Automatic and Reliable Leaf Disease Detection Using Deep Learning Techniques
Abstract
:1. Introduction
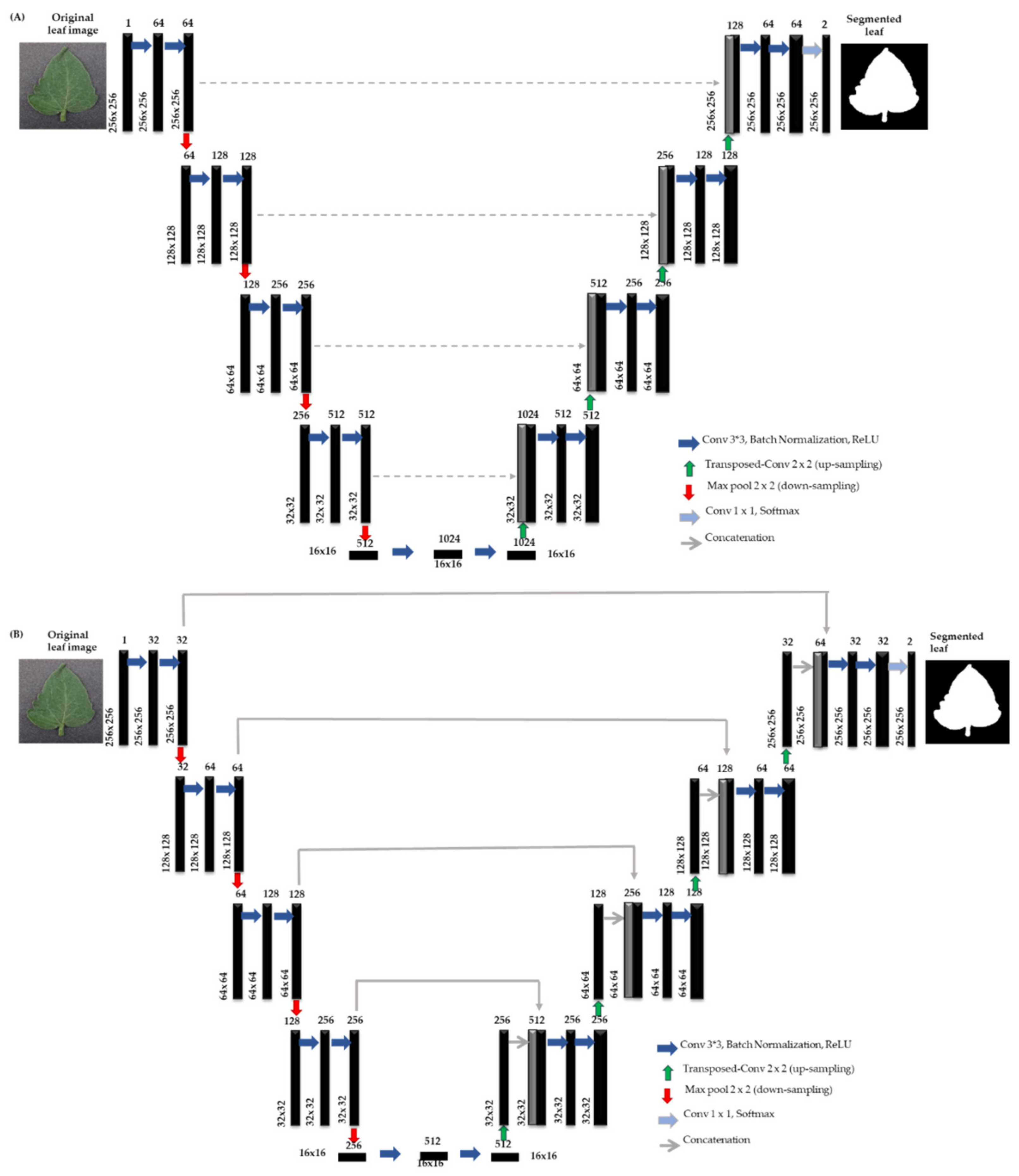
- Different variants of U-net architecture are investigated to propose the best segmentation model by comparing the model predictions to the ground truth segmented images.
- Investigation of the classification tasks for different variants of CNN architecture for binary and different multi-class classifications of tomato diseases. Several experiments employing different CNN architectures were conducted. Three different types of classifications were done in this work: (a) Binary classification of healthy and diseased leaves, (b) Five-class classification of healthy and four diseased leaves, and finally, (c) Ten-class classification with healthy and nine different diseases classes.
- The performance achieved in this work outperforms the existing state-of-the-art works in this domain.
