom. Intell. Lab. Syst. 2006, 82, 200–209. [Google Scholar] [CrossRef]
- Oprea, T.I.; Matter, H. Integrating virtual screening in lead discovery. Curr. Opin. Chem. Biol. 2004, 8, 349–358. [Google Scholar] [CrossRef]
- Rocha, J.A.; Rego, N.C.S.; Carvalho, B.T.S.; Silva, F.I.; Sousa, J.A.; Ramos, R.M.; Passos, I.N.G.; de Moraes, J.; Leite, J.R.S.A.; Lima, F.C.A. Computational quantum chemistry, molecular docking, and ADMET predictions of imidazole alkaloids of Pilocarpus microphyllus with schistosomicidal properties. PLoS ONE 2018, 13, e0198476. [Google Scholar] [CrossRef]
- Abe, R.; Ohtani, K. An ethnobotanical study of medicinal plants and traditional therapies on Batan Island, the Philippines. J. Ethnopharmacol. 2013, 145, 554–565. [Google Scholar] [CrossRef]
- Leone, A.; Spada, A.; Battezzati, A.; Schiraldi, A.; Aristil, J.; Bertoli, S. Cultivation, Genetics, Ethnopharmacology, Phytochemistry and Pharmacology of Moringa oleifera Leaves: An Overview. Int. J. Mol. Sci. 2015, 16, 12791. [Google Scholar] [CrossRef]
- Razis, A.F.A.; Ibrahim, M.D.; Kntayya, S.B. Health Benefits of Moringa oleifera. Asian Pac. J. Cancer Prev. 2014, 15, 8571–8576. [Google Scholar] [CrossRef]
- Padayachee, B.; Baijnath, H. An updated comprehensive review of the medicinal, phytochemical and pharmacological properties of Moringa oleifera. S. Afr. J. Bot. 2020, 129, 304–316. [Google Scholar] [CrossRef]
- Muratov, E.N.; Amaro, R.; Andrade, C.H.; Brown, N.; Ekins, S.; Fourches, D.; Isayev, O.; Kozakov, D.; Medina-Franco, J.L.; Merz, K.M.; et al. A critical overview of computational approaches employed for COVID-19 drug discovery. Chem. Soc. Rev. 2021, 50, 9121–9151. [Google Scholar] [CrossRef]
- Mishra, B.B.; Tiwari, V.K. Natural products: An evolving role in future drug discovery. Eur. J. Med. Chem. 2011, 46, 4769–4807. [Google Scholar] [CrossRef] [PubMed]
- Mehyar, N. Coronaviruses SARS-CoV, MERS-CoV, and SARS-CoV-2 helicase inhibitors: A systematic review of in vitro studies. J. Virus Erad. 2023, 9, 100327. [Google Scholar] [CrossRef] [PubMed]
- Power, H.; Wu, J.; Turville, S.; Aggarwal, A.; Valtchev, P.; Schindeler, A.; Dehghani, F. Virtual screening and in vitro validation of natural compound inhibitors against SARS-CoV-2 spike protein. Bioorg. Chem. 2022, 119, 105574. [Google Scholar] [CrossRef] [PubMed]
- Guedes, I.A.; de Magalhães, C.S.; Dardenne, L.E. Receptor-ligand molecular docking. Biophys. Rev. 2014, 6, 75–87. [Google Scholar] [CrossRef]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Barros, R.O.; Junior, F.L.C.C.; Pereira, W.S.; Oliveira, N.M.N.; Ramos, R.M. Interaction of Drug Candidates with Various SARS-CoV-2 Receptors: An in Silico Study to Combat COVID-19. J. Proteome Res. 2020, 19, 4567–4575. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. Ligplot: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Anand, K.; Ziebuhr, J.; Wadhwani, P.; Mesters, J.R.; Hilgenfeld, R. Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science 2003, 300, 1763–1767. [Google Scholar] [CrossRef]
- Kumar, S.B.; Krishna, S.; Pradeep, S.; Mathews, D.E.; Pattabiraman, R.; Murahari, M.; Murthy, T.K. Screening of natural compounds from Cyperus rotundus Linn against SARS-CoV-2 main protease (Mpro): An integrated computational approach. Comput. Biol. Med. 2021, 134, 104524. [Google Scholar] [CrossRef] [PubMed]
- Glaab, E.; Manoharan, G.B.; Abankwa, D. Pharmacophore Model for SARS-CoV-2 3CLpro Small-Molecule Inhibitors and In Vitro Experimental Validation of Computationally Screened Inhibitors. J. Chem. Inf. Model 2021, 61, 4082–4096. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Bidon, M.; Jaimes, J.A.; Whittaker, G.R.; Daniel, S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020, 178, 104792. [Google Scholar] [CrossRef]
- Santos, E.S.; Silva, P.C.; Sousa, P.S.A.; Aquino, C.C.; Pacheco, G.; Teixeira, L.F.L.S.; Araujo, A.R.; Sousa, F.B.M.; Barros, R.O.; Ramo, R.M.; et al. Antiviral potential of diminazene aceturate against SARS-CoV-2 proteases using computational and in vitro approaches. Chem. Biol. Interact. 2022, 367, 110161. [Google Scholar] [CrossRef]
- Ho, C.; Nazarie, W.F.W.M.; Lee, P.-C. An In Silico Design of Peptides Targeting the S1/S2 Cleavage Site of the SARS-CoV-2 Spike Protein. Viruses 2023, 15, 1930. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Pokhrel, S.; Bouback, T.A.; Samad, A.; Nur, S.M.; Alam, R.; Abdullah-Al-Mamun, M.; Nain, Z.; Imon, R.R.; Talukder, E.K.; Tareq, M.I.; et al. Spike protein recognizer receptor ACE2 targeted identification of potential natural antiviral drug candidates against SARS-CoV-2. Int. J. Biol. Macromol. 2021, 30, 1114–1125. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Barreto, M.B.; De Freitas, J.V.B.; Silveira, E.R.; Bezerra, A.M.E.; Nunes, E.P.; Gramosa, N.V. Volatile and non-volatile chemical constituents of Moringa oleifera Lam., Moringaceae. Braz. J. Pharmacogn. 2009, 19, 893–897. [Google Scholar] [CrossRef]
- Ferreira, P.M.P.; Farias, D.F.; de Abreu Oliveira, J.T.; de Fátima Urano Carvalho, A. Moringa oleifera: Bioactive compounds and nutritional potential. Rev. Nutr. 2008, 21, 431–437. [Google Scholar] [CrossRef]
- Bicas, T.C. Effects of Hydroalcoholic Extract of Syzygium Leaves Malaccense and Moringa oleifera under Oxidative Stress in Streptozotocin—Induced Diabetic Rats. 26 August 2019. Available online: http://repositorio.utfpr.edu.br:8080/jspui/handle/1/4590 (accessed on 8 August 2023).
