Modelling the Distribution of the Red Macroalgae Asparagopsis to Support Sustainable Aquaculture Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Species and Area
2.2. Data Collection
2.3. Data Analysis
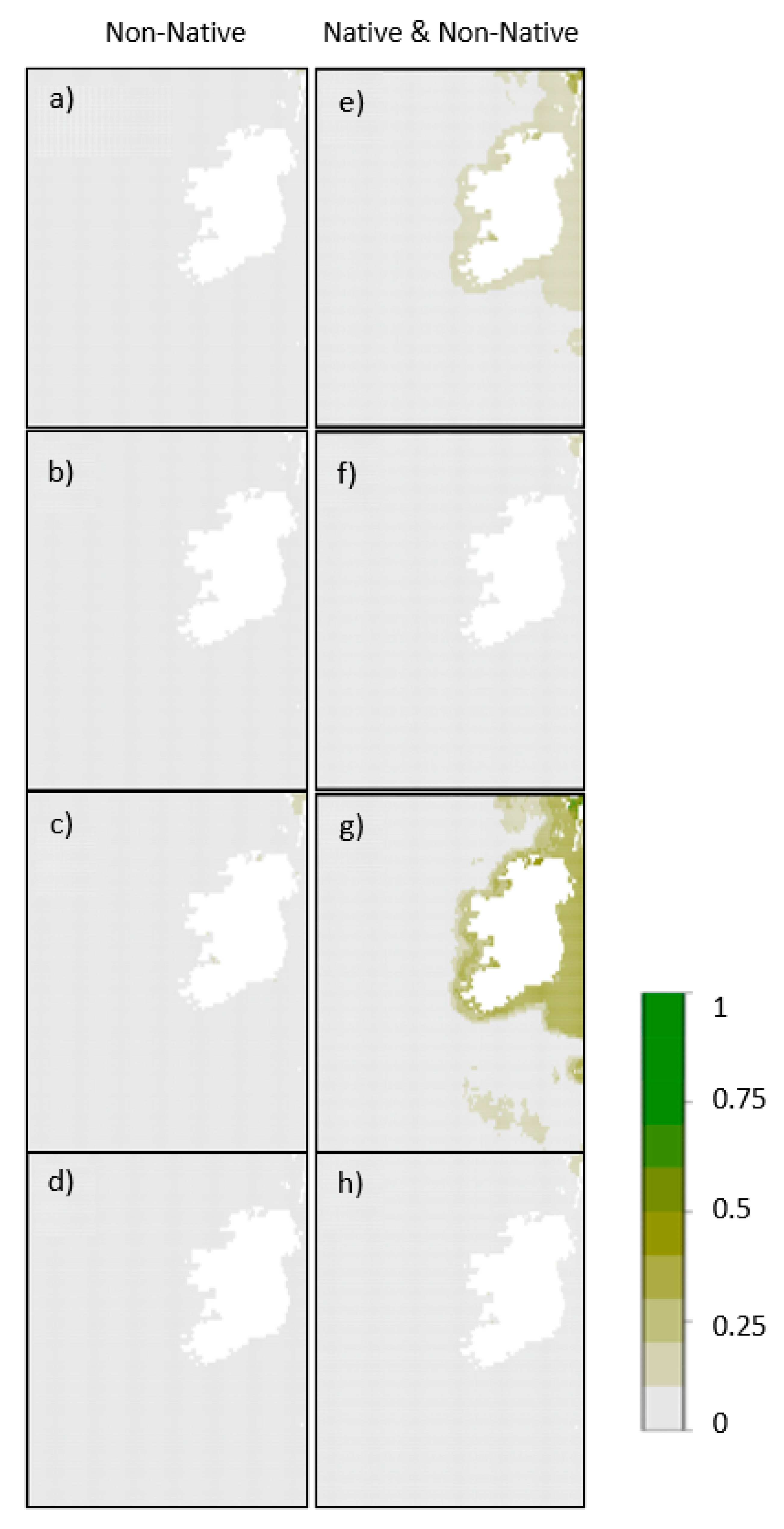
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, P.; Bustamante, M.; Ahammad, H.; Clark, H.; Dong, E.A.; Elsiddig, H.; Haberl, R.; Harper, J.; House, M.; Jafari, O.; et al. Agriculture, forestry and other land use (AFOLU). In Climate Change 2014: Mitigation of Climate Change. Contribution of Working Group III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Edenhofer, O., Pichs-Madruga, R., Sokona, Y., Farahani, S., Kadner, K., Seyboth, A., Adler, I., Baum, S., Brunner, P., Eickemeier, B., et al., Eds.; Cambridge University Press: Cambridge, UK, 2014; pp. 811–922. [Google Scholar]
- Herrero, M.; Thornton, P. Livestock and global change: Emerging issues for sustainable food systems. Proc. Natl. Acad. Sci. USA 2013, 110, 20878–20881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrero, M.; Henderson, B.; Havlík, P.; Thornton, P.K.; Conant, R.T.; Smith, P.; Wirsenius, S.; Hristov, A.N.; Gerber, P.; Gill, M.; et al. Greenhouse gas mitigation potentials in the livestock sector. Nat. Clim. Chang. 2016, 6, 452–461. [Google Scholar] [CrossRef] [Green Version]
- Myhre, G.; Shindell, D.; Bréon, F.M.; Collins, W.; Fuglestvedt, J.; Huang, J.; Koch, D.; Lamarque, J.F.; Lee, D.; Mendoza, B.; et al. Anthropogenic and natural radiative forcing. In Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK, 2013. [Google Scholar]
- Saunois, M.; Jackson, R.; Bousquet, P.; Poulter, B.; Canadell, J. The growing role of methane in anthropogenic climate change. Environ. Res. Lett. 2016, 11, 120207. [Google Scholar] [CrossRef] [Green Version]
- Smith, P.; Olesen, J. Synergies between the mitigation of, and adaptation to, climate change in agriculture. J. Agric. Sci. 2010, 148, 543–552. [Google Scholar] [CrossRef] [Green Version]
- Kinley, R.; Martinez-Fernandez, G.; Matthews, M.; de Nys, R.; Magnusson, M.; Tomkins, N. Mitigating the carbon footprint and improving productivity of ruminant livestock agriculture using a red seaweed. J. Clean. Prod. 2020, 259, 120836. [Google Scholar] [CrossRef]
