Sustainable Strategy to Boost Legumes Growth under Salinity and Drought Stress in Semi-Arid and Arid Regions
Abstract
:1. Introduction
2. Legume–Rhizobium Symbiosis: Evolution, Mechanisms and Concerns
3. The Effect of Drought and Salinity on Legume Growth and Their Symbiotic Interactions
4. The PGPB: Effective Candidates to Improve the Agricultural System in Drylands
5. Rhizobia Application in Legume Cultures under Arid Environments
5.1. Drought Stress
5.2. Salinity
6. Non-Rhizobial Endophytes: Plant Biofertilizers in Arid and Semi-Arid Regions
6.1. Application under Drought Stress
6.2. Application in Saline Soils
7. PGPB Consortia for Alleviating Drought and Salinity Stresses in Legumes
8. PGPB Interactions in the Rhizosphere: Cell-to-Cell Communication
9. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mirzabaev Mirzabaev, A.; Wu, J.; Evans, J.; García-Oliva, F.; Hussein, I.A.; Iqbal, M.H.; Kimutai, J.; Knowles, T.; Meza, F.; Nedjroaoui, D.; et al. Desertification. In Climate Change and Land: An IPCC Special Report on Climate Change, Desertification, Land Degradation, Sustainable Land Management, Food Security, and Greenhouse Gas Fluxes in Terrestrial Ecosystems; Shukla, P.R., Skeg, J., Calvo Buendia, E., Masson-Delmotte, V., Pörtner, H.-O., Roberts, D.C., Zhai, P., Slade, R., Connors, S., van Diemen, R., et al., Eds.; IPCC: Geneva, Switzerland, 2019. [Google Scholar]
- Ma, Z.; Yang, Q. Global patterns of aridity trends and time regimes in transition. In Aridity Trend in Northern China; World Scientific: Singapore, 2017; pp. 67–90. [Google Scholar]
- Yadav, S.; Modi, P.; Dave, A.; Vijapura, A.; Patel, D.; Patel, M. Effect of abiotic stress on crops. Sustain. Crop Prod. 2020, 3, 5–16. [Google Scholar]
- Gull, A.; Lone, A.A.; Wani, N.U.I. Biotic and Abiotic Stresses in Plants; IntechOpen Publishing: London, UK, 2019; pp. 1–19. [Google Scholar]
- Seleiman, M.F.; Al-Suhaibani, N.; Ali, N.; Akmal, M.; Alotaibi, M.; Refay, Y.; Dindaroglu, T.; Abdul-Wajid, H.H.; Battaglia, M.L. Drought stress impacts on plants and different approaches to alleviate its adverse effects. Plants 2021, 10, 259. [Google Scholar] [CrossRef]
- Wang, L.; Kaseke, K.F.; Seely, M.K. Effects of non-rainfall water inputs on ecosystem functions. Wiley Interdiscip. Rev. Water 2017, 4, e1179. [Google Scholar] [CrossRef]
- Etesami, H.; Beattie, G. Plant-Microbe Interactions in Adaptation of Agricultural Crops to Abiotic Stress Conditions; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Corwin, D.L. Climate change impacts on soil salinity in agricultural areas. Eur. J. Soil Sci. 2021, 72, 842–862. [Google Scholar] [CrossRef]
- Dutta, T.; Neelapu, N.R.R.; Wani, S.H.; Challa, S. Response of pulses to drought and salinity stress response: A physiological perspective. In Pulse Improvement: Physiological, Molecular and Genetic Perspectives; Springer: Berlin/Heidelberg, Germany, 2018; pp. 77–98. [Google Scholar]
- Wang, Y.; Hao, Y.; Cui, X.Y.; Zhao, H.; Xu, C.; Zhou, X.; Xu, Z. Responses of soil respiration and its components to drought stress. J. Soils Sediments 2014, 14, 99–109. [Google Scholar] [CrossRef]
- Yosef, B.A.; Asmamaw, D.K. Rainwater harvesting: An option for dry land agriculture in arid and semi-arid Ethiopia. Int. J. Water Resour. Environ. Eng. 2015, 7, 17–28. [Google Scholar]
- Wang, W.; Zhuo, L.; Li, M.; Liu, Y.; Wu, P. The effect of development in water-saving irrigation techniques on spatial-temporal variations in crop water footprint and benchmarking. J. Hydrol. 2019, 577, 123916. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, Z.; Cheng, X.; Wang, Q. Effects of Surface Mulching on the Growth and Water Consumption of Maize. Agriculture 2022, 12, 1868. [Google Scholar] [CrossRef]
- Mdlambuzi, T.; Tsubo, M.; Muchaonyerwa, P. Maize (Zea mays L.) Production from Co-application of Biogas Slurry with Chemical Fertilizer and Effects on Soil Quality in a Semi-arid Region of South Africa. Commun. Soil Sci. Plant Anal. 2022, 53, 2574–2583. [Google Scholar] [CrossRef]
- Li, D.-P.; Wu, Z.-J. Impact of chemical fertilizers application on soil ecological environment. Ying Yong Sheng Tai Xue Bao J. Appl. Ecol. 2008, 19, 1158–1165. [Google Scholar]
- Dercon, S.; Christiaensen, L. Consumption risk, technology adoption and poverty traps: Evidence from Ethiopia. J. Dev. Econ. 2011, 96, 159–173. [Google Scholar] [CrossRef]
- Atafar, Z.; Mesdaghinia, A.; Nouri, J.; Homaee, M.; Yunesian, M.; Ahmadimoghaddam, M.; Mahvi, A.H. Effect of fertilizer application on soil heavy metal concentration. Environ. Monit. Assess. 2010, 160, 83–89. [Google Scholar] [CrossRef]
- Ceccarelli, S. Efficiency of plant breeding. Crop Sci. 2015, 55, 87–97. [Google Scholar] [CrossRef]
- Chialva, M.; Lanfranco, L.; Bonfante, P. The plant microbiota: Composition, functions, and engineering. Curr. Opin. Biotechnol. 2022, 73, 135–142. [Google Scholar] [CrossRef]
- Müller, D.B.; Vogel, C.; Bai, Y.; Vorholt, J.A. The plant microbiota: Systems-level insights and perspectives. Annu. Rev. Genet. 2016, 50, 211–234. [Google Scholar] [CrossRef]
- Compant, S.; Cambon, M.C.; Vacher, C.; Mitter, B.; Samad, A.; Sessitsch, A. The plant endosphere world–bacterial life within plants. Environ. Microbiol. 2021, 23, 1812–1829. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Wang, X.; Wang, M.; Xie, X.; Guo, S.; Zhou, Y.; Zhang, X.; Yu, N.; Wang, E. An amplification-selection model for quantified rhizosphere microbiota assembly. Sci. Bull. 2020, 65, 983–986. [Google Scholar] [CrossRef]
- Truyens, S.; Weyens, N.; Cuypers, A.; Vangronsveld, J. Bacterial seed endophytes: Genera, vertical transmission and interaction with plants. Environ. Microbiol. Rep. 2015, 7, 40–50. [Google Scholar] [CrossRef]
- Leveau, J.H. A brief from the leaf: Latest research to inform our understanding of the phyllosphere microbiome. Curr. Opin. Microbiol. 2019, 49, 41–49. [Google Scholar] [CrossRef]
- Mendes, L.W.; de Lima Brossi, M.J.; Kuramae, E.E.; Tsai, S.M. Land-use system shapes soil bacterial communities in Southeastern Amazon region. Appl. Soil Ecol. 2015, 95, 151–160. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Hawkes, C.V.; Kjøller, R.; Raaijmakers, J.M.; Riber, L.; Christensen, S.; Rasmussen, S.; Christensen, J.H.; Dahl, A.B.; Westergaard, J.C.; Nielsen, M. Extension of plant phenotypes by the foliar microbiome. Annu. Rev. Plant Biol. 2021, 72, 823–846. [Google Scholar] [CrossRef]
- Zhang, J.; Cook, J.; Nearing, J.T.; Zhang, J.; Raudonis, R.; Glick, B.R.; Langille, M.G.; Cheng, Z. Harnessing the plant microbiome to promote the growth of agricultural crops. Microbiol. Res. 2021, 245, 126690. [Google Scholar] [CrossRef]
- Orozco-Mosqueda, M.; Flores, A.; Rojas-Sánchez, B.; Urtis-Flores, C.A.; Morales-Cedeño, L.R.; Valencia-Marin, M.F.; Chávez-Avila, S.; Rojas-Solis, D.; Santoyo, G. Plant growth-promoting bacteria as bioinoculants: Attributes and challenges for sustainable crop improvement. Agronomy 2021, 11, 1167. [Google Scholar] [CrossRef]
- Abd-Alla, M.H.; Al-Amri, S.M.; El-Enany, A.-W.E. Enhancing Rhizobium–Legume Symbiosis and Reducing Nitrogen Fertilizer Use Are Potential Options for Mitigating Climate Change. Agriculture 2023, 13, 2092. [Google Scholar] [CrossRef]