- Özcan, M.M. Moringa spp: Composition and bioactive properties. S. Afr. J. Bot. 2020, 129, 25–31. [Google Scholar] [CrossRef]
- Ahmadu, T.; Ahmad, K.; Ismail, S.I.; Rashed, O.; Asib, N.; Omar, D. Antifungal efficacy of Moringa oleifera leaf and seed extracts against Botrytis cinerea causing gray mold disease of tomato (Solanum lycopersicum L.). Braz. J. Biol. 2021, 81, 1007–1022. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Li, B.; Olayanju, J.B.; Drake, J.M.; Chen, N. Nutraceutical or Pharmacological Potential of Moringa oleifera Lam. Nutrients 2018, 10, 343. [Google Scholar] [CrossRef] [PubMed]
- Anzano, A.; de Falco, B.; Ammar, M.; Ricciardelli, A.; Grauso, L.; Sabbah, M.; Capparelli, R.; Lanzotti, V. Chemical Analysis and Antimicrobial Activity of Moringa oleifera Lam. Leaves and Seeds. Molecules 2022, 27, 8920. [Google Scholar] [CrossRef] [PubMed]
- MSS Calf. Identification of Organic Compounds Present in Ethanolic and Hexanic Extracts of Moringa oleifera LAM Leaves; State University of Piauí: Floriano, Brazil, 2020. [Google Scholar]
- Approved Drugs-National Health Surveillance Agency-Anvisa. Available online: https://www.gov.br/anvisa/pt-br/assuntos/paf/coronavirus/medicamentos (accessed on 8 August 2023).
- Ferreira, L.L.G.; Andricopulo, A.D. Medicines and treatments for COVID-19. Adv. Stud. 2020, 34, 7–27. [Google Scholar]
- Neto, I.F.D.S.; Ricardino, I.E.F.; dos Santos, Í.T.; de Lima, E.V.M.; Souza, M.N.C.; Marques, A.E.F.; Silva, M.R. A review of the antiviral activity of the Indian Nim and its potential in front of the new coronavirus (SARS-CoV-2). J. Biol. Pharm. Agric. Manag. 2021, 17, 108–126. [Google Scholar]
- García-Niño, W.R.; Zazueta, C. Ellagic acid: Pharmacological activities and molecular mechanisms involved in liver protection. Pharmacol. Res. 2015, 97, 84–103. [Google Scholar] [CrossRef]
- Vattem, D.; Shetty, K. Biological functionality of ellagic acid: A review. J. Food Biochem. 2005, 29, 234–266. [Google Scholar] [CrossRef]
- Xu, Y.-M.; Deng, J.-Z.; Ma, J.; Chen, S.-N.; Marshall, R.; Jones, S.H.; Johnson, R.K.; Hecht, S.M. DNA damaging activity of ellagic acid derivatives. Bioorg. Med. Chem. 2003, 11, 1593–1596. [Google Scholar] [CrossRef]
- Ibrahim, A.K.; Youssef, A.I.; Arafa, A.S.; Ahmed, S.A. Anti-H5N1 virus flavonoids from Capparis sinaica Veill. Nat. Prod. Res. 2013, 27, 2149–2153. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.H.; Lee, J.Y.; Cho, C.H.; Kim, C.J. Anti-asthmatic action of quercetin and rutin in conscious guinea-pigs challenged with aerosolized ovalbumin. Arch. Pharmacal Res. 2007, 30, 1599–1607. [Google Scholar] [CrossRef]
- Araruna, M.K.; Brito, S.A.; Morais-Braga, M.F.; Santos, K.K.; Souza, T.M.; Leite, T.R.; Costa, J.G.; Coutinho, H.D. Evaluation of antibiotic & antibiotic modifying activity of pilocarpine & rutin. Indian J. Med. Res. 2012, 135, 252–254. [Google Scholar] [PubMed]
- Umar, S.; Mishra, N.K.; Pal, K.; Sajad, M.; Neha; Ansari, M.; Ahmad, S.; Katiyar, C.K.; Khan, H.A. Protective effect of rutin in attenuation of collagen-induced arthritis in Wistar rat by inhibiting inflammation and oxidative stress. Indian J. Rheumatol. 2012, 7, 191–198. [Google Scholar] [CrossRef]
- Leong, C.N.A.; Tako, M.; Hanashiro, I.; Tamaki, H. Antioxidant flavonoid glycosides from the leaves of Ficus pumila L. Food Chem. 2008, 109, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Semwal, D.K.; Semwal, R.B.; Combrinck, S.; Viljoen, A. Myricetin: A dietary molecule with diverse biological activities. Nutrients 2016, 8, 90. [Google Scholar] [CrossRef]