- Crosson, P.; Shalloo, L.; O’Brien, D.; Lanigan, G.; Foley, P.; Boland, T.; Kenny, D. A review of whole farm systems models of greenhouse gas emissions from beef and dairy cattle production systems. Anim. Feed Sci. Technol. 2011, 166–167, 29–45. [Google Scholar] [CrossRef]
- Pickering, N.K.; Oddy, V.H.; Basarab, J.; Cammack, K.; Hayes, B.; Hegarty, R.S.; Lassen, J.; McEwan, J.C.; Miller, S.; Pinares-Patiño, C.S.; et al. Animal board invited review: Genetic possibilities to reduce enteric methane emissions from ruminants. Animal 2015, 9, 1431–1440. [Google Scholar] [CrossRef] [Green Version]
- Lassen, J.; Løvendahl, P. Heritability estimates for enteric methane emissions from Holstein cattle measured using noninvasive methods. J. Dairy Sci. 2016, 99, 1959–1967. [Google Scholar] [CrossRef] [Green Version]
- González-Recio, O.; López-Paredes, J.; Ouatahar, L.; Charfeddine, N.; Ugarte, E.; Alenda, R.; Jiménez-Montero, J.A. Mitigation of greenhouse gases in dairy cattle via genetic selection: 2. Incorporating methane emissions into the breeding goal. J. Dairy Sci. 2020, 103, 7210–7221. [Google Scholar] [CrossRef]
- Mayberry, D.; Bartlett, H.; Moss, J.; Davison, T.; Herrero, M. Pathways to carbon-neutrality for the Australian red meat sector. Agric. Syst. 2019, 175, 13–21. [Google Scholar] [CrossRef]
- Machado, L.; Magnusson, M.; Paul, N.; de Nys, R.; Tomkins, N. Effects of Marine and Freshwater Macroalgae on In Vitro Total Gas and Methane Production. PLoS ONE 2014, 9, e85289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinley, R.; Vucko, M.; Machado, L.; Tomkins, N. In Vitro: Evaluation of the Antimethanogenic Potency and Effects on Fermentation of Individual and Combinations of Marine Macroalgae. Am. J. Plant Sci. 2016, 7, 2038–2054. [Google Scholar] [CrossRef] [Green Version]
- Kinley, R.; de Nys, R.; Vucko, M.; Machado, L.; Tomkins, N. The red macroalgae Asparagopsis taxiformis is a potent natural anti-methanogenic that reduces methane production during in vitro fermentation with rumen fluid. Anim. Prod. Sci. 2016, 56, 282. [Google Scholar] [CrossRef]
- Environmental Protection Agency (EPA). Irelands Final Greenhouse Gas Emissions 1990–2018; Environmental Protection Agency (EPA): Wexford, Ireland, 2020.