- Ben Gaied, R.; Sbissi, I.; Tarhouni, M.; Brígido, C. Enhancing Pisum sativum growth and symbiosis under heat stress: The synergistic impact of co-inoculated bacterial consortia and ACC deaminase-lacking Rhizobium. Arch. Microbiol. 2024, 206, 203. [Google Scholar] [CrossRef]
- Yanni, Y.; Zidan, M.; Dazzo, F.; Rizk, R.; Mehesen, A.; Abdelfattah, F.; Elsadany, A. Enhanced symbiotic performance and productivity of drought stressed common bean after inoculation with tolerant native rhizobia in extensive fields. Agric. Ecosyst. Environ. 2016, 232, 119–128. [Google Scholar] [CrossRef]
- Abd-Alla, M.H.; Issa, A.A.; Ohyama, T. Impact of harsh environmental conditions on nodule formation and dinitrogen fixation of legumes. Adv. Biol. Ecol. Nitrogen Fixat. 2014, 7, 131–187. [Google Scholar]
- Chinnaswamy, A.; Coba De La Peña, T.; Stoll, A.; De La Peña Rojo, D.; Bravo, J.; Rincón, A.; Lucas, M.; Pueyo, J. A nodule endophytic Bacillus megaterium strain isolated from Medicago polymorpha enhances growth, promotes nodulation by Ensifer medicae and alleviates salt stress in alfalfa plants. Ann. Appl. Biol. 2018, 172, 295–308. [Google Scholar] [CrossRef]
- Oliveira, R.S.; Carvalho, P.; Marques, G.; Ferreira, L.; Pereira, S.; Nunes, M.; Rocha, I.; Ma, Y.; Carvalho, M.F.; Vosátka, M. Improved grain yield of cowpea (Vigna unguiculata) under water deficit after inoculation with Bradyrhizobium elkanii and Rhizophagus irregularis. Crop Pasture Sci. 2017, 68, 1052–1059. [Google Scholar] [CrossRef]
- Stagnari, F.; Maggio, A.; Galieni, A.; Pisante, M. Multiple benefits of legumes for agriculture sustainability: An overview. Chem. Biol. Technol. Agric. 2017, 4, 2. [Google Scholar] [CrossRef]
- Shah, A.; Nazari, M.; Antar, M.; Msimbira, L.A.; Naamala, J.; Lyu, D.; Rabileh, M.; Zajonc, J.; Smith, D.L. PGPR in agriculture: A sustainable approach to increasing climate change resilience. Front. Sustain. Food Syst. 2021, 5, 667546. [Google Scholar] [CrossRef]
- Mishra, P.; Mishra, J.; Arora, N.K. Plant growth promoting bacteria for combating salinity stress in plants–recent developments and prospects: A review. Microbiol. Res. 2021, 252, 126861. [Google Scholar] [CrossRef]
- Álvarez-Aragón, R.; Palacios, J.M.; Ramírez-Parra, E. Rhizobial symbiosis promotes drought tolerance in Vicia sativa and Pisum sativum. Environ. Exp. Bot. 2023, 208, 105268. [Google Scholar] [CrossRef]
- Ben Gaied, R.; Sbissi, I.; Tarhouni, M.; Brígido, C. Bacterial Endophytes from Legumes Native to Arid Environments Are Promising Tools to Improve Mesorhizobium–Chickpea Symbiosis under Salinity. Biology 2024, 13, 96. [Google Scholar] [CrossRef]
- Zahran, H.H. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol. Mol. Biol. Rev. 1999, 63, 968–989. [Google Scholar] [CrossRef]
- Sprent, J.I.; Gehlot, H.S. Nodulated legumes in arid and semi-arid environments: Are they important? Plant Ecol. Divers. 2010, 3, 211–219. [Google Scholar] [CrossRef]
- Ayangbenro, A.S.; Babalola, O.O. Reclamation of arid and semi-arid soils: The role of plant growth-promoting archaea and bacteria. Curr. Plant Biol. 2021, 25, 100173. [Google Scholar] [CrossRef]
- Soltis, D.E.; Soltis, P.S.; Morgan, D.R.; Swensen, S.M.; Mullin, B.C.; Dowd, J.M.; Martin, P.G. Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc. Natl. Acad. Sci. USA 1995, 92, 2647–2651. [Google Scholar] [CrossRef]
- Wang, H.; Moore, M.J.; Soltis, P.S.; Bell, C.D.; Brockington, S.F.; Alexandre, R.; Davis, C.C.; Latvis, M.; Manchester, S.R.; Soltis, D.E. Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc. Natl. Acad. Sci. USA 2009, 106, 3853–3858. [Google Scholar] [CrossRef]
- Bell, C.D.; Soltis, D.E.; Soltis, P.S. The age and diversification of the angiosperms re-revisited. Am. J. Bot. 2010, 97, 1296–1303. [Google Scholar] [CrossRef]
- Cathebras, C.; Gong, X.; Andrade, R.E.; Vondenhoff, K.; Keller, J.; Delaux, P.-M.; Hayashi, M.; Griesmann, M.; Parniske, M. A novel cis-element enabled bacterial uptake by plant cells. bioRxiv 2022. [Google Scholar] [CrossRef]
- Doyle, J.J. Phylogenetic perspectives on the origins of nodulation. Mol. Plant-Microbe Interact. 2011, 24, 1289–1295. [Google Scholar] [CrossRef]
- Doyle, J.J. Chasing unicorns: Nodulation origins and the paradox of novelty. Am. J. Bot. 2016, 103, 1865–1868. [Google Scholar] [CrossRef]
- Parniske, M. Uptake of bacteria into living plant cells, the unifying and distinct feature of the nitrogen-fixing root nodule symbiosis. Curr. Opin. Plant Biol. 2018, 44, 164–174. [Google Scholar] [CrossRef]
- Remigi, P.; Zhu, J.; Young, J.P.W.; Masson-Boivin, C. Symbiosis within Symbiosis: Evolving Nitrogen-Fixing Legume Symbionts. Trends Microbiol. 2016, 24, 63–75. [Google Scholar] [CrossRef]
- Werner, G.D.; Cornwell, W.K.; Sprent, J.I.; Kattge, J.; Kiers, E.T. A single evolutionary innovation drives the deep evolution of symbiotic N2-fixation in angiosperms. Nat. Commun. 2014, 5, 4087. [Google Scholar] [CrossRef]
- Pawlowski, K.; Demchenko, K.N. The diversity of actinorhizal symbiosis. Protoplasma 2012, 249, 967–979. [Google Scholar] [CrossRef] [PubMed]
- Masson-Boivin, C.; Sachs, J.L. Symbiotic nitrogen fixation by rhizobia-the roots of a success story. Curr. Opin. Plant Biol. 2018, 44, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Suzaki, T.; Kawaguchi, M. Root nodulation: A developmental program involving cell fate conversion triggered by symbiotic bacterial infection. Curr. Opin. Plant Biol. 2014, 21, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Shumilina, J.; Soboleva, A.; Abakumov, E.; Shtark, O.Y.; Zhukov, V.A.; Frolov, A. Signaling in Legume-Rhizobia Symbiosis. Int. J. Mol. Sci. 2023, 24, 17397. [Google Scholar] [CrossRef] [PubMed]
- Compton, K.K.; Scharf, B.E. Rhizobial Chemoattractants, the Taste and Preferences of Legume Symbionts. Front. Plant Sci. 2021, 12, 686465. [Google Scholar] [CrossRef] [PubMed]
- D’Haeze, W.; Holsters, M. Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology 2002, 12, 79r–105r. [Google Scholar] [CrossRef]
- Jones, K.M.; Kobayashi, H.; Davies, B.W.; Taga, M.E.; Walker, G.C. How rhizobial symbionts invade plants: The Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 2007, 5, 619–633. [Google Scholar] [CrossRef]
- Ibáñez, F.; Wall, L.; Fabra, A. Starting points in plant-bacteria nitrogen-fixing symbioses: Intercellular invasion of the roots. J. Exp. Bot. 2017, 68, 1905–1918. [Google Scholar] [CrossRef]
- Kohlen, W.; Ng, J.L.P.; Deinum, E.E.; Mathesius, U. Auxin transport, metabolism, and signalling during nodule initiation: Indeterminate and determinate nodules. J. Exp. Bot. 2017, 69, 229–244. [Google Scholar] [CrossRef]
- Perret, X.; Staehelin, C.; Broughton, W.J. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 2000, 64, 180–201. [Google Scholar] [CrossRef]
- Madsen, L.H.; Tirichine, L.; Jurkiewicz, A.; Sullivan, J.T.; Heckmann, A.B.; Bek, A.S.; Ronson, C.W.; James, E.K.; Stougaard, J. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 2010, 1, 10. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.; Harrison, M.J.; Paszkowski, U. Reprogramming plant cells for endosymbiosis. Science 2009, 324, 753–754. [Google Scholar] [CrossRef]