- Jiang, M.; Zhu, M.; Wang, L.; Yu, S. Anti-tumor effects and associated molecular mechanisms of myricetin. Biomed. Pharmacother. 2019, 120, 109506. [Google Scholar] [CrossRef]
- Jiang, S.; Tang, X.; Chen, M.; He, J.; Su, S.; Liu, L.; He, M.; Xue, W. Design, synthesis and antibacterial activities against Xanthomonas oryzae pv. oryzae, Xanthomonas axonopodis pv. Citri and Ralstonia solanacearum of novel myricetin derivatives containing sulfonamide moiety. Pest Manag. Sci. 2020, 76, 853–860. [Google Scholar] [CrossRef]
- Ortega, J.T.; Suárez, A.I.; Serrano, M.L.; Baptista, J.; Pujol, F.H.; Rangel, H.R. The role of the glycosyl moiety of myricetin derivatives in anti-HIV-1 activity in vitro. AIDS Res. Ther. 2017, 14, 57. [Google Scholar] [CrossRef]
- Ren, R.; Yin, S.; Lai, B.; Ma, L.; Wen, J.; Zhang, X.; Lai, F.; Liu, S.; Li, L. Myricetin antagonizes semen-derived enhancer of viral infection (SEVI) formation and influences its infection-enhancing activity. Retrovirology 2018, 15, 49. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yao, J.; Han, C.; Yang, J.; Chaudhry, M.T.; Wang, S.; Liu, H.; Yin, Y. Quercetin, inflammation and immunity. Nutrients 2016, 8, 167. [Google Scholar] [CrossRef] [PubMed]
- Rahaman, S.T.; Mondal, S. Flavonoids: A vital resource in healthcare and medicine. Pharm. Pharmacol. Int. J. 2020, 8, 91–104. [Google Scholar] [CrossRef]
- Wang, W.; Sun, C.; Mao, L.; Ma, P.; Liu, F.; Yang, J.; Gao, Y. The biological activities, chemical stability, metabolism and delivery systems of quercetin: A review. Trends Food Sci. Technol. 2016, 56, 21–38. [Google Scholar] [CrossRef]
- López-Lázaro, M. Distribution and biological activities of the flavonoid luteolin. Mini Rev. Med. Chem. 2009, 9, 31–59. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Peng, W.H.; Tsai, K.D.; Hsu, S.L. Luteolin suppresses inflammation-associated gene expression by blocking NF-κB and AP-1 activation pathway in mouse alveolar macrophages. Life Sci. 2007, 81, 1602–1614. [Google Scholar] [CrossRef]
- Manju, V.; Balasubramaniyan, V.; Nalini, N. Rat colonic lipid peroxidation and antioxidant status: The effects of dietary luteolin on 1,2-dimethylhydrazine challenge. Cell. Mol. Biol. Lett. 2005, 10, 535. [Google Scholar]
- Han, D.-H.; Denison, M.S.; Tachibana, H.; Yamada, K. Relationship between estrogen receptor-binding and estrogenic activities of environmental estrogens and suppression by flavonoids. Biosci. Biotechnol. Biochem. 2002, 66, 1479–1487. [Google Scholar] [CrossRef]
- Zhu, X.; Zhang, H.; Lo, R. Phenolic compounds from the leaf extract of artichoke (Cynara scolymus L.) and their antimicrobial activities. J. Agric. Food Chem. 2004, 52, 7272–7278. [Google Scholar] [CrossRef]
- Yi, L.; Li, Z.; Yuan, K.; Qu, X.; Chen, J.; Wang, G.; Zhang, H.; Luo, H.; Zhu, L.; Jiang, P.; et al. Small Molecules Blocking the Entry of Severe Acute Respiratory Syndrome Coronavirus into Host Cells. J. Virol. 2004, 78, 11334–11339. [Google Scholar] [CrossRef]
- Da Costa, J.A.; de Oliveira Lima, D.; dos Santos Carvalho, A.G.; Martins, J.A.; dos Santos, M.D.S.; de Sousa, D.G.; da Silva Carvalho, G.; Barros, F.R.; Ferreira, K.R.; de Barros, G.M.; et al. Compostos bioativos derivados de matrizes alimentares com potencial terapêutico para a infecção por SARS-CoV-2: Uma revisão de estudos in silico. Res. Soc. Dev. 2021, 10, e17810817178. [Google Scholar] [CrossRef]
- Aini, N.S.; Kharisma, V.D.; Widyananda, M.H.; Murtadlo, A.A.A.; Probojati, R.T.; Turista, D.D.R.; Tamam, M.B.; Jakhmola, V.; Sari, D.P.; Albari, M.T.; et al. In silico screening of bioactive compounds from Syzygium cumini L. and Moringa oleifera L. against SARS-CoV-2 via tetra inhibitors. Pharmacogn. J. 2022, 14, 267–272. [Google Scholar] [CrossRef]