- Franklin, J. Mapping Species Distributions: Spatial Inference and Prediction; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Peterson, A.T.; Soberon, J.; Pearson, R.G.; Anderson, R.P.; Martinez-Meyer, E.; Nakamura, M.; Araujo, M.B. Ecological Niches and Geographic Distributions; Princeton University Press: Princeton, NJ, USA, 2011; p. 315. [Google Scholar]
- Miller, J.A.; Holloway, P. Niche theory and models. In International Encyclopedia of Geography: People, the Earth, Environment and Technology; John Wiley & Sons: Hoboken, NJ, USA, 2017; pp. 1–10. [Google Scholar]
- Verbruggen, H.; Tyberghein, L.; Belton, G.S.; Mineur, F.; Jueter-Bock, A.; Hoarau, G.; Gurgel, C.F.D.; De Clerck, O. Improving transferability of introduced species’ distribution models: New tools to forecast the spread of a highly invasive seaweed. PLoS ONE 2013, 8, e68337. [Google Scholar] [CrossRef] [Green Version]
- Marcelino, V.R.; Verbruggen, H. Ecological Niche Models of invasive seaweeds. J. Phycol. 2015, 51, 606–620. [Google Scholar] [CrossRef] [PubMed]
- Westmeijer, G.; Everaert, G.; Pirlet, H.; De Clerck, O.; Vandegehuchte, M. Mechanistic niche modelling to identify favourable growth sites of temperate macroalgae. Algal Res. 2019, 41, 101529. [Google Scholar] [CrossRef]
- Dijoux, L.; Viard, F.; Payri, C. The more we search, the more we find: Discovery of a new lineage and a new species complex in the genus Asparagopsis. PLoS ONE 2014, 9, e103826. [Google Scholar] [CrossRef] [Green Version]
- Elith, J.; Kearney, M.; Phillips, S. The art of modelling range-shifting species. Methods Ecology Evolution Novel methods improve prediction of species’ distributions from occurrence data. Methods Ecol. Evol. 2020, 1, 330–342. [Google Scholar] [CrossRef]
- Jimenez-Valverde, A.; Lobo, J.M.; Hortal, J.; Jimenez-Valverde, A. Not as good as they seem: The importance of concepts in species distribution modelling. Divers. Distrib. 2008, 14, 885–890. [Google Scholar] [CrossRef]
- Peterson, A.T.; Soberon, J.; Sanchez-Cordero, V. Conservatism of ecological niches in evolutionary time. Science 1999, 285, 1265–1267. [Google Scholar] [CrossRef]
- Peterson, A.T. Predicting the geography of species’ invasions via ecological niche modeling. Quant. Reverse Biol. 2003, 78, 419–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, J.; Verlaque, M. The Caulerpa racemosa invasion: A critical review. Mar. Pollut. Bull. 2008, 56, 205–225. [Google Scholar] [CrossRef]
- Katsanevakis, S.; Salomidi, M.; Panou, A. Modelling distribution patterns and habitat preference of the invasive green alga Caulerpa racemosa in the Saronikos Gulf (Eastern Mediterranean). Aquat. Biol. 2010, 10, 57–67. [Google Scholar] [CrossRef]
- Whitney, K.D.; Gabler, C.A. Rapid evolution in introduced species, “invasive traits” and recipient communities: Challenges for predicting invasive potential. Divers. Distrib. 2008, 14, 569–580. [Google Scholar] [CrossRef]
- Barbet-Massin, M.; Jiguet, F.; Albert, C.; Thuiller, W. Selecting pseudo-absences for species distribution models: How, where and how many? Methods Ecol. Evol. 2012, 3, 327–338. [Google Scholar] [CrossRef]