- Walker, L.; Lagunas, B.; Gifford, M.L. Determinants of Host Range Specificity in Legume-Rhizobia Symbiosis. Front. Microbiol. 2020, 11, 585749. [Google Scholar] [CrossRef]
- Mendoza-Suárez, M.; Andersen, S.U.; Poole, P.S.; Sánchez-Cañizares, C. Competition, Nodule Occupancy, and Persistence of Inoculant Strains: Key Factors in the Rhizobium-Legume Symbioses. Front. Plant Sci. 2021, 12, 690567. [Google Scholar] [CrossRef]
- Irisarri, P.; Cardozo, G.; Tartaglia, C.; Reyno, R.; Gutiérrez, P.; Lattanzi, F.A.; Rebuffo, M.; Monza, J. Selection of Competitive and Efficient Rhizobia Strains for White Clover. Front. Microbiol. 2019, 10, 768. [Google Scholar] [CrossRef] [PubMed]
- Onishchuk, O.P.; Vorobyov, N.I.; Provorov, N.A. Nodulation competitiveness of nodule bacteria: Genetic control and adaptive significance: Review. Appl. Biochem. Microbiol. 2017, 53, 131–139. [Google Scholar] [CrossRef]
- Atieno, M.; Lesueur, D. Opportunities for improved legume inoculants: Enhanced stress tolerance of rhizobia and benefits to agroecosystems. Symbiosis 2019, 77, 191–205. [Google Scholar] [CrossRef]
- da-Silva, J.R.; Alexandre, A.; Brígido, C.; Oliveira, S. Can stress response genes be used to improve the symbiotic performance of rhizobia? AIMS Microbiol. 2017, 3, 365–382. [Google Scholar] [CrossRef]
- Mousavi-Derazmahalleh, M.; Bayer, P.E.; Hane, J.K.; Valliyodan, B.; Nguyen, H.T.; Nelson, M.N.; Erskine, W.; Varshney, R.K.; Papa, R.; Edwards, D. Adapting legume crops to climate change using genomic approaches. Plant Cell Environ. 2019, 42, 6–19. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P. Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef]
- Harrison, S. Plant community diversity will decline more than increase under climatic warming. Philos. Trans. R. Soc. B 2020, 375, 20190106. [Google Scholar] [CrossRef] [PubMed]
- Omae, N.; Tsuda, K. Plant-microbiota interactions in abiotic stress environments. Mol. Plant-Microbe Interact. 2022, 35, 511–526. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S. Plant drought stress: Effects, mechanisms and management. In Sustainable Agriculture; Springer: Berlin/Heidelberg, Germany, 2009; pp. 153–188. [Google Scholar]
- Mansour, E.; Desoky, E.-S.M.; Ali, M.M.; Abdul-Hamid, M.I.; Ullah, H.; Attia, A.; Datta, A. Identifying drought-tolerant genotypes of faba bean and their agro-physiological responses to different water regimes in an arid Mediterranean environment. Agric. Water Manag. 2021, 247, 106754. [Google Scholar] [CrossRef]
- Hussain, S.; Rao, M.J.; Anjum, M.A.; Ejaz, S.; Zakir, I.; Ali, M.A.; Ahmad, N.; Ahmad, S. Oxidative stress and antioxidant defense in plants under drought conditions. In Plant Abiotic Stress Tolerance: Agronomic, Molecular and Biotechnological Approaches; Springer: Berlin/Heidelberg, Germany, 2019; pp. 207–219. [Google Scholar]
- Lawlor, D.W.; Tezara, W. Causes of decreased photosynthetic rate and metabolic capacity in water-deficient leaf cells: A critical evaluation of mechanisms and integration of processes. Ann. Bot. 2009, 103, 561–579. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Gogoi, N.; Barthakur, S.; Baroowa, B.; Bharadwaj, N.; Alghamdi, S.S.; Siddique, K.H. Drought stress in grain legumes during reproduction and grain filling. J. Agron. Crop Sci. 2017, 203, 81–102. [Google Scholar] [CrossRef]
- Dietz, K.J.; Zörb, C.; Geilfus, C.M. Drought and crop yield. Plant Biol. 2021, 23, 881–893. [Google Scholar] [CrossRef] [PubMed]
- Cherif-Silini, H.; Silini, A.; Chenari Bouket, A.; Alenezi, F.N.; Luptakova, L.; Bouremani, N.; Nowakowska, J.A.; Oszako, T.; Belbahri, L. Tailoring next generation plant growth promoting microorganisms as versatile tools beyond soil desalinization: A road map towards field application. Sustainability 2021, 13, 4422. [Google Scholar] [CrossRef]
- Arif, Y.; Singh, P.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salinity induced physiological and biochemical changes in plants: An omic approach towards salt stress tolerance. Plant Physiol. Biochem. 2020, 156, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Shaukat, M.; Ashraf, M.; Zhu, C.; Jin, Q.; Zhang, J. Salinity stress in arid and semi-arid climates: Effects and management in field crops. Clim. Chang. Agric. 2019, 13, 201–226. [Google Scholar]
- Mickan, B.S.; Abbott, L.K.; Solaiman, Z.M.; Mathes, F.; Siddique, K.H.; Jenkins, S.N. Soil disturbance and water stress interact to influence arbuscular mycorrhizal fungi, rhizosphere bacteria and potential for N and C cycling in an agricultural soil. Biol. Fertil. Soils 2019, 55, 53–66. [Google Scholar] [CrossRef]
- Zhou, Z.; Yu, M.; Ding, G.; Gao, G.; He, Y. Diversity and structural differences of bacterial microbial communities in rhizocompartments of desert leguminous plants. PLoS ONE 2020, 15, e0241057. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Zhang, G.; Yu, Z.; Ding, H.; Xu, Y.; Zhang, Z. Effect of drought stress and developmental stages on microbial community structure and diversity in peanut rhizosphere soil. Int. J. Mol. Sci. 2019, 20, 2265. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Xu, H.; Shi, J.; Wang, Z.; Lv, J.; Li, L.; Wang, X. Soil microbial composition, diversity, and network stability in intercropping versus monoculture responded differently to drought. Agric. Ecosyst. Environ. 2024, 365, 108915. [Google Scholar] [CrossRef]
- Dollete, D.; Lumactud, R.A.; Carlyle, C.N.; Szczyglowski, K.; Hill, B.; Thilakarathna, M.S. Effect of drought stress on symbiotic nitrogen fixation, soil nitrogen availability and soil microbial diversity in forage legumes. Plant Soil 2024, 495, 445–467. [Google Scholar] [CrossRef]
- Schimel, J.P. Life in dry soils: Effects of drought on soil microbial communities and processes. Annu. Rev. Ecol. Evol. Syst. 2018, 49, 409–432. [Google Scholar] [CrossRef]
- Dubey, S.; Khatri, S.; Bhattacharjee, A.; Sharma, S. Multiple passaging of rhizospheric microbiome enables mitigation of salinity stress in Vigna radiata. Plant Growth Regul. 2022, 97, 537–549. [Google Scholar] [CrossRef]
- Zheng, Y.; Cao, X.; Zhou, Y.; Li, Z.; Yang, Y.; Zhao, D.; Li, Y.; Xu, Z.; Zhang, C.-S. Effect of planting salt-tolerant legumes on coastal saline soil nutrient availability and microbial communities. J. Environ. Manag. 2023, 345, 118574. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, L.; Singh, R.P.; Meng, C.; Ma, S.; Jing, C.; Li, Y.; Zhang, C. Nodule and root zone microbiota of salt-tolerant wild soybean in coastal sand and saline-alkali soil. Front. Microbiol. 2020, 11, 2178. [Google Scholar] [CrossRef]
- Yan, N.; Marschner, P.; Cao, W.; Zuo, C.; Qin, W. Influence of salinity and water content on soil microorganisms. Int. Soil Water Conserv. Res. 2015, 3, 316–323. [Google Scholar] [CrossRef]
- Frankenberger Jr, W.; Bingham, F. Influence of salinity on soil enzyme activities. Soil Sci. Soc. Am. J. 1982, 46, 1173–1177. [Google Scholar] [CrossRef]
- Jones, J.M.; Boehm, E.L.; Kahmark, K.; Lau, J.; Evans, S. Microbial community response to drought depends on crop. Elem. Sci. Anth. 2022, 10, 00110. [Google Scholar] [CrossRef]