- Mawaddani, N.; Sutiyanti, E.; Widyananda, M.H.; Kharisma, V.D.; Turista, D.D.R.; Tamam, M.B.; Jakhmola, V.; Syamsurizal; Fajri, B.R.; Ghifari, M.R.; et al. In silico study of entry inhibitor from Moringa oleifera bioactive compounds against SARS-CoV-2 infection. Pharmacogn. J. 2022, 14, 565–574. [Google Scholar] [CrossRef]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef]
- Zumla, A.; Chan, J.F.; Azhar, E.I.; Hui, D.S.; Yuen, K.Y. Coronaviruses—Drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016, 15, 327–347. [Google Scholar] [CrossRef]
- Sanders, J.M.; Monogue, M.L.; Jodlowski, T.Z.; Cutrell, J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): A review. JAMA 2020, 323, 1824–1836. [Google Scholar] [CrossRef]
- Maurya, V.K.; Kumar, S.; Prasad, A.K.; Bhatt, M.L.B.; Saxena, S.K. Structure-based drug designing for potential antiviral activity of selected natural products from Ayurveda against SARS-CoV-2 spike glycoprotein and its cellular receptor. Virusdisease 2020, 31, 179–193. [Google Scholar] [CrossRef]
- Ashburn, T.T.; Thor, K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef]
- Araújo, J.L.; Azevedo, V.S.; Araújo, A.D.S.; Araújo, J.L.; Araújo, L.; Sousa, L.A.D.; Silva, G.T.; de Freitas Pinheir, W.; Santos, A.M.A.; Cruz, G.T.; et al. SARS-CoV-2 or COVID-19: The search for alternative treatment for the new coronavirus. Int. J. Dev. Res. 2020, 10, 37117–37122. [Google Scholar]
- Bastos, R.S.; Sousa, C.S.; Oliveira, J.S.; da Silva MH, V.; Lima FD, C.A.; Rocha, J.A. Prospecção de Proteínas do Novo Coronavírus COVID-2019 e Potencial da Bioinformática na Busca de Novas Drogas Promissoras. Cad. Prospecção 2020, 13, 347–358. [Google Scholar] [CrossRef]
- Araújo, J.L.; de Sousa, L.A.; Sousa, A.O.; Bastos, R.S.; Santos, G.T.; Lage, M.R.; Stoyanov, S.R.; Passos, I.N.G.; Azevedo, R.B.d.; Rocha, J.A.; et al. DFT, molecular docking, and ADME/Tox screening investigations of Market-Available drugs against SARS-CoV-2. J. Braz. Chem. Soc. 2021, 32, 1628–1641. [Google Scholar] [CrossRef]
- Dos Santos, A.F.; Ortiz, M.M.; Montagner, G.E.; Schultz, J.V.; Gomes, P.; da Silva, I.Z.; Fagan, S.B. In-Silico study of antivirals and non-antivirals for the treatment of SARS-CoV-2. Discip. Sci. Nat. Tecnol. 2022, 23, 57–83. [Google Scholar] [CrossRef]
- Islam, T.; Hasan, M.; Rahman, M.S.; Islam, R. Comparative evaluation of authorized drugs for treating COVID-19 patients. Health Sci. Rep. 2022, 5, e671. [Google Scholar] [CrossRef] [PubMed]
- Belal, A. Drug likeness, targets, molecular docking and ADMET studies for some indolizine derivatives. Int. J. Pharm. Sci. 2018, 73, 635–642. [Google Scholar]
- De Souza, J.; Freitas, Z.M.F.; Storpirtis, S. In vitro models for the determination of drug absorption and a prediction of dissolution/absorption relationships. Braz. J. Pharm. Sci. 2007, 43, 515–527. [Google Scholar]
- Zhao, Y.H.; Le, J.; Abraham, M.H.; Hersey, A.; Eddershaw, P.J.; Luscombe, C.N.; Boutina, D.; Beck, G.; Sherborne, B.; Cooper, I.; et al. Evaluation of human intestinal absorption data and subsequent derivation of a quantitative structure-activity relationship (QSAR) with the Abraham descriptors. J. Pharm. Sci. 2001, 90, 749–784. [Google Scholar] [CrossRef]
- Yee, S. In vitro permeability across Caco-2 cells (colonic) can predict in vivo (small intestinal) absorption in man—Fact or myth. Pharm. Res. 1997, 14, 763–766. [Google Scholar] [CrossRef]