- Guiry, M.; Guiry, G. AlgaeBase [Internet]; National University of Ireland: Galway, Ireland, 2020. [Google Scholar]
- Guiry, M.D.; Dawes, C.J. Daylength, temperature and nutrient control of tetrasporogenesis in Asparagopsis armata (Rhodophyta). J. Exp. Mar. Biol. Ecol. 1992, 158, 197–219. [Google Scholar] [CrossRef]
- Andreakis, N.; Procanni, G.; Maggs, C.; Kooistra, W. Phylogeography of the invasive seaweed Asparagopsis (Bonnemaisoniales, Rhodophyta) reveals cryptic diversity. Mol. Ecol. 2007, 16, 2285–2299. [Google Scholar] [CrossRef]
- Bonin, D.R.; Hawkes, M.W. Systematics and life histories of New Zealand Bonnemaisoniaceae (Bonnemaisoniales, Rhodophyta): I. The genus Asparagopsis. N. Z. J. Bot. 1987, 25, 577–590. [Google Scholar] [CrossRef] [Green Version]
- Feldmann, J.; Feldmann, G. Sur le d éveloppement des carpospores et l’alternance de gènérations de l’Asparagopsis armata Harvey. C. R. Hebd. Séanc. Acad. Sci. Paris 1939, 208, 1240–1242. [Google Scholar]
- Andreakis, N.; Procaccini, G.; Kooistra, W.H.C.F. Asparagopsis taxiformis and Asparagopsis armata (Bonnemaisoniales, Rhodophyta): Genetic and morphological identification of Mediterranean populations. Eur. J. Phycol. 2004, 39, 273–283. [Google Scholar] [CrossRef]
- Womersley, H.B.S. The Marine Benthic Flora of Southern Australia. Rhodophyta -Part IIIB. Gracilariales, Rhodymeniales, Corallinales and Bonnemaisoniales; Australian Biological Resources Study: Camberra, Australia, 1996; p. 392. [Google Scholar]
- Altamirano, M.; Muñoz, A.; De la Rosa, J.; Barrajón-Mínguez, A.; Barrajón-Domenech, A.; Moreno-Robledo, C.; Arroyo, M. The invasive species Asparagopsis taxiformis (Bonnemaisoniales, Rhodophyta) on Andalusian coast (Southern Spain): Reproductive stages, new records and invaded communities. Acta Bot. Malacit. 2020, 33, 5. [Google Scholar] [CrossRef]
- McCarthy, G.D.; Gleeson, E.; Walsh, S. The influence of ocean variations on the climate of Ireland. Weather 2015, 70, 242–245. [Google Scholar] [CrossRef] [Green Version]
- Guiry, M.D.; Hession, C. Eat up your seaweed! Ireland of the Welcomes. Glob. Ecol. Biogeogr. Lett. 1996, 45, 22–25. [Google Scholar]
- Devoy, R.J.N. Coastal Vulnerability and the implications of Sea-Level Rise for Ireland. J. Coast. Res. 2008, 24, 325–341. [Google Scholar] [CrossRef]
- Cronin, K.M.; Devoy, R.J.N.; Gault, J. Modelling Estuarine Morphodynamics on the South Coast of Ireland. J. Coast. Res. 2007, 50, 474–479. [Google Scholar]
- Kraan, S.; Barrington, K. Commercial farming of Asparagopsis armata (Bonnemaisoniceae, Rhodophyta) in Ireland, maintenance of an introduced species? J. Appl. Phycol. 2005, 17, 103–110. [Google Scholar] [CrossRef]
- Chamberlain, S.; Barve, V.; Mcglinn, D.; Oldoni, D.; Desmet, P.; Geffert, L.; Ram, K. rgbif: Interface to the Global Biodiversity Information Facility API. R Package Version 3.5.2. Available online: https://CRAN.R-project.org/package=rgbif (accessed on 1 June 2020).