- Liu, X.; Lu, X.; Zhao, W.; Yang, S.; Wang, J.; Xia, H.; Wei, X.; Zhang, J.; Chen, L.; Chen, Q. The rhizosphere effect of native legume Albizzia julibrissin on coastal saline soil nutrient availability, microbial modulation, and aggregate formation. Sci. Total Environ. 2022, 806, 150705. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Ma, Q.; Chen, Y.; Tian, B.; Xu, L.; Bai, Y.; Chen, W.; Li, X. Variation in rhizosphere microbial communities and its association with the symbiotic efficiency of rhizobia in soybean. ISME J. 2020, 14, 1915–1928. [Google Scholar] [CrossRef]
- Preece, C.; Peñuelas, J. Rhizodeposition under drought and consequences for soil communities and ecosystem resilience. Plant Soil 2016, 409, 1–17. [Google Scholar] [CrossRef]
- Ahmed, A.A.Q.; Odelade, K.A.; Babalola, O.O. Microbial inoculants for improving carbon sequestration in agroecosystems to mitigate climate change. In Handbook of Climate Change Resilience; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–21. [Google Scholar]
- Abdalla, M.; Bitterlich, M.; Jansa, J.; Püschel, D.; Ahmed, M.A. The role of arbuscular mycorrhizal symbiosis in improving plant water status under drought. J. Exp. Bot. 2023, 74, 4808–4824. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.; de Vries, F.T. Plant root exudation under drought: Implications for ecosystem functioning. New Phytol. 2020, 225, 1899–1905. [Google Scholar] [CrossRef] [PubMed]
- Carvalhais, L.C.; Dennis, P.G.; Fedoseyenko, D.; Hajirezaei, M.R.; Borriss, R.; von Wirén, N. Root exudation of sugars, amino acids, and organic acids by maize as affected by nitrogen, phosphorus, potassium, and iron deficiency. J. Plant Nutr. Soil Sci. 2011, 174, 3–11. [Google Scholar] [CrossRef]
- Tătar, A.-S. Early comunication between plants and their symbiont nitrogen fixing bacteria-a minireview. Extrem. Life Biospeol. Astrobiol. 2013, 5, 117–121. [Google Scholar]
- Liu, C.-W.; Murray, J.D. The role of flavonoids in nodulation host-range specificity: An update. Plants 2016, 5, 33. [Google Scholar] [CrossRef]
- Tian, T.; Reverdy, A.; She, Q.; Sun, B.; Chai, Y. The role of rhizodeposits in shaping rhizomicrobiome. Environ. Microbiol. Rep. 2020, 12, 160–172. [Google Scholar] [CrossRef]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; Da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Canarini, A.; Merchant, A.; Dijkstra, F.A. Drought effects on Helianthus annuus and Glycine max metabolites: From phloem to root exudates. Rhizosphere 2016, 2, 85–97. [Google Scholar] [CrossRef]
- Bobille, H.; Fustec, J.; Robins, R.J.; Cukier, C.; Limami, A.M. Effect of water availability on changes in root amino acids and associated rhizosphere on root exudation of amino acids in Pisum sativum L. Phytochemistry 2019, 161, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Dardanelli, M.S.; Manyani, H.; González-Barroso, S.; Rodríguez-Carvajal, M.A.; Gil-Serrano, A.M.; Espuny, M.R.; López-Baena, F.J.; Bellogín, R.A.; Megías, M.; Ollero, F.J. Effect of the presence of the plant growth promoting rhizobacterium (PGPR) Chryseobacterium balustinum Aur9 and salt stress in the pattern of flavonoids exuded by soybean roots. Plant Soil 2010, 328, 483–493. [Google Scholar] [CrossRef]
- Rubia, M.I.; Ramachandran, V.K.; Arrese-Igor, C.; Larrainzar, E.; Poole, P.S. A novel biosensor to monitor proline in pea root exudates and nodules under osmotic stress and recovery. Plant Soil 2020, 452, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Cesari, A.; Paulucci, N.; López-Gómez, M.; Hidalgo-Castellanos, J.; Plá, C.L.; Dardanelli, M.S. Restrictive water condition modifies the root exudates composition during peanut-PGPR interaction and conditions early events, reversing the negative effects on plant growth. Plant Physiol. Biochem. 2019, 142, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Dardanelli, M.S.; de Córdoba, F.J.F.; Estévez, J.; Contreras, R.; Cubo, M.T.; Rodríguez-Carvajal, M.Á.; Gil-Serrano, A.M.; López-Baena, F.J.; Bellogín, R.; Manyani, H. Changes in flavonoids secreted by Phaseolus vulgaris roots in the presence of salt and the plant growth-promoting rhizobacterium Chryseobacterium balustinum. Appl. Soil Ecol. 2012, 57, 31–38. [Google Scholar] [CrossRef]
- L’taief, B.; Sifi, B.; Zaman-Allah, M.; Drevon, J.-J.; Lachaâl, M. Effect of salinity on root-nodule conductance to the oxygen diffusion in the Cicer arietinum–Mesorhizobium ciceri symbiosis. J. Plant Physiol. 2007, 164, 1028–1036. [Google Scholar] [CrossRef] [PubMed]
- Serraj, R.; Roy, G.; Drevon, J.J. Salt stress induces a decrease in the oxygen uptake of soybean nodules and in their permeability to oxygen diffusion. Physiol. Plant. 1994, 91, 161–168. [Google Scholar] [CrossRef]
- Babber, S.; Sheokand, S.; Malik, S. Nodule structure and functioning in chickpea (Cicer arietinum) as affected by salt stress. Biol. Plant 2000, 43, 269–273. [Google Scholar] [CrossRef]
- Ashraf, M.; Iram, A. Drought stress induced changes in some organic substances in nodules and other plant parts of two potential legumes differing in salt tolerance. Flora-Morphol. Distrib. Funct. Ecol. Plants 2005, 200, 535–546. [Google Scholar] [CrossRef]
- An, S.; Couteau, C.; Luo, F.; Neveu, J.; DuBow, M.S. Bacterial diversity of surface sand samples from the Gobi and Taklamaken deserts. Microb. Ecol. 2013, 66, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Soussi, A.; Ferjani, R.; Marasco, R.; Guesmi, A.; Cherif, H.; Rolli, E.; Mapelli, F.; Ouzari, H.I.; Daffonchio, D.; Cherif, A. Plant-associated microbiomes in arid lands: Diversity, ecology and biotechnological potential. Plant Soil 2016, 405, 357–370. [Google Scholar] [CrossRef]
- Sharma, R.; Manda, R.; Gupta, S.; Kumar, S.; Kumar, V. Isolation and characterization of osmotolerant bacteria from Thar Desert of Western Rajasthan (India). Rev. Biol. Trop. 2013, 61, 1551–1562. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Huang, Y.; Liu, Y.; Mohamed, O.A.A.; Fan, X.; Wang, L.; Li, L.; Ma, J. Bacterial community structure and potential microbial coexistence mechanism associated with three halophytes adapting to the extremely hypersaline environment. Microorganisms 2022, 10, 1124. [Google Scholar] [CrossRef] [PubMed]
- Sorty, A.M.; Meena, K.K.; Choudhary, K.; Bitla, U.M.; Minhas, P.; Krishnani, K. Effect of plant growth promoting bacteria associated with halophytic weed (Psoralea corylifolia L) on germination and seedling growth of wheat under saline conditions. Appl. Biochem. Biotechnol. 2016, 180, 872–882. [Google Scholar] [CrossRef] [PubMed]
- Jorquera, M.A.; Maruyama, F.; Ogram, A.V.; Navarrete, O.U.; Lagos, L.M.; Inostroza, N.G.; Acuña, J.J.; Rilling, J.I.; de La Luz Mora, M. Rhizobacterial community structures associated with native plants grown in Chilean extreme environments. Microb. Ecol. 2016, 72, 633–646. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, W.S.; Akhkha, A.; El-Naggar, M.Y.; Elbadry, M. In vitro antagonistic activity, plant growth promoting traits and phylogenetic affiliation of rhizobacteria associated with wild plants grown in arid soil. Front. Microbiol. 2014, 5, 651. [Google Scholar] [CrossRef]