- Chtita, S.; Belaidi, S.; Qais, F.A.; Ouassaf, M.; AlMogren, M.M.; Al-Zahrani, A.A.; Bakhouch, M.; Belhassan, A.; Zaki, H.; Bouachrine, M.; et al. Unsymmetrical aromatic disulfides as SARS-CoV-2 Mpro inhibitors: Molecular docking, molecular dynamics, and ADME scoring investigations. J. King Saud. Univ. Sci. 2022, 34, 102226. [Google Scholar] [CrossRef]
- Flores-Holguín, N.; Frau, J.; Glossman-Mitnik, D. Computational Pharmacokinetics Report, ADMET Study and Conceptual DFT-Based Estimation of the Chemical Reactivity Properties of Marine Cyclopeptides. ChemistryOpen 2021, 10, 1142. [Google Scholar] [CrossRef]
- Mortelmans, K.; Zeiger, E. The Ames Salmonella/microsome mutagenicity assay. Mutat. Res. Mol. Mech. Mutagen. 2000, 455, 29–60. [Google Scholar] [CrossRef]


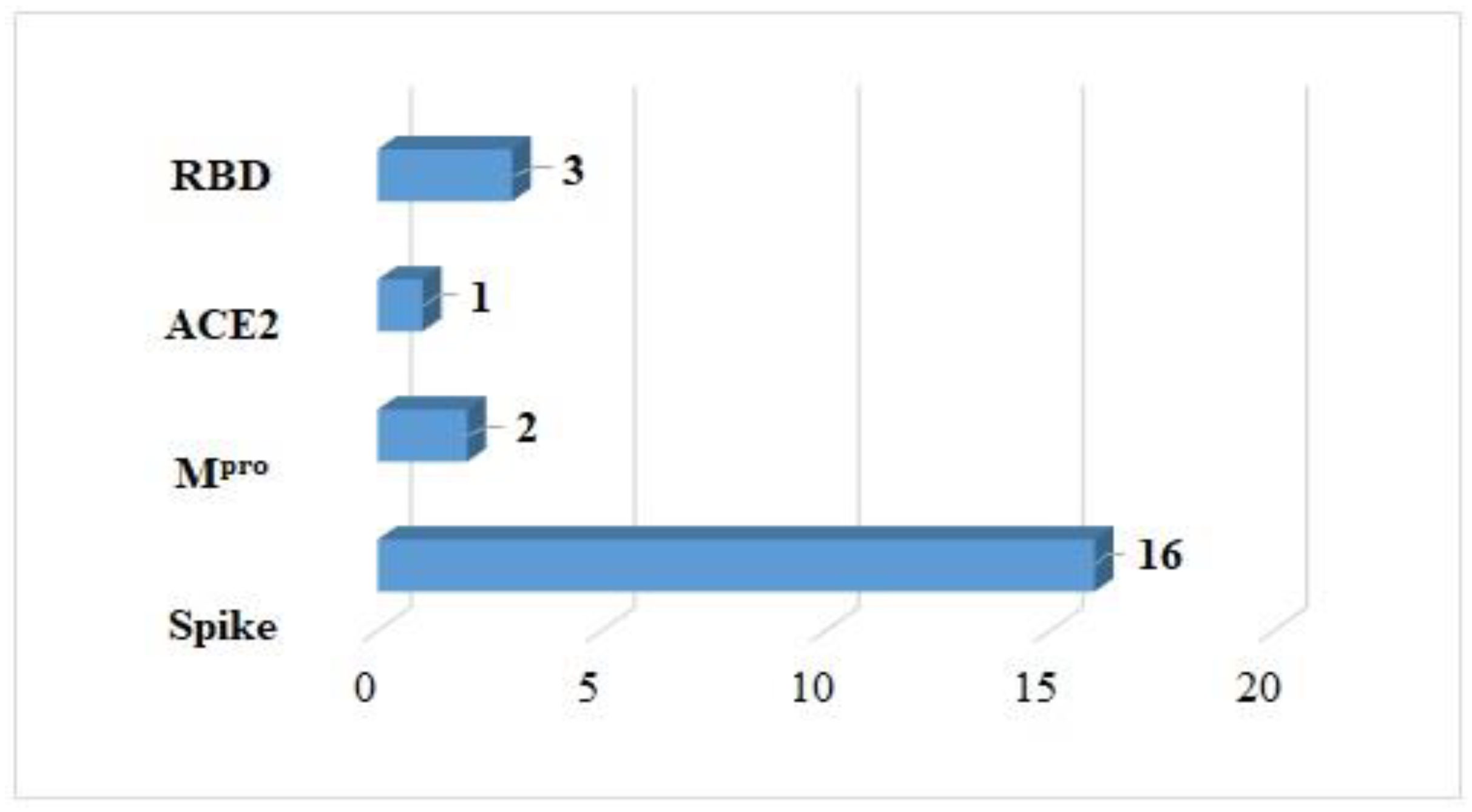

| Receptor | Reference Amino Acid | Coordinates of Grid | Center Grid Box Size |
|---|---|---|---|
| 6VXX | Gly548(A) | center_x = 180.306 | size_x = 30 size_y = 30 size_z = 30 |
| center_y = 211.382 | |||
| center_z = 224.580 | |||
| 1R42 | His374(A) | center_x = 51.467 | |
| center_y = 73.108 | |||
| center_z = 34.037 | |||
| 6LU7 | Gly143(A) | center_x = −8.918 | |
| center_y = 17.918 | |||
| center_z = 62.905 | |||
| Receptor 5 | Phe32(B) | center_x = 0.804 | |
| center_y = −7.902 | |||
| center_z = −5.193 |
| Plant Parts | Isolated Phytoconstituents | CID | Molecular Targets | References | |||
|---|---|---|---|---|---|---|---|
| ACE2 | Mpro | Receptor 5 (RBD) | Spike | ||||
| ΔG bind a (kcal/mol) | |||||||
| Flower | Niazirin | 129556 | −6.5 | −6.8 | −6.3 | −7.3 | Barreto et al., 2009 [42] |
| Sheet | Linalool | 6549 | −3.9 | −4.3 | −5.3 | −5.3 | |
| Geraniol | 637566 | −4.2 | −3.8 | −4.7 | −5.2 | ||
| Thymol | 6989 | −5.1 | −4.7 | −6.4 | −5.8 | ||
| Spathulenol | 92231 | −6.1 | −5.6 | −6.4 | −7.1 | ||
| Flower | Pentadecanol | 12397 | −3.8 | −3.9 | −5.3 | −5.0 | |
| Seed | Palmitic acid | 985 | −3.9 | −4.3 | −5.6 | −4.9 | Ferreira et al., 2008 [43] |
| Flower | Quercetin | 5280343 | −7.4 | −7.5 | −7.0 | −9.0 | |
| Kaempferol | 5280863 | −6.9 | −7.8 | −6.9 | −8.7 | ||
| Seed | Oleic acid | 445639 | −4.2 | −4.2 | −5.2 | −5.8 | |
| Sheet | Isoquercetrin | 5480505 | −7.8 | −8.9 | −8.0 | −8.6 | Bicas et al., 2019 [44] |
| Chlorogenic Acid | 1794427 | −7.3 | −7.6 | −7.3 | −8.7 | ||
| Lutein | 5281243 | −7.5 | −6.6 | −8.7 | −7.8 | ||
| Rutin | 5280805 | −8.2 | −8.8 | −8.0 | −9.1 | ||
| Seed | Lauric acid | 3893 | −3.9 | −4.1 | −4.9 | −4.9 | Ozcan, 2020 [45] |
| Myristic acid | 11005 | −4.0 | −4.2 | −4.9 | −4.8 | ||
| Linolenic acid | 5280934 | −4.6 | −4.6 | −6.4 | −5.8 | ||
| Brassicasterol | 5281327 | −7.5 | −7.0 | −7.7 | −8.0 | ||
| Campesterol | 173183 | −7.2 | −6.8 | −6.9 | −7.8 | ||
| Campestanol | 119394 | −6.9 | −6.9 | −7.0 | −7.9 | ||
| Stigmasterol | 5280794 | −7.3 | −7.0 | −7.4 | −8.0 | ||
| Ergosterol | 444679 | −7.6 | −7.3 | −7.3 | −8.0 | ||
| B-sitosterol | 222284 | −7.0 | −6.8 | −7.2 | −7.9 | ||
| Clerosterol | 5283638 | −6.7 | −6.3 | −7.2 | −7.6 | ||
| Stigmastanol | 241572 | −6.5 | −6.8 | −7.1 | −7.9 | ||
| Sheet | Zeatin | 449093 | −5.9 | −5.5 | −5.9 | −6.4 | Ahmadu et al., 2020 [46] |
| Myricetin | 5281672 | −7.4 | −7.4 | −7.3 | −9.1 | ||
| Niazin | 4472 | −7.3 | −6.9 | −7.3 | −7.6 | ||
| 2-Furancarboxaldehyde | 7362 | −3.7 | −4.1 | −4.2 | −4.1 | ||
| Malonic acid | 867 | −3.7 | −4.4 | −4.4 | −4.3 | ||
| Phenylvaleric acid | 16757 | −5.1 | −5.0 | −5.5 | −5.8 | ||
| Caffeic acid | 689,043 | −5.7 | −5.7 | −6.2 | −7.2 | ||
| Quinic acid | 6508 | −5.2 | −5.5 | −6.0 | −6.5 | ||
| Sheet | Ellagic acid | 5281855 | −7.3 | −7.5 | −7.5 | −9.3 | Kou et al., 2018 [47] |
| Ferulic acid | 445858 | −5.3 | −5.5 | −5.8 | −7.0 | ||
| Epicatechin | 72276 | −6.8 | −7.1 | −6.3 | −8.4 | ||
| Catechin | 9064 | −7.0 | −7.2 | −6.6 | −8.6 | ||