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: https://www.R-project.org (accessed on 1 June 2020).
- GBIF. GBIF Occurrence Download. 2020. Available online: https://doi.org/10.15468/dl.extoyj (accessed on 20 February 2020).
- GBIF. GBIF Occurrence Download. 2020. Available online: https://doi.org/10.15468/dl.tvxp24 (accessed on 13 September 2020).
- Assis, J.; Tyberghein, L.; Bosch, S.; Verbruggen, H.; Serrão, E.; De Clerck, O. Bio-ORACLE v2.0: Extending marine data layers for bioclimatic modelling. Glob. Ecol. Biogeogr. 2017, 27, 277–284. [Google Scholar] [CrossRef]
- Tyberghein, L.; Verbruggen, H.; Pauly, K.; Troupin, C.; Mineur, F.; De Clerck, O. Bio-ORACLE: A global environmental dataset for marine species distribution modelling. Glob. Ecol. Biogeogr. 2012, 21, 272–281. [Google Scholar] [CrossRef]
- Sbrocco, E.; Barber, P. MARSPEC: Ocean climate layers for marine spatial ecology. Ecology 2013, 94, 979. [Google Scholar] [CrossRef]
- Bosch, S.; Tyberghein, L.; De Clerck, O.; Fernandez, S.; Schepers, L. sdmpredictors: Species Distribution Modelling Predictor Datasets. R Package Version 0.2.9. Available online: https://CRAN.R-project.org/package=sdmpredictors (accessed on 1 June 2020).
- Dray, S.; Dufour, A.; Thioulouse, J. ade4: Analysis of Ecological Data: Exploratory and Euclidean Methods in Environmental Sciences. R Package Version 1.7-16. Available online: https://CRAN.R-project.org/package=ade4 (accessed on 1 June 2020).
- Wilson, K.; Skinner, M.; Lotze, H. Projected 21st-century distribution of canopy-forming seaweeds in the Northwest Atlantic with climate change. Divers. Distrib. 2018, 25, 582–602. [Google Scholar] [CrossRef] [Green Version]
- Roleda, M.; Hurd, C. Seaweed nutrient physiology: Application of concepts to aquaculture and bioremediation. Phycologia 2019, 58, 552–562. [Google Scholar] [CrossRef] [Green Version]
- Barbet-Massin, M.; Rome, Q.; Villemant, C.; Courchamp, F. Can species distribution models really predict the expansion of invasive species? PLoS ONE 2018, 13, e0193085. [Google Scholar] [CrossRef] [Green Version]
- Thuiller, W.; Georges, D.; Engler, R.; Breiner, F.; Georges, M.D.; Thuiller, C.W. Biomod2: Ensemble Platform for Species Distribution Modelling. R Package Version 3.4.6. Available online: https://CRAN.R-project.org/package=biomod2 (accessed on 26 February 2020).