- Zia, R.; Nawaz, M.S.; Yousaf, S.; Amin, I.; Hakim, S.; Mirza, M.S.; Imran, A. Seed inoculation of desert-plant growth-promoting rhizobacteria induce biochemical alterations and develop resistance against water stress in wheat. Physiol. Plant. 2021, 172, 990–1006. [Google Scholar] [CrossRef]
- Getahun, A.; Muleta, D.; Assefa, F.; Kiros, S. Plant Growth-Promoting Rhizobacteria Isolated from Degraded Habitat Enhance Drought Tolerance of Acacia (Acacia abyssinica Hochst. ex Benth.) Seedlings. Int. J. Microbiol. 2020, 2020, 8897998. [Google Scholar] [CrossRef]
- Sood, N.; Prajapat, S.P.; Shaikh, N.S.; Gokhale, T.; Thushar, S. Screening of Plant-Growth-Promoting Bacterial Isolates from Rhizosphere Soil of Prosopis cineraria from UAE. Environ. Sci. Proc. 2022, 16, 69. [Google Scholar] [CrossRef]
- Bonatelli, M.L.; Lacerda-Júnior, G.V.; dos Reis Junior, F.B.; Fernandes-Júnior, P.I.; Melo, I.S.; Quecine, M.C. Beneficial plant-associated microorganisms from semiarid regions and seasonally dry environments: A review. Front. Microbiol. 2021, 11, 553223. [Google Scholar] [CrossRef] [PubMed]
- Bueno Batista, M.; Dixon, R. Manipulating nitrogen regulation in diazotrophic bacteria for agronomic benefit. Biochem. Soc. Trans. 2019, 47, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Saranraj, P.; Sayyed, R.; Sivasakthivelan, P.; Kokila, M.; Al-Tawaha, A.R.M.; Amala, K.; Yasmin, H. Symbiotic effectiveness of rhizobium strains in agriculture. In Plant Growth Promoting Microorganisms of Arid Region; Springer: Berlin/Heidelberg, Germany, 2023; pp. 389–421. [Google Scholar]
- Li, P.; Teng, C.-c.; Ding, B.-j.; Liu, Y.-j.; Hou, W.-w.; He, T. A study on drought tolerance of rhizobia strains of faba bean (Vicia faba L.) isolated from drought regions in Qinghai plateau. Acta Agric. Univ. Jiangxiensis 2021, 43, 1241–1249. [Google Scholar]
- Athar, A. Drought tolerance by lentil rhizobia (Rhizobium leguminosarum) from arid and semiarid areas of Pakistan. Lett. Appl. Microbiol. 1998, 26, 38–42. [Google Scholar] [CrossRef]
- Shoushtari, N.H.; Pepper, I.L. Mesquite rhizobia isolated from the Sonoran desert: Competitiveness and survival in soil. Soil Biol. Biochem. 1985, 17, 803–806. [Google Scholar] [CrossRef]
- Aserse, A.A.; Markos, D.; Getachew, G.; Yli-Halla, M.; Lindström, K. Rhizobial inoculation improves drought tolerance, biomass and grain yields of common bean (Phaseolus vulgaris L.) and soybean (Glycine max L.) at Halaba and Boricha in Southern Ethiopia. Arch. Agron. Soil Sci. 2020, 66, 488–501. [Google Scholar] [CrossRef]
- Bano, A.; Batool, R.; Dazzo, F. Adaptation of chickpea to desiccation stress is enhanced by symbiotic rhizobia. Symbiosis 2010, 50, 129–133. [Google Scholar] [CrossRef]
- Kaschuk, G.; Hungria, M.; Leffelaar, P.; Giller, K.; Kuyper, T. Differences in photosynthetic behaviour and leaf senescence of soybean (Glycine max [L.] Merrill) dependent on N2 fixation or nitrate supply. Plant Biol. 2010, 12, 60–69. [Google Scholar] [CrossRef]
- Belane, A.; Dakora, F. Assessing the relationship between photosynthetic C accumulation and symbiotic N nutrition in leaves of field-grown nodulated cowpea (Vigna unguiculata L. Walp.) Genotypes. Photosynthetica 2015, 53, 562–571. [Google Scholar] [CrossRef]
- Cerezini, P.; Kuwano, B.H.; Grunvald, A.K.; Hungria, M.; Nogueira, M.A. Soybean tolerance to drought depends on the associated Bradyrhizobium strain. Braz. J. Microbiol. 2020, 51, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, A.; Khan, M.; Ahemad, M.; Oves, M. Plant growth promotion by phosphate solubilizing bacteria. Acta Microbiol. Immunol. Hung. 2009, 56, 263–284. [Google Scholar] [CrossRef] [PubMed]
- Ben Romdhane, S.; De Lajudie, P.; Fuhrmann, J.J.; Mrabet, M. Potential role of rhizobia to enhance chickpea-growth and yield in low fertility-soils of Tunisia. Antonie Van Leeuwenhoek 2022, 115, 921–932. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, M.; Turkyilmaz Unal, B.; García-Caparrós, P.; Khursheed, A.; Gul, A.; Hasanuzzaman, M. Osmoregulation and its actions during the drought stress in plants. Physiol. Plant. 2021, 172, 1321–1335. [Google Scholar] [CrossRef]
- Poudel, M.; Mendes, R.; Costa, L.A.; Bueno, C.G.; Meng, Y.; Folimonova, S.Y.; Garrett, K.A.; Martins, S.J. The role of plant-associated bacteria, fungi, and viruses in drought stress mitigation. Front. Microbiol. 2021, 12, 743512. [Google Scholar] [CrossRef]
- Abdul Rahman, N.S.N.; Abdul Hamid, N.W.; Nadarajah, K. Effects of abiotic stress on soil microbiome. Int. J. Mol. Sci. 2021, 22, 9036. [Google Scholar] [CrossRef]
- Duan, J.; Jiang, W.; Cheng, Z.; Heikkila, J.J.; Glick, B.R. The complete genome sequence of the plant growth-promoting bacterium Pseudomonas sp. UW4. PLoS ONE 2013, 8, e58640. [Google Scholar] [CrossRef]
- Vardharajula, S.; Zulfikar Ali, S.; Grover, M.; Reddy, G.; Bandi, V. Drought-tolerant plant growth promoting Bacillus spp.: Effect on growth, osmolytes, and antioxidant status of maize under drought stress. J. Plant Interact. 2011, 6, 1–14. [Google Scholar] [CrossRef]
- Amine-Khodja, I.R.; Boscari, A.; Riah, N.; Kechid, M.; Maougal, R.T.; Belbekri, N.; Djekoun, A. Impact of Two Strains of Rhizobium leguminosarum on the Adaptation to Terminal Water Deficit of Two Cultivars Vicia faba. Plants 2022, 11, 515. [Google Scholar] [CrossRef]
- Khadraji, A.; Ghoulam, C. Effect of drought on growth, physiological and biochemical processes of chickpea-rhizobia symbiosis. Legume Res. Int. J. 2017, 40, 94–99. [Google Scholar]
- Owino, W.; Manabe, Y.; Mathooko, F.; Kubo, Y.; Inaba, A. Regulatory mechanisms of ethylene biosynthesis in response to various stimuli during maturation and ripening in fig fruit (Ficus carica L.). Plant Physiol. Biochem. 2006, 44, 335–342. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Brígido, C.; Glick, B.R.; Rossi, M.J. The role of rhizobial ACC deaminase in the nodulation process of leguminous plants. Int. J. Agron. 2016, 2016, 1369472. [Google Scholar] [CrossRef]
- DHULL, S.; SHEORAN, H.S.; KAKAR, R.; GERA, R. Screening and characterisation of ACC deaminase producing rhizobacteria from root nodules of clusterbean (Cyamopsis tetragonoloba). Ann. Plant Soil Res. 2018, 20, 254–257. [Google Scholar]
- Belimov, A.A.; Zinovkina, N.Y.; Safronova, V.I.; Litvinsky, V.A.; Nosikov, V.V.; Zavalin, A.A.; Tikhonovich, I.A. Rhizobial ACC deaminase contributes to efficient symbiosis with pea (Pisum sativum L.) under single and combined cadmium and water deficit stress. Environ. Exp. Bot. 2019, 167, 103859. [Google Scholar] [CrossRef]
- Sarapat, S.; Songwattana, P.; Longtonglang, A.; Umnajkitikorn, K.; Girdthai, T.; Tittabutr, P.; Boonkerd, N.; Teaumroong, N. Effects of increased 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity in Bradyrhizobium sp. SUTN9-2 on mung bean symbiosis under water deficit conditions. Microbes Environ. 2020, 35, ME20024. [Google Scholar] [CrossRef]
- FAO; ITPS. Status of the World’s Soil Resources (SWSR)—Main Report; Food and Agriculture Organization of the United Nations and Intergovernmental Technical Panel on Soils: Rome, Italy, 2015; Available online: http://www.fao.org/3/a-i5199e.pdf (accessed on 17 July 2024).