| Leaf and Seed | Glucomoringin | 162639104 | −7.2 | −7.9 | −7.9 | −8.5 | Anzano et al., 2022 [48] |
| Trigonelline | 5570 | −4.5 | −4.4 | −5.2 | −5.1 | ||
| Sheet | Isorhamnetin | 5281654 | −7.0 | −7.2 | −6.9 | −8.8 | Bezerra, 2020 [49] |
| Cysteine | 5862 | −3.4 | −3.7 | −3.9 | −4.1 | ||
| Methionine | 6137 | −3.8 | −4.0 | −4.2 | −4.5 | ||
| Tryptophan | 6305 | −5.8 | −5.8 | −5.7 | −6.6 | ||
| Lysine | 5962 | −3.7 | −4.3 | −4.5 | −5.1 | ||
| Serine | 5951 | −3.6 | −4.3 | −4.5 | −4.3 | ||
| Proline | 145742 | −4.0 | −4.6 | −4.2 | −4.8 | ||
| Glutamic acid | 33032 | −4.2 | −4.8 | −4.8 | −5.4 | ||
| Glycine | 750 | −3.8 | −3.6 | −3.6 | −3.8 | ||
| Arginine | 6322 | −4.8 | −4.8 | −5.6 | −5.8 | ||
| Histidine | 6274 | −4.5 | −5.1 | −5.3 | −5.8 | ||
| Valine | 1182 | −3.8 | −4.2 | −4.2 | −4.8 | ||
| Leucine | 6106 | −4.2 | −4.1 | −4.3 | −4.9 | ||
| Isoleucine | 6306 | −3.8 | −4.2 | −4.4 | −5.0 | ||
| Threonine | 6288 | −3.9 | −4.3 | −4.6 | −4.7 | ||
| Alanine | 602 | −4.0 | −3.8 | −4.1 | −4.1 | ||
| Aspartic acid | 5960 | −4.0 | −4.7 | −4.9 | −5.2 | ||
| 2,2-Dimethyl-1-pentanol | 16911 | −3.5 | −3.8 | −4.3 | −4.3 | ||
| 3,4-Dimethyl-2-Hexanol | 140547 | −3.7 | −3.8 | −4.2 | −4.6 | ||
| 4-Methyl-2,3-hexadien-1-ol | 566111 | −3.6 | −3.7 | −4.6 | −4.4 | ||
| Luteolin | 5280445 | −7.1 | −7.5 | −7.2 | −9.0 | ||
| Apigenin | 5280443 | −6.7 | −7.7 | −7.0 | −8.6 | ||
| Complex (Protein Binding) | ΔG bind a (kcal/mol) | Amino Acids That Interacted by Hydrogen Bonding | Amino Acids That Interacted by Hydrophobic Bonding |
|---|---|---|---|
| Elagic acid/spike | −9.3 | Asn978, Leu977, Arg1000, Tyr741, Met740, Thr549 | Phe541, Val976, Gly744, Gly548 |
| Rutin/spike | −9.1 | Ser967, Ser968, Leu754, Ser50, His49, Thr51, Gln52, Asn969, Ser975, Asp568, Ile569 | Asp571, Gly757, Arg567, Gln755, His519 |
| Myricitin/spike | −9.1 | Thr549, Gly744, Arg1000, Tyr741 | Phe541, Gly548, Leu977, Leu966, Asn856, Met740, Thr572, Thr573, Pro589, Ile587 |
| Quercetin/spike | −9.0 | Thr549, Gly744, Tyr741, Arg1000, Ile742, Met740 | Phe541, Gly548, Ile587, Thr572, Pro589, Thr573 |
| Luteolin/spike | −9.0 | Met740, Phe855, Thr573, Arg1000, Tyr741 | Gly744, Asn856, Gly548, Thr547, Leu546, Asn978, Val976, Thr572, Leu966 |
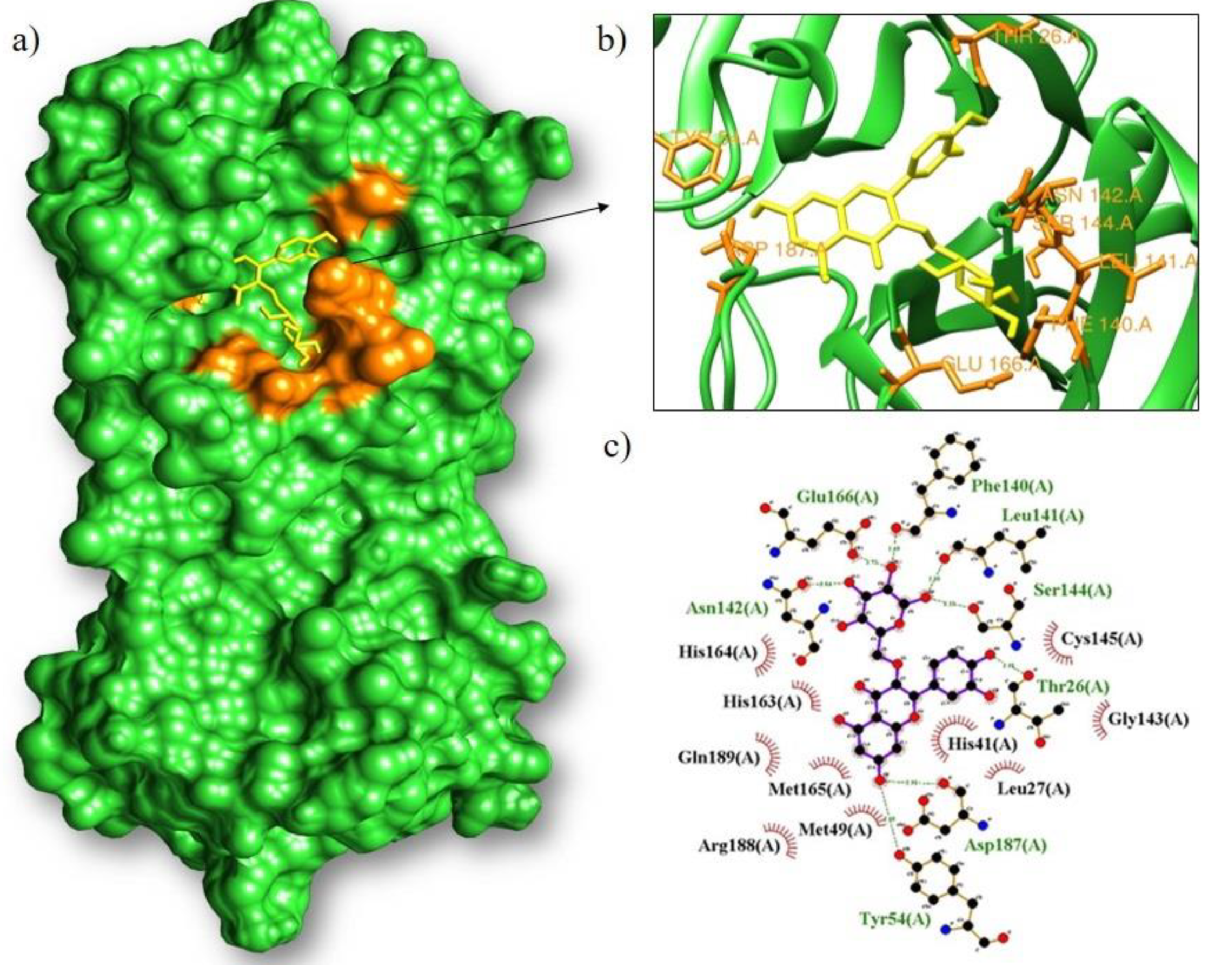
| Isoquercetrin/Mpro | −8.9 | Phe140, Leu141, Ser144, Thr26, Asp187, Tyr54, Asn142, Glu166 | Cys145, Gly143, Leu27, His41, Met49, Arg188, Met165, Gln189, His163, His164 |
| Rutin/Mpro | −8.8 | Leu141, Phe140, Asn142, Gly143, His41, Thr26, Thr190, Glu166, Ser144, His163 | Leu27, Thr25, Cys145, Arg188, Met165, Gln189, His164 |
| Isorhamnetin/spike | −8.8 | Arg1000, tyr741, Gly744, Thr549 | Ile587, Thr572, Pro589, Thr573, Ser975, Leu977, Val976, Met740, Leu966, Phe541, Gly548 |
| Kaempferol/spike | −8.7 | Tyr741, Arg1000, Leu977, Thr573, Phe855 | Leu966, Val976, Leu546, Thr547, Asn978, Thr572, Asn856, Met740, Gly744 |
| Chlorogenic acid/spike | −8.7 | Tyr741, Gly744, Asn978, Thr573, Asp568 | Ile587, Lys854, Pro589, Phe855, Leu966, Arg1000, Leu977, Thr572, Asp574, |
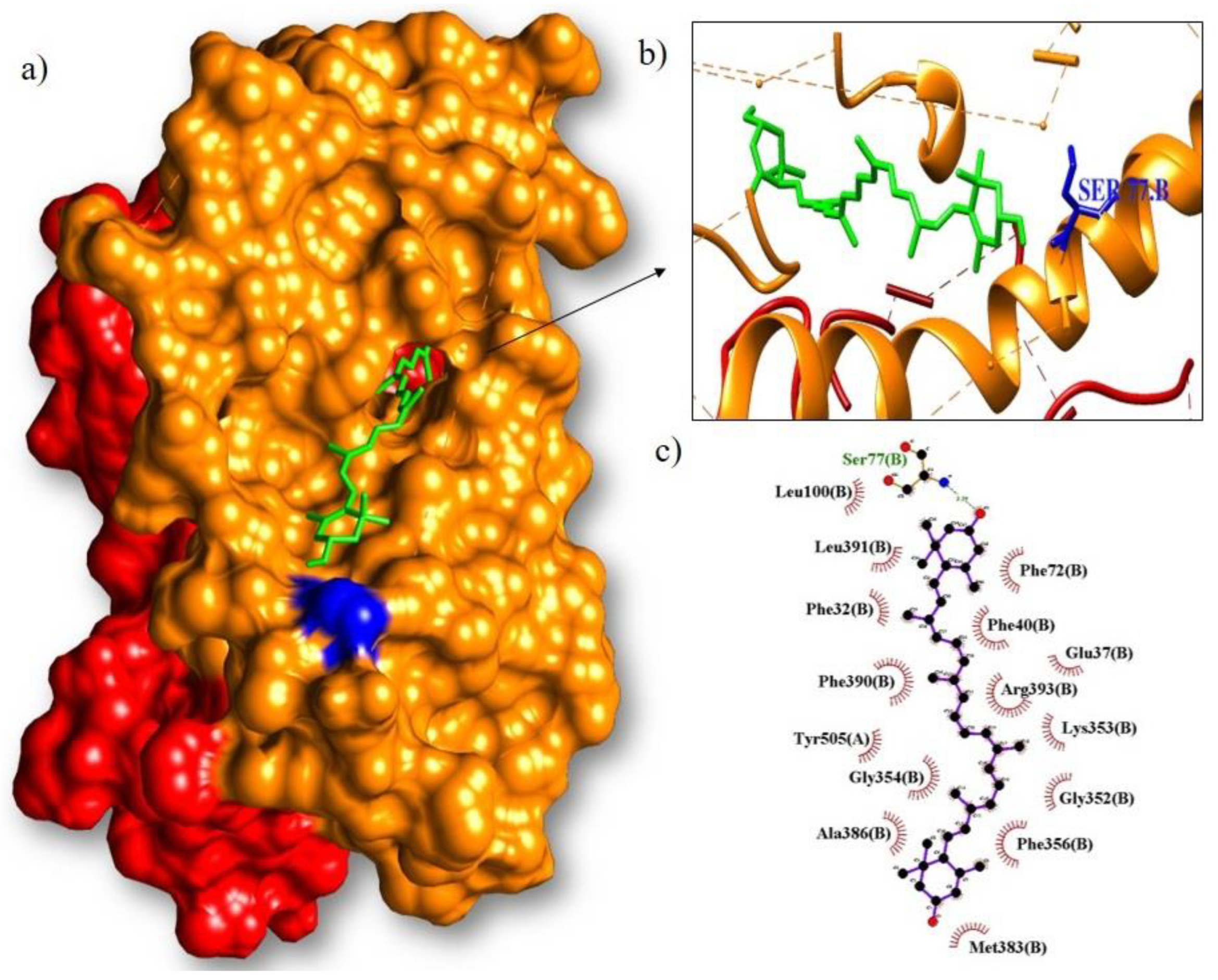
| Lutein/RBD | −8.7 | Ser77 | Phe72, Phe40, Glu37, Arg393, Lys353, Gly352, Phe356, Met383, Ala386, Gly354, Tyr505, Phe390, Phe32, Leu391, Leu100 |
| Isoquercetrin/spike | −8.6 | Arg567, Asp568, Asp571, Gly757, Ser50, His49 | Val47, Ile569, Arg44, Ser967, Ser968, Leu754, Gln755, Lys964 |
| Catechin/spike | −8.6 | Thr549, Arg1000, Ile742, Tyr741, Asn856 | Leu546, Thr573, Thr547, Asn978, Gly744, Leu966, Phe541, Gly548 |
| Apigenin/spike | −8.6 | Gly744, Tyr741, Ile742 | Thr573, Asn978, Val976, Leu977, Arg1000, Thr572, Ile587 |