- Thuiller, W. Patterns and uncertainties of species’ range shifts under climate change. Glob. Chang. Biol. 2004, 10, 2020–2027. [Google Scholar] [CrossRef]
- Araujo, M.; Pearson, R.; Thuiller, W.; Erhard, M. Validation of species-climate impact models under climate change. Glob. Chang. Biol. 2005, 11, 1504–1513. [Google Scholar] [CrossRef] [Green Version]
- Araujo, M.B.; Guisan, A. Five (or so) challenges for species distribution modelling. J. Biogeogr. 2006, 33, 1677–1688. [Google Scholar] [CrossRef]
- Hijmans, R.J. Cross-validation of species distribution models: Removing spatial sorting bias and calibration with a null model. Ecology 2012, 93, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Hastings, R.; Cummins, V.; Holloway, P. Assessing the Impact of Physical and Anthropogenic Environmental Factors in Determining the Habitat Suitability of Seagrass Ecosystems. Sustainability 2020, 12, 8302. [Google Scholar] [CrossRef]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. distributions and projecting potential future shifts under. Radiology 1982, 143, 29–33. [Google Scholar] [CrossRef] [Green Version]
- McHugh, M. Interrater reliability: The kappa statistic. Biochem. Med. 2012, 15, 276–282. [Google Scholar] [CrossRef]
- Lüning, K.; Yarish, C.; Kirkman, H. Seaweeds: Their Environment, Biogeography, and Ecophysiology; John and Wiley & Sons: Hoboken, NJ, USA, 1990. [Google Scholar]
- Eggert, A. Seaweed responses to temperature. In Seaweed Biology: Novel Insights into Eco-Physiology, Ecology and Utilization; Wiencke, C., Bischof, K., Eds.; Springer: Heidelberg/Berlin, Germany, 2012; pp. 47–66. [Google Scholar]
- Dormann, C.F.; Bobrowski, M.; Dehling, D.M.; Harris, D.J.; Hartig, F.; Lischke, H.; Moretti, M.D.; Pagel, J.; Pinkert, S.; Schleuning, M.; et al. Biotic interactions in species distribution modelling: 10 questions to guide interpretation and avoid false conclusions. Glob. Ecol. Biogeogr. 2018, 27, 1004–1016. [Google Scholar] [CrossRef] [Green Version]
- Peterson, A.T.; Soberón, J.; Ramsey, J.; Osorio-Olvera, L. Co-occurrence networks do not support identification of biotic interactions. Biodivers. Inform. 2020, 15, 1–10. [Google Scholar] [CrossRef]
- De la Torre Cerro, R.; Holloway, P. A review of the methods for studying biotic interactions in phenological analyses. Methods Ecol. Evol. 2021, 12, 227–244. [Google Scholar] [CrossRef]
- Wisz, M.S.; Pottier, J.; Kissling, W.D.; Pellissier, L.; Lenoir, J.; Damgaard, C.F.; Dormann, C.F.; Forchhammer, M.C.; Grytnes, J.A.; Guisan, A.; et al. The role of biotic interactions in shaping distributions and realised assemblages of species: Implications for species distribution modelling. Biol. Rev. 2013, 88, 15–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, J.A.; Holloway, P. Incorporating movement in species distribution models. Prog. Phys. Geogr. 2015, 39, 837–849. [Google Scholar] [CrossRef]
- Engler, R.; Guisan, A.; Rechsteiner, L. An improved approach for predicting the distribution of rare and endangered species from occurrence and pseudo-absence data. J. Appl. Ecol. 2004, 41, 263–274. [Google Scholar] [CrossRef]
- Guisan, A.; Thuiller, W.; Zimmerman, N.E. Habitat Suitability and Distribution Models: With Applications in R; Cambridge University Press: Cambridge, UK, 2017. [Google Scholar]
- Flanders Marine Institute. Maritime Boundaries Geodatabase: Maritime Boundaries and Exclusive Economic Zones (200NM), Version 11. Available online: https://www.marineregions.org (accessed on 1 June 2020).