- Dong, R.; Zhang, J.; Huan, H.; Bai, C.; Chen, Z.; Liu, G. High salt tolerance of a Bradyrhizobium strain and its promotion of the growth of Stylosanthes guianensis. Int. J. Mol. Sci. 2017, 18, 1625. [Google Scholar] [CrossRef]
- Arora, N.K.; Fatima, T.; Mishra, J.; Mishra, I.; Verma, S.; Verma, R.; Verma, M.; Bhattacharya, A.; Verma, P.; Mishra, P. Halo-tolerant plant growth promoting rhizobacteria for improving productivity and remediation of saline soils. J. Adv. Res. 2020, 26, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Wdowiak-Wróbel, S.; Leszcz, A.; Małek, W. Salt tolerance in Astragalus cicer microsymbionts: The role of glycine betaine in osmoprotection. Curr. Microbiol. 2013, 66, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Luo, Y.; Li, Z.; Wang, J.; Wei, G. Role of exopolysaccharide in salt stress resistance and cell motility of Mesorhizobium alhagi CCNWXJ12–2T. Appl. Microbiol. Biotechnol. 2017, 101, 2967–2978. [Google Scholar] [CrossRef]
- Donot, F.; Fontana, A.; Baccou, J.; Schorr-Galindo, S. Microbial exopolysaccharides: Main examples of synthesis, excretion, genetics and extraction. Carbohydr. Polym. 2012, 87, 951–962. [Google Scholar] [CrossRef]
- Fujishige, N.A.; Kapadia, N.N.; De Hoff, P.L.; Hirsch, A.M. Investigations of Rhizobium biofilm formation. FEMS Microbiol. Ecol. 2006, 56, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Vanderlinde, E.M.; Harrison, J.J.; Muszyński, A.; Carlson, R.W.; Turner, R.J.; Yost, C.K. Identification of a novel ABC transporter required for desiccation tolerance, and biofilm formation in Rhizobium leguminosarum bv. viciae 3841. FEMS Microbiol. Ecol. 2010, 71, 327–340. [Google Scholar] [CrossRef]
- Mushtaq, Z.; Faizan, S.; Gulzar, B.; Hakeem, K.R. Inoculation of Rhizobium alleviates salinity stress through modulation of growth characteristics, physiological and biochemical attributes, stomatal activities and antioxidant defence in Cicer arietinum L. J. Plant Growth Regul. 2021, 40, 2148–2163. [Google Scholar] [CrossRef]
- Azevedo Neto, A.D.d.; Prisco, J.T.; Enéas-Filho, J.; Lacerda, C.F.d.; Silva, J.V.; Costa, P.H.A.d.; Gomes-Filho, E. Effects of salt stress on plant growth, stomatal response and solute accumulation of different maize genotypes. Braz. J. Plant Physiol. 2004, 16, 31–38. [Google Scholar] [CrossRef]
- Javot, H.; Lauvergeat, V.; Santoni, V.; Martin-Laurent, F.; Güçlü, J.; Vinh, J.; Heyes, J.; Franck, K.I.; Schaffner, A.R.; Bouchez, D. Role of a single aquaporin isoform in root water uptake. Plant Cell 2003, 15, 509–522. [Google Scholar] [CrossRef]
- Kapilan, R.; Vaziri, M.; Zwiazek, J.J. Regulation of aquaporins in plants under stress. Biol. Res. 2018, 51, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wallace, I.S.; Choi, W.-G.; Roberts, D.M. The structure, function and regulation of the nodulin 26-like intrinsic protein family of plant aquaglyceroporins. Biochim. Biophys. Acta (BBA)-Biomembr. 2006, 1758, 1165–1175. [Google Scholar] [CrossRef]
- Sujkowska, M.; Górska-Czekaj, M.; Bederska, M.; Borucki, W. Vacuolar organization in the nodule parenchyma is important for the functioning of pea root nodules. Symbiosis 2011, 54, 1–16. [Google Scholar] [CrossRef]
- Singh, R.K.; Deshmukh, R.; Muthamilarasan, M.; Rani, R.; Prasad, M. Versatile roles of aquaporin in physiological processes and stress tolerance in plants. Plant Physiol. Biochem. 2020, 149, 178–189. [Google Scholar] [CrossRef]
- Franzini, V.I.; Azcón, R.; Ruiz-Lozano, J.M.; Aroca, R. Rhizobial symbiosis modifies root hydraulic properties in bean plants under non-stressed and salinity-stressed conditions. Planta 2019, 249, 1207–1215. [Google Scholar] [CrossRef]
- Chakraborty, S.; Driscoll, H.E.; Abrahante, J.E.; Zhang, F.; Fisher, R.F.; Harris, J.M. Salt stress enhances early symbiotic gene expression in Medicago truncatula and induces a stress-specific set of rhizobium-responsive genes. Mol. Plant-Microbe Interact. 2021, 34, 904–921. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Hidalgo, P.; Hirsch, A.M. The nodule microbiome: N2-fixing rhizobia do not live alone. Phytobiomes J. 2017, 1, 70–82. [Google Scholar] [CrossRef]
- Chavoshi, S.; Nourmohamadi, G.; Madani, H.; Heidari Sharif Abad, H.; Alavi Fazel, M. The effects of biofertilizers on physiological traits and biomass accumulation of red beans (Phaseolus vulgaris cv. Goli) Under Water Stress. Iran. J. Plant Physiol. 2018, 8, 2555–2562. [Google Scholar]
- Amara, U.; Khalid, R.; Hayat, R. Soil bacteria and phytohormones for sustainable crop production. In Bacterial Metabolites in Sustainable Agroecosystem; Springer: Berlin/Heidelberg, Germany, 2015; pp. 87–103. [Google Scholar] [CrossRef]
- Kaushal, M.; Wani, S.P. Plant-growth-promoting rhizobacteria: Drought stress alleviators to ameliorate crop production in drylands. Ann. Microbiol. 2016, 66, 35–42. [Google Scholar] [CrossRef]
- Mohanty, P.; Singh, P.K.; Chakraborty, D.; Mishra, S.; Pattnaik, R. Insight into the role of PGPR in sustainable agriculture and environment. Front. Sustain. Food Syst. 2021, 5, 667150. [Google Scholar] [CrossRef]