| Glucomoringin/spike | −8.5 | Ser974, Ser975, Asp571, Thr430, Arg983, Ser514, Ile973, Asn969 | His519, Arg567, Val976, Asp979, Phe429, Pro426, Phe515, Phe464, Tyr200, Leu518, Glu516, Leu517, |
| Epicatechin/spike | −8.4 | Thr547, Arg1000, Tyr741, Met740, Asp745 | Gly548, Asn978, Thr572, Ile742, Gly744, Asn856, Thr549, Pro589, Phe541, Ile587 |
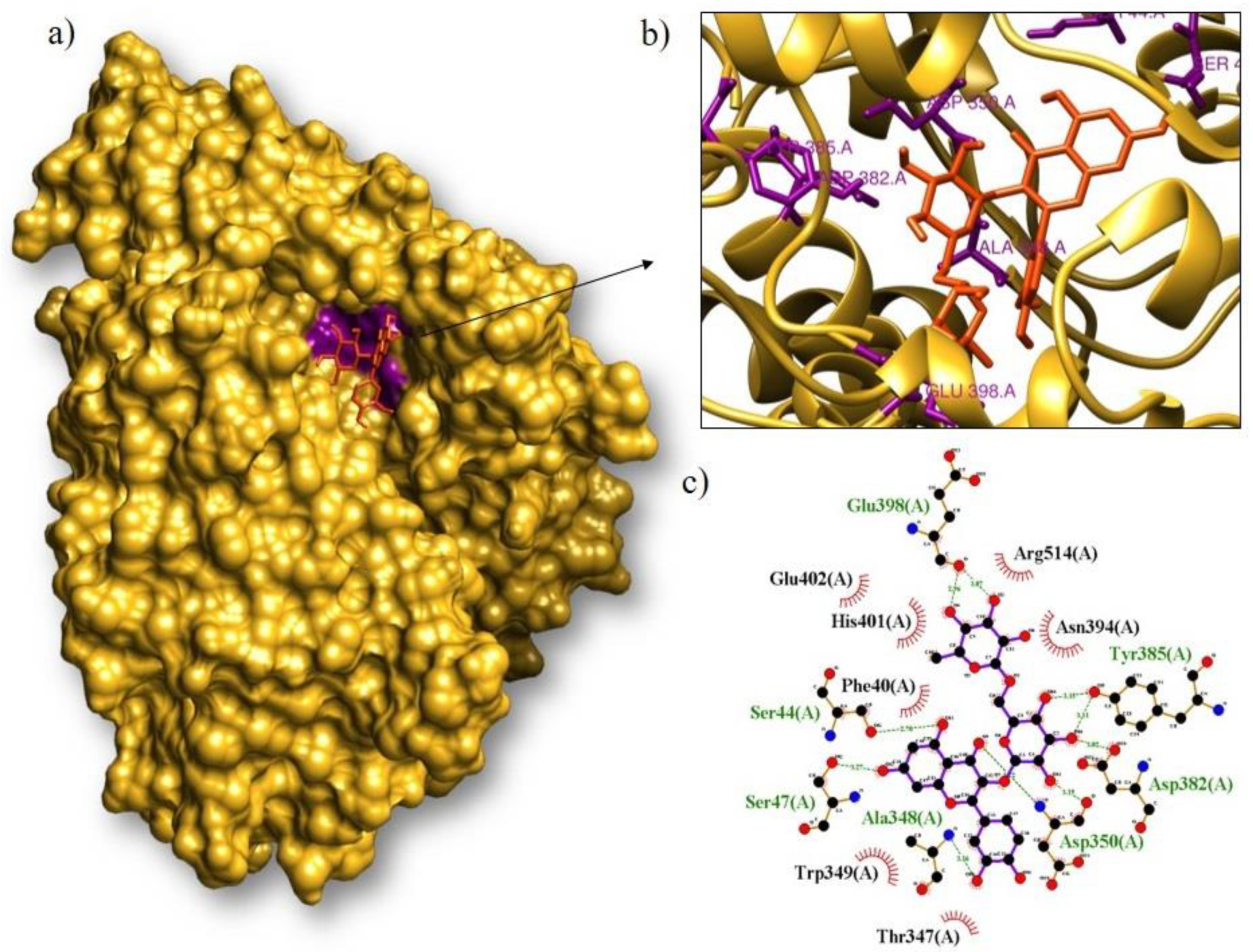
| Rutin/ACE2 | −8.2 | Glu398, Tyr385, Asp382, Asp350, Ala348, Ser47, Ser44 | Arg514, Asn394, Thr347, Trp349, Phe40, His401, Glu402 |
| Isoquercitrin/RBD | −8.0 | Arg393, Glu37, Tyr505, Asp405, Lys417, Asp30, Asn33, His34, Arg403, Tyr453 | Gln409, Ile418, Gly416, Leu455 |
| Rutin/RBD | −8.0 | Glu37, Tyr453, His34, Ala386, Arg393, Gln388, Asp405, Arg403 | Glu406, Lys417, Ile418, Gln409, Asn33, Leu455, Ala387, Tyr505 |
| Brassicasterol/spike | −8.0 | Ile973, Ser974, Arg983, Leu518, Thr430, Glu516, Phe515, Tyr200, Leu517 | |
| Stigmasterol/spike | −8.0 | Asp571 | Val976, Asp979, His519, Leu517, Glu516, Ser514, Pro426, Phe429, Phe515, Phe464, Thr430, Leu518, Ile973, Ser974, Arg567 |
| Ergosterol/spike | −8.0 | Ser974 | Tyr200, Glu516, Leu518, Ile973, Arg983, Leu517, Phe515, Thr430 |
| Drugs | CID | ACE2 Protein | Mpro Protein | RBD | Spike Protein |
|---|---|---|---|---|---|
| Baricitinib | 44,205,240 | −6.8 | −7.9 | −7.8 | −8.0 |
| Molnupiravir | 145,996,610 | −7.2 | −6.7 | −6.8 | −7.9 |
| Paxlovid (Nirmatrelvir + Ritonavir) | 155,903,259 | −7.1 | −7.6 | −7.0 | −7.3 |
| Remdesivir | 121,304,016 | −7.3 | −7.9 | −7.6 | −7.5 |
| Compounds | Absorption | Distribution | ||||||
|---|---|---|---|---|---|---|---|---|
| Solubility in Water (log mol/L) | P Caco2 (Log Papp at 10−6 cm/s) | AIH% | Skin Permeability (log Kp) | P-glycoprotein I Inhibitor | P-glycoprotein II Inhibitor | VDss (huma) (log L/kg) | PBH (BB) | |
| Apigenin | −3.178 | 1.076 | 91.856 | −2.736 | No | No | −0.105 | −0.951 |
| Brassicasterol | −6.635 | 1.209 | 94.138 | −2.798 | Yes | Yes | 0.232 | 0.767 |
| Catechin | −3.024 | −0.41 | 72.539 | −2.735 | No | No | 0.589 | −1.278 |
| Chlorogenic acid | −2.823 | −0.607 | 18.192 | −2.735 | No | No | −1.359 | −1.737 |
| Ellagic acid | −3.181 | 0.371 | 73.933 | −2.735 | No | No | 0.442 | −1.426 |
| Epicatechin | −3.024 | −0.41 | 72.539 | −2.735 | No | No | 0.589 | −1.278 |
| Ergosterol | −6.612 | 1.21 | 94.285 | −2.799 | Yes | Yes | 0.231 | 0.77 |
| Glucomoringin | −2.901 | −0.726 | 0 | −2.735 | No | No | −0.598 | −2.303 |
| Isoquercitrin | −3.028 | −0.755 | 38.939 | −2.735 | No | No | −0.287 | −2.417 |
| Isorhamnetin | −3.551 | 0.497 | 79.101 | −2.735 | No | No | 0.399 | −1.283 |
| Kaempferol | −3.332 | 0.627 | 81.862 | −2.735 | No | No | 0.078 | −1.143 |
| Lutein | −6.838 | 1.284 | 88.333 | −2.749 | No | Yes | −0.29 | −0.238 |
| Luteolin | −3.173 | 0.762 | 81.082 | −2.735 | No | No | 0.071 | −1.199 |
| Myricetin | −2.941 | −0.649 | 65.116 | −2.735 | No | No | 0.209 | −1.739 |
| Quercetin | −2.982 | 0.694 | 74.84 | −2.735 | No | No | 0.31 | −1.377 |
| Rutin | −2.909 | −0.662 | 25.454 | −2.735 | No | No | −0.155 | −2.556 |
| Stigmasterol | −6.671 | 1.21 | 94.73 | −2.781 | Yes | Yes | 0.176 | 0.79 |
| Compounds | Metabolism | Excretion | ||||||
|---|---|---|---|---|---|---|---|---|
| CYP2D6 Substrate | CYP3A4 Substrate | CYP1A2 Inhibitor | CYP2C19 Inhibitor | CYP2C9 Inhibitor | CYP2D6 Inhibitor | CYP3A4 Inhibitor | OCT2 Renal Substrate | |
| Apigenin | No | No | Yes | Yes | Yes | No | No | No |
| Brassicasterol | No | Yes | No | No | No | No | No | No |
| Catechin | No | No | No | No | No | No | No | No |
| Chlorogenic acid | No | No | No | No | No | No | No | No |
| Ellagic acid | No | No | Yes | No | No | No | No | No |