| Layer | Justification |
|---|---|
| Mean of diffuse attenuation | Diffuse attenuation, which is an indicator of light availability [54]; this light availability is important as it drives photosynthesis and growth of seaweeds [55]. |
| Dissolved oxygen | Significant contributor in PCA analysis (Supplementary Information 1). |
| Nitrate | The nutrient Nitrogen limits seaweed growth [55]. |
| pH | Significant contributor in PCA analysis (Supplementary Information 1). |
| Phosphate | The nutrient phosphorous limits seaweed growth (Roleda and Hurd, 2019). |
| Sea surface temperature range | Temperature is a primary range limiting factor [33]. |
| Temperature of warmest month | Temperature is a primary range limiting factor [33]. |
| Mean sea surface salinity | Significant contributor in PCA analysis (Supplementary Information 1). |
| Distance from shore | Distance to shore as A.armata is mainly found in the sublittoral zone [44]. |
| Bathymetry | Bathymetry as A.armata is mainly found in the sublittoral zone [44]. |
| Ulva lactuca species distribution | A.armata is an epiphyte that attaches to other seaweeds utilising its barbs [32]. |
| Model | Presence | Pseudo-Absence |
|---|---|---|
| A. armata (non-native only) | Ireland | Ireland (10,000) |
| A. armata (native and non-native) | Ireland New Zealand | Ireland (5000) New Zealand (5000) |
| A. taxiformis (non-native only) | Portugal | Portugal (10,000) |
| A. taxiformis (native and non-native) | Portugal Australia | Portugal (5000) Australia (5000) |
| Model | Metric | Mean Ensemble | CI (Lower) Ensemble | CI (Upper) Ensemble | Median Ensemble |
|---|---|---|---|---|---|
| A. armata (non-native) | AUC | 0.999 | 0.999 | 0.999 | 0.998 |
| A. armata (non-native) | TSS | 0.985 | 0.985 | 0.984 | 0.984 |
| A. armata (non-native and native) | AUC | 0.998 | 0.994 | 0.998 | 0.995 |
| A. armata (non-native and native) | TSS | 0.968 | 0.961 | 0.971 | 0.957 |
| A. taxiformis (non-native) | AUC | 1.000 | 1.000 | 1.000 | 0.999 |
| A. taxiformis (non-native) | TSS | 0.994 | 0.994 | 0.994 | 0.994 |
| A. taxiformis (non-native and native) | AUC | 0.998 | 0.979 | 0.998 | 0.994 |
| A. taxiformis (non-native and native) | TSS | 0.978 | 0.947 | 0.975 | 0.918 |
| Environmental Variables | A. armata (Non-Native Only) | A. armata (Native and Non-Native) | A. taxiformis (Non-Native Only) | A. taxiformis (Native and Non-Native) |
|---|---|---|---|---|
| Mean of diffuse attenuation | 0.08(±0.02) | 0.16(±0.04) | 0.59(±0.06) | 0.47(±0.05) |
| Dissolved oxygen | 0.08(±0.02) | 0.10(±0.05) | ||
| Nitrate | 0.05(±0.02) | 0.08(±0.04) | 0.74(±0.09) | 0.30(±0.06) |
| pH | 0.09(±0.05) | 0.07(±0.03) | 0.21(±0.06) | 0.20(±0.05) |
| Phosphate | 0.1(±0.06) | 0.13(±0.06) | ||
| SST range | 0.1(±0.05) | 0.03(±0.01) | 0.33(±0.03) | 0.07(±0.02) |
| Temperature of warmest month | 0.06(±0.05) | 0.06(±0.02) | 0.60(±0.06) | 0.14(±0.01) |
| Mean sea surface salinity | 0.1(±0.03) | 0.07(±0.02) | 0.43(±0.06) | 0.06(±0.06) |
| Distance from shore | 0.2(±0.07) | 0.62(±0.01) | ||
| Bathymetry | 0.2(±0.08) | 0.33(±0.04) | ||
| Ulva lactuca species distribution | 0.61(±0.09) | 0.19(±0.04) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Mahony, J.; de la Torre Cerro, R.; Holloway, P. Modelling the Distribution of the Red Macroalgae Asparagopsis to Support Sustainable Aquaculture Development. AgriEngineering 2021, 3, 251-265. https://doi.org/10.3390/agriengineering3020017
O’Mahony J, de la Torre Cerro R, Holloway P. Modelling the Distribution of the Red Macroalgae Asparagopsis to Support Sustainable Aquaculture Development. AgriEngineering. 2021; 3(2):251-265. https://doi.org/10.3390/agriengineering3020017
Chicago/Turabian StyleO’Mahony, James, Rubén de la Torre Cerro, and Paul Holloway. 2021. "Modelling the Distribution of the Red Macroalgae Asparagopsis to Support Sustainable Aquaculture Development" AgriEngineering 3, no. 2: 251-265. https://doi.org/10.3390/agriengineering3020017
APA StyleO’Mahony, J., de la Torre Cerro, R., & Holloway, P. (2021). Modelling the Distribution of the Red Macroalgae Asparagopsis to Support Sustainable Aquaculture Development. AgriEngineering, 3(2), 251-265. https://doi.org/10.3390/agriengineering3020017