- Uzma, M.; Iqbal, A.; Hasnain, S. Drought tolerance induction and growth promotion by indole acetic acid producing Pseudomonas aeruginosa in Vigna radiata. PLoS ONE 2022, 17, e0262932. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, C.; Saleem, A.R.; Della Rocca, G.; Emiliani, G.; De Carlo, A.; Balestrini, R.; Khalid, A.; Mahmood, T.; Centritto, M. Effects of plant growth-promoting rhizobacteria strains producing ACC deaminase on photosynthesis, isoprene emission, ethylene formation and growth of Mucuna pruriens (L.) DC. in response to water deficit. J. Biotechnol. 2021, 331, 53–62. [Google Scholar] [CrossRef]
- Saleem, A.R.; Brunetti, C.; Khalid, A.; Della Rocca, G.; Raio, A.; Emiliani, G.; De Carlo, A.; Mahmood, T.; Centritto, M. Drought response of Mucuna pruriens (L.) DC. inoculated with ACC deaminase and IAA producing rhizobacteria. PLoS ONE 2018, 13, e0191218. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Canseco, J.; Bautista-Cruz, A.; Sánchez-Mendoza, S.; Aquino-Bolaños, T.; Sánchez-Medina, P.S. Plant growth-promoting halobacteria and their ability to protect crops from abiotic stress: An eco-friendly alternative for saline soils. Agronomy 2022, 12, 804. [Google Scholar] [CrossRef]
- Hmaeid, N.; Wali, M.; Mahmoud, O.M.-B.; Pueyo, J.J.; Ghnaya, T.; Abdelly, C. Efficient rhizobacteria promote growth and alleviate NaCl-induced stress in the plant species Sulla carnosa. Appl. Soil Ecol. 2019, 133, 104–113. [Google Scholar] [CrossRef]
- Mufti, R.; Amna Rafique, M.; Haq, F.; Hussain, M.; Munis Masood, S.; Mumtaz, A.S.; Chaudhary, H.J. Genetic diversity and metal resistance assessment of endophytes isolated from Oxalis corniculata. Soil Environ. 2015, 34, 89–99. [Google Scholar]
- Khan, A.A.; Wang, T.; Hussain, T.; Ali, F.; Shi, F.; Latef, A.A.H.A.; Ali, O.M.; Hayat, K.; Mehmood, S.; Zainab, N. Halotolerant-Koccuria rhizophila (14asp)-induced amendment of salt stress in pea plants by limiting Na+ uptake and elevating production of antioxidants. Agronomy 2021, 11, 1907. [Google Scholar] [CrossRef]
- Agami, R.A.; Ghramh, H.A.; Hasheem, M. Seed inoculation with Azospirillum lipoferum alleviates the adverse effects of drought stress on wheat plants. J. Appl. Bot. Food Qual. 2017, 90, 165–173. [Google Scholar]
- Yousefi, S.; Kartoolinejad, D.; Bahmani, M.; Naghdi, R. Effect of Azospirillum lipoferum and Azotobacter chroococcum on germination and early growth of hopbush shrub (Dodonaea viscosa L.) under salinity stress. J. Sustain. For. 2017, 36, 107–120. [Google Scholar] [CrossRef]
- El-Esawi, M.A.; Al-Ghamdi, A.A.; Ali, H.M.; Alayafi, A.A. Azospirillum lipoferum FK1 confers improved salt tolerance in chickpea (Cicer arietinum L.) by modulating osmolytes, antioxidant machinery and stress-related genes expression. Environ. Exp. Bot. 2019, 159, 55–65. [Google Scholar] [CrossRef]
- Abd_Allah, E.F.; Alqarawi, A.A.; Hashem, A.; Radhakrishnan, R.; Al-Huqail, A.A.; Al-Otibi, F.O.N.; Malik, J.A.; Alharbi, R.I.; Egamberdieva, D. Endophytic bacterium Bacillus subtilis (BERA 71) improves salt tolerance in chickpea plants by regulating the plant defense mechanisms. J. Plant Interact. 2018, 13, 37–44. [Google Scholar] [CrossRef]
- Abd El-Ghany, M.F.; Attia, M. Effect of exopolysaccharide-producing bacteria and melatonin on faba bean production in saline and non-saline soil. Agronomy 2020, 10, 316. [Google Scholar] [CrossRef]
- Abdiev, A.; Khaitov, B.; Toderich, K.; Park, K.W. Growth, nutrient uptake and yield parameters of chickpea (Cicer arietinum L.) enhance by Rhizobium and Azotobacter inoculations in saline soil. J. Plant Nutr. 2019, 42, 2703–2714. [Google Scholar] [CrossRef]
- Gritli, T.; Boubakri, H.; Essahibi, A.; Hsouna, J.; Ilahi, H.; Didier, R.; Mnasri, B. Salt stress mitigation in Lathyrus cicera by combining different microbial inocula. Physiol. Mol. Biol. Plants 2022, 28, 1191–1206. [Google Scholar] [CrossRef]
- Abdela, A.A.; Barka, G.D.; Degefu, T. Co-inoculation effect of Mesorhizobium ciceri and Pseudomonas fluorescens on physiological and biochemical responses of Kabuli chickpea (Cicer arietinum L.) during drought stress. Plant Physiol. Rep. 2020, 25, 359–369. [Google Scholar] [CrossRef]
- Mansour, E.; Mahgoub, H.A.; Mahgoub, S.A.; El-Sobky, E.-S.E.; Abdul-Hamid, M.I.; Kamara, M.M.; AbuQamar, S.F.; El-Tarabily, K.A.; Desoky, E.-S.M. Enhancement of drought tolerance in diverse Vicia faba cultivars by inoculation with plant growth-promoting rhizobacteria under newly reclaimed soil conditions. Sci. Rep. 2021, 11, 24142. [Google Scholar] [CrossRef]
- Venturi, V.; Bez, C. A call to arms for cell–cell interactions between bacteria in the plant microbiome. Trends Plant Sci. 2021, 26, 1126–1132. [Google Scholar] [CrossRef]
- Taha, K.; El Attar, I.; Hnini, M.; Raif, A.; Béna, G.; Aurag, J. Beneficial effect of Rhizobium laguerreae co-inoculated with native Bacillus sp. and Enterobacter aerogenes on lentil growth under drought stress. Rhizosphere 2022, 22, 100523. [Google Scholar] [CrossRef]
- Michie, K.L.; Cornforth, D.M.; Whiteley, M. Bacterial tweets and podcasts# signaling# eavesdropping# microbialfightclub. Mol. Biochem. Parasitol. 2016, 208, 41–48. [Google Scholar]