| Epicatechin | No | No | No | No | No | No | No | No |
| Ergosterol | No | Yes | No | No | No | No | No | No |
| Glucomoringin | No | No | No | No | No | No | No | No |
| Isoquercitrin | No | No | No | No | No | No | No | No |
| Isorhamnetin | No | No | Yes | No | No | No | No | No |
| Kaempferol | No | No | Yes | No | No | No | No | No |
| Lutein | No | Yes | No | No | No | No | No | No |
| Luteolin | No | No | Yes | No | No | No | No | No |
| Myricetin | No | No | Yes | No | No | No | No | No |
| Quercetin | No | No | Yes | No | No | No | No | No |
| Rutin | No | No | No | No | No | No | No | No |
| Stigmasterol | No | Yes | No | No | No | No | No | No |
| Compounds | Toxicity | |||||||
|---|---|---|---|---|---|---|---|---|
| AMES Toxicity | DMT (Human) (Log mg/kg/day) | hERG I Inhibitor | hERG II Inhibitor | TAO (Rats) (LD50) (mol/kg) | TAO (Rats) (LOAEL) (log mg/kg.bw/Day) | Hepatotoxicity | S-Skin | |
| Apigenin | No | 0.931 | No | Yes | 2.376 | 1.461 | No | No |
| Brassicasterol | No | −0.725 | No | Yes | 2.286 | 0.825 | No | No |
| Catechin | Yes | 0.516 | No | No | 2011 | 2.919 | No | No |
| Chlorogenic acid | No | 1.327 | No | No | 2.229 | 3.618 | No | No |
| Ellagic acid | No | 0.806 | No | No | 2.45 | 2.555 | No | No |
| Epicatechin | Yes | 0.516 | No | No | 2011 | 2.919 | No | No |
| Ergosterol | No | −0.731 | No | Yes | 2.28 | 0.824 | No | No |
| Glucomoringin | No | 0.416 | No | No | 2.473 | 4.372 | No | No |
| Isoquercitrin | Yes | 0.814 | No | Yes | 2.812 | 3.382 | No | No |
| Isorhamnetin | No | 0.882 | No | No | 2.358 | 2.804 | No | No |
| Kaempferol | No | 1.020 | No | No | 2.228 | 2.662 | Yes | No |
| Lutein | No | −1.237 | No | Yes | 2.590 | 2.543 | No | No |
| Luteolin | No | 0.975 | No | No | 2.450 | 1833 | No | No |
| Myricetin | Yes | 0.621 | No | No | 2.645 | 3.475 | No | No |
| Quercetin | Yes | 0.954 | No | No | 2.308 | 3.134 | No | No |
| Rutin | Yes | 0.550 | No | Yes | 2.523 | 4.415 | No | No |
| Stigmasterol | No | −0.639 | No | Yes | 2.345 | 0.802 | No | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Souza, H.C.A.; Souza, M.D.A.; Sousa, C.S.; Viana, E.K.A.; Alves, S.K.S.; Marques, A.O.; Ribeiro, A.S.N.; de Sousa do Vale, V.; Islam, M.T.; de Miranda, J.A.L.; et al. Molecular Docking and ADME-TOX Profiling of Moringa oleifera Constituents against SARS-CoV-2. Adv. Respir. Med. 2023, 91, 464-485. https://doi.org/10.3390/arm91060035
Souza HCA, Souza MDA, Sousa CS, Viana EKA, Alves SKS, Marques AO, Ribeiro ASN, de Sousa do Vale V, Islam MT, de Miranda JAL, et al. Molecular Docking and ADME-TOX Profiling of Moringa oleifera Constituents against SARS-CoV-2. Advances in Respiratory Medicine. 2023; 91(6):464-485. https://doi.org/10.3390/arm91060035
Chicago/Turabian StyleSouza, Hellen Cris Araújo, Maycon Douglas Araújo Souza, Cássio Silva Sousa, Edilanne Katrine Amparo Viana, Sabrina Kelly Silva Alves, Alex Oliveira Marques, Arthur Serejo Neves Ribeiro, Vanessa de Sousa do Vale, Muhammad Torequl Islam, João Antônio Leal de Miranda, and et al. 2023. "Molecular Docking and ADME-TOX Profiling of Moringa oleifera Constituents against SARS-CoV-2" Advances in Respiratory Medicine 91, no. 6: 464-485. https://doi.org/10.3390/arm91060035
APA StyleSouza, H. C. A., Souza, M. D. A., Sousa, C. S., Viana, E. K. A., Alves, S. K. S., Marques, A. O., Ribeiro, A. S. N., de Sousa do Vale, V., Islam, M. T., de Miranda, J. A. L., da Costa Mota, M., & Rocha, J. A. (2023). Molecular Docking and ADME-TOX Profiling of Moringa oleifera Constituents against SARS-CoV-2. Advances in Respiratory Medicine, 91(6), 464-485. https://doi.org/10.3390/arm91060035