- Majdura, J.; Jankiewicz, U.; Gałązka, A.; Orzechowski, S. The role of quorum sensing molecules in bacterial–plant interactions. Metabolites 2023, 13, 114. [Google Scholar] [CrossRef]
- Lupp, C.; Ruby, E.G. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol. 2005, 187, 3620–3629. [Google Scholar] [CrossRef]
- Kim, C.S.; Gatsios, A.; Cuesta, S.; Lam, Y.C.; Wei, Z.; Chen, H.; Russell, R.M.; Shine, E.E.; Wang, R.; Wyche, T.P. Characterization of Autoinducer-3 Structure and Biosynthesis in E. coli. ACS Cent. Sci. 2020, 6, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yu, Z.; Ding, T. Quorum-sensing regulation of antimicrobial resistance in bacteria. Microorganisms 2020, 8, 425. [Google Scholar] [CrossRef] [PubMed]
- Parsek, M.R.; Greenberg, E. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005, 13, 27–33. [Google Scholar] [CrossRef]
- Barber, C.; Tang, J.; Feng, J.; Pan, M.; Wilson, T.; Slater, H.; Dow, J.; Williams, P.; Daniels, M. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 1997, 24, 555–566. [Google Scholar] [CrossRef]
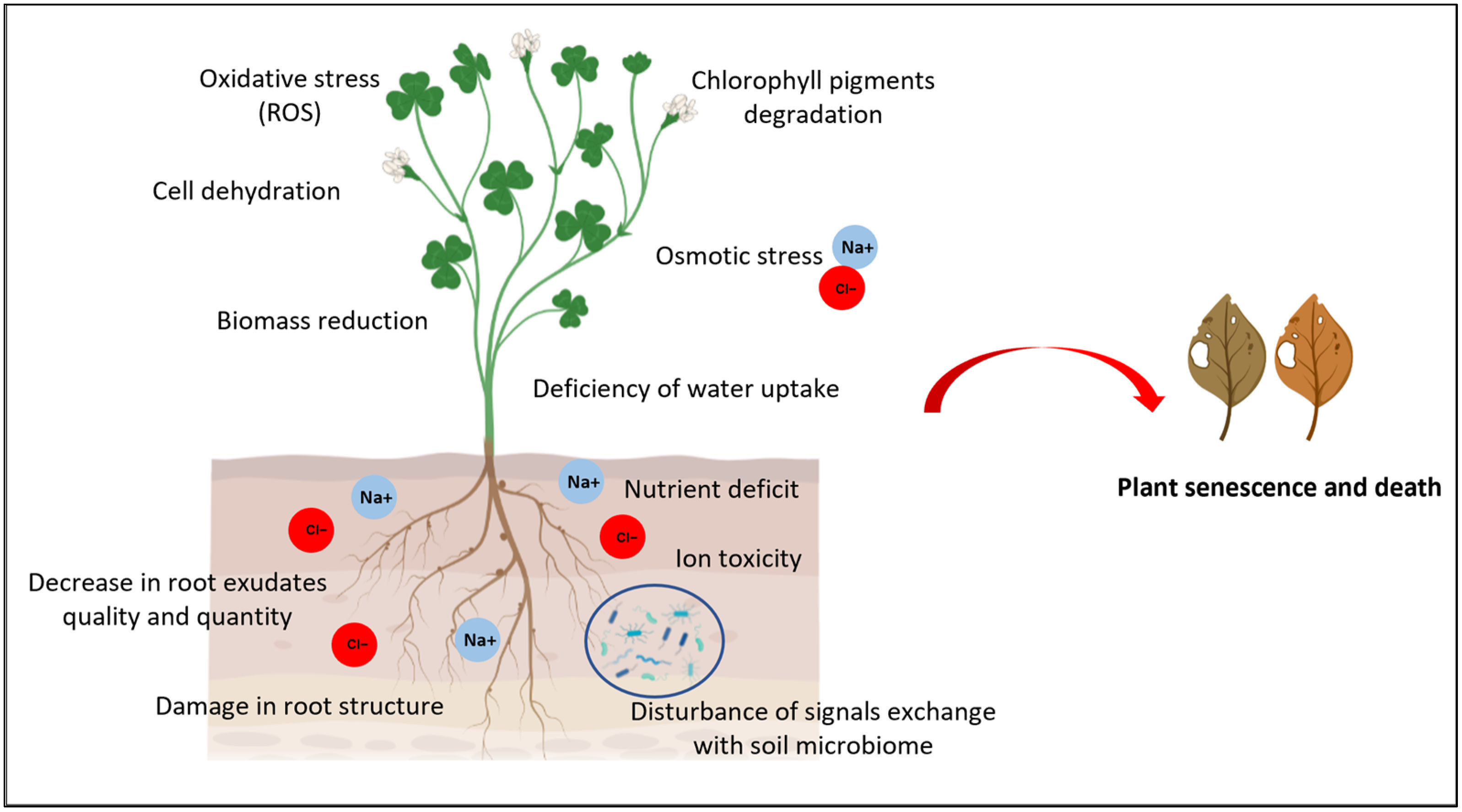
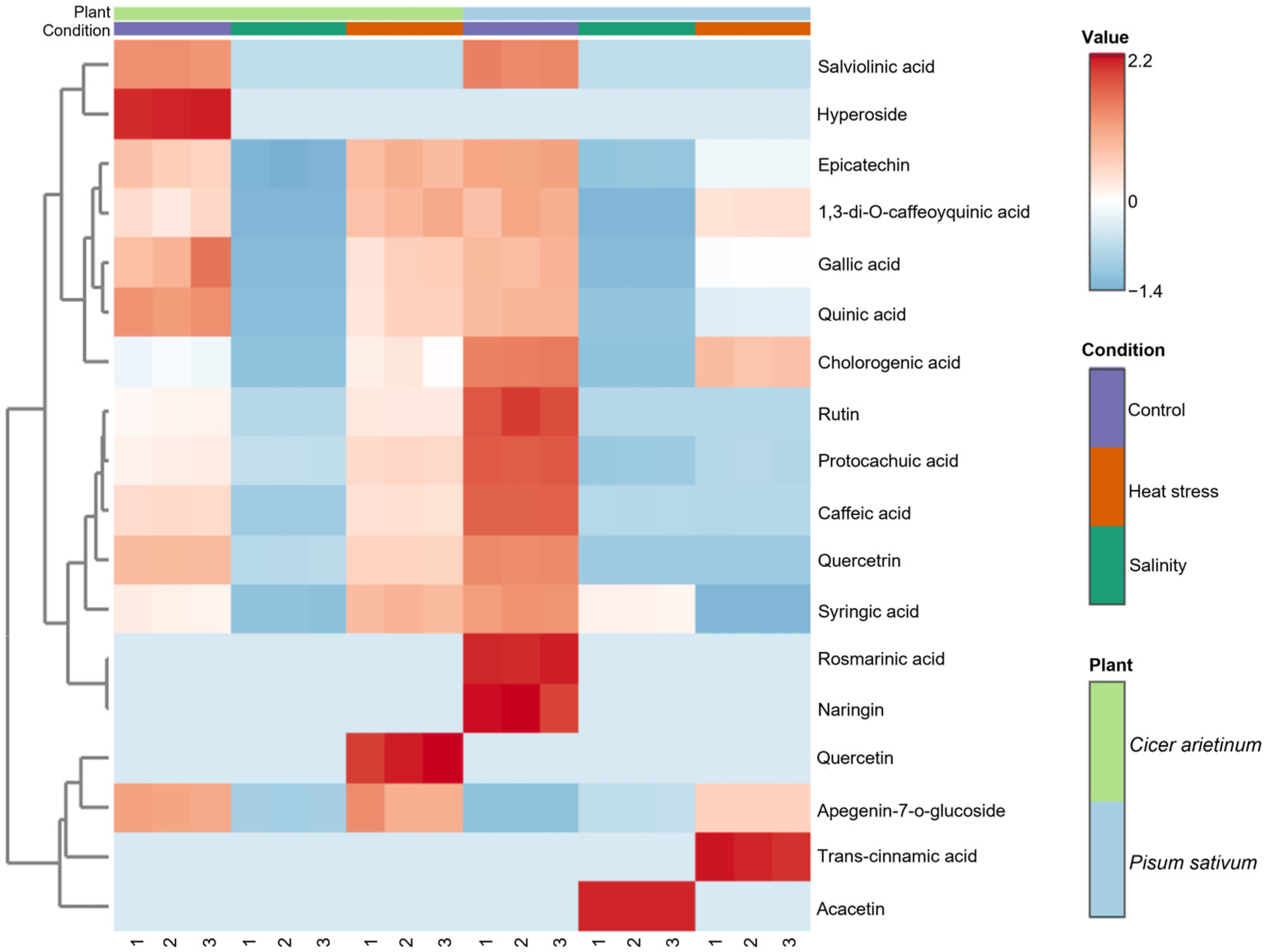

| Plant Host | Microbial Taxa | Type of Stress | Soil Type | Study |
|---|---|---|---|---|
| Caragana microphylla Hedysarum mongolicum Hedysarum scoparium | Rhizobiales, Xanthomonadales, Burkholderiales, Sphingomonadales, Solirubrobacterales, and Nitrosomonadales | Drought (Ningxia Province, northwest China, natural dry area) | Rhizosphere soil | [88] |
| Arachis hypogaea | Actinobacteriota, Planctomycetes, and Cyanobacteria | 45% FC (drought stress) and 85% FC (control) | Rhizosphere soil | [89] |
| Arachis hypogaea | Actinobacteriota, Proteobacteria, Chloroflexi, Acidobacteriota, and Firmicutes | Short-term drought treatment | Rhizosphere soil | [90] |
| Medicago sativa Trifolium pratense | Actinobacteriota, Proteobacteria, Firmicutes, Acidobacteriota, and Gemmatimonadetes | 20% FC (severe drought), 40% FC (moderate drought), and 80% FC (control) for three weeks | Rhizosphere soil | [91] |
| Glycine max | Acidobacteria, Bacteroidetes, Gemmatimonadetes, and Verrucomicrobia | Drought (plants were watered once per 6–10 days) | Rhizosphere soil | [98] |
| Albizzia julibrissin | Chloroflexi, Acidobacteria, Gemmatimonadetes, Proteobacteria, and Bacteroidetes | Salinity (natural saline field; salt content 4.1 ± 3.2 g kg−1) | Rhizosphere soil | [99] |
| Glycine max | Actinobacteria, Proteobacteria, Firmicutes, and Gemmatimonadetes | Salinity (natural saline soil) | Rhizosphere soil | [100] |
| Glycine soja (Wild soybean) Sesbania cannabina | Proteobacteria, Actinobacteriota, Chloroflexi, Acidobacteriota, Firmicutes, Gemmatimonadota and Crenarchaeota (archaea) | Salinity (natural saline soil; EC above 1500 µs cm−1) | Rhizosphere soil | [94] |
| Vigna radiata | Proteobacteria, Planctomycetes, Actinobacteria and Firmicutes | Salinity (150 to 180 mM NaCl) | Rhizosphere soil | [93] |
| Glycine soja (Wild soybean) | Proteobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Acidobacteriota, Gemmatimonadetes, Planctomycetes, and Firmicutes | Salinity (natural saline soil; EC of 565 ± 33 μS/cm) | Rhizosphere soil | [95] |
| Plant Species | Type of Stress | Identified Compound (s) | Reference |
|---|---|---|---|
| Pisum sativum var. Avola | Drought Salinity | Proline | [113] |
| Glycine max | Drought | Proline and pinitol | [110] |
| Arachis hypogaea | Drought | Apigenin, Genistein, Luteolin, Naringenin, Naringin, Rutin, IAA, and Tryptophane | [114] |
| Pisum sativum | Drought | Amino acids (proline, alanine, glutamate, and homoserine) | [111] |
| Glycine max | Salinity | 7,4-Dihydroxyflavone, Apigenin, Quercetin, Naringenin, Isoliquiritigenin (4, 2′, 4′-trihydroxychalcone), and Umbelliferone | [112] |
| Phaseolus vulgaris | Salinity | 7,4-Dihydroxyflavone, Quercetin, Naringenin, Hesperetin, Isoliquiritigenin, Umbelliferone | [115] |
| Cicer arietinum | Salinity | Quinic acid, Gallic acid, Cafeic acid, Syringic acid, Epicatechin, Quercitrin, and Apegenin-7-o-glucoside | [43] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben Gaied, R.; Brígido, C.; Sbissi, I.; Tarhouni, M. Sustainable Strategy to Boost Legumes Growth under Salinity and Drought Stress in Semi-Arid and Arid Regions. Soil Syst. 2024, 8, 84. https://doi.org/10.3390/soilsystems8030084
Ben Gaied R, Brígido C, Sbissi I, Tarhouni M. Sustainable Strategy to Boost Legumes Growth under Salinity and Drought Stress in Semi-Arid and Arid Regions. Soil Systems. 2024; 8(3):84. https://doi.org/10.3390/soilsystems8030084
Chicago/Turabian StyleBen Gaied, Roukaya, Clarisse Brígido, Imed Sbissi, and Mohamed Tarhouni. 2024. "Sustainable Strategy to Boost Legumes Growth under Salinity and Drought Stress in Semi-Arid and Arid Regions" Soil Systems 8, no. 3: 84. https://doi.org/10.3390/soilsystems8030084
APA StyleBen Gaied, R., Brígido, C., Sbissi, I., & Tarhouni, M. (2024). Sustainable Strategy to Boost Legumes Growth under Salinity and Drought Stress in Semi-Arid and Arid Regions. Soil Systems, 8(3), 84. https://doi.org/10.3390/soilsystems8030084