2. Background Study
2.1. Deep Convolutional Neural Networks (CNN)
Width, W = bφ
Resolution, R = cφ
a ≥ 1, b ≥ 1, c ≥ 1
2.2. Segmentation
2.3. Visualization Techniques
2.4. Pathogens of Tomato Leaves
3. Methodology
3.1. Datasets Description
3.2. Preprocessing
3.3. Experiments
3.4. Performance Matrix
4. Results
4.1. Tomato Leaf Segmentation
4.2. Tomato Leaf Disease Classification
4.3. Visualization Using Score-Cam
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chowdhury, M.E.; Khandakar, A.; Ahmed, S.; Al-Khuzaei, F.; Hamdalla, J.; Haque, F.; Mamun, B.I.R.; Ahmed, A.S.; Nasser, A.E. Design, construction and testing of iot based automated indoor vertical hydroponics farming test-bed in Qatar. Sensors 2020, 20, 5637. [Google Scholar] [CrossRef] [PubMed]
- Strange, R.N.; Scott, P.R. Plant disease: A threat to global food security. Annu. Rev. Phytopathol. 2005, 43, 83–116. [Google Scholar] [CrossRef]
- Oerke, E.-C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Touati, F.; Khandakar, A.; Chowdhury, M.E.; Antonio, S., Jr.; Sorino, C.K.; Benhmed, K. Photo-Voltaic (PV) monitoring system, performance analysis and power prediction models in Doha, Qatar. In Renewable Energy; IntechOpen: London, UK, 2020. [Google Scholar]
- Khandakar, A.; Chowdhury, M.E.H.; Kazi, M.K.; Benhmed, K.; Touati, F.; Al-Hitmi, M.; Gonzales, A.S.P., Jr. Machine learning based photovoltaics (PV) power prediction using different environmental parameters of Qatar. Energies 2019, 12, 2782. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, M.H.; Shuzan, M.N.I.; Chowdhury, M.E.; Mahbub, Z.B.; Uddin, M.M.; Khandakar, A.; Mamun, B.I.R. Estimating blood pressure from the photoplethysmogram signal and demographic features using machine learning techniques. Sensors 2020, 20, 3127. [Google Scholar] [CrossRef]
- Chowdhury, M.E.; Khandakar, A.; Alzoubi, K.; Mansoor, S.; Tahir, A.M.; Reaz, M.B.I.; Nasser, A.-E. Real-time smart-digital stethoscope system for heart diseases monitoring. Sensors 2019, 19, 2781. [Google Scholar]
- Rahman, T.; Khandakar, A.; Kadir, M.A.; Islam, K.R.; Islam, K.F.; Mazhar, R.; Tahir, R.; Mohammad, T.I.; Saad, B.A.K.; Mohamed, A.A.; et al. Reliable tuberculosis detection using chest X-ray with deep learning, segmentation and visualization. IEEE Access 2020, 8, 191586–191601. [Google Scholar] [CrossRef]
- Tahir, A.; Qiblawey, Y.; Khandakar, A.; Rahman, T.; Khurshid, U.; Musharavati, F.; Islam, M.T.; Kiranyaz, S.; Chowdhury, M.E.H. Coronavirus: Comparing COVID-19, SARS and MERS in the eyes of AI. arXiv 2020, arXiv:2005.11524. [Google Scholar]
- Chowdhury, M.E.; Rahman, T.; Khandakar, A.; Mazhar, R.; Kadir, M.A.; Mahbub, Z.B.; Atif, I.; Nasser, A.-E.; Khan, M.S.; Islam, K.R. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access 2020, 8, 132665–132676. [Google Scholar] [CrossRef]
- Rahman, T.; Chowdhury, M.E.; Khandakar, A.; Islam, K.R.; Islam, K.F.; Mahbub, Z.B.; Muhammad, A.K.; Saad, K. Transfer learning with deep convolutional neural network (CNN) for pneumonia detection using chest X-ray. Appl. Sci. 2020, 10, 3233. [Google Scholar] [CrossRef]
- Chowdhury, M.E.; Rahman, T.; Khandakar, A.; Al-Madeed, S.; Zughaier, S.M.; Hassen, H.; Mohammad, T.I. An early warning tool for predicting mortality risk of COVID-19 patients using machine learning. arXiv 2020, arXiv:2007.15559. [Google Scholar]
- Chouhan, S.S.; Kaul, A.; Singh, U.P.; Jain, S. Bacterial foraging optimization based radial basis function neural network (BRBFNN) for identification and classification of plant leaf diseases: An automatic approach towards plant pathology. IEEE Access 2018, 6, 8852–8863. [Google Scholar] [CrossRef]
- LeCun, Y.; Haffner, P.; Bottou, L.; Bengio, Y. Object recognition with gradient-based learning. In Shape, Contour and Grouping in Computer Vision; Springer: Berlin/Heidelberg, Germany, 1999; pp. 319–345. [Google Scholar]
- Arya, S.; Singh, R. A Comparative Study of CNN and AlexNet for Detection of Disease in Potato and Mango leaf. In Proceedings of the 2019 International Conference on Issues and Challenges in Intelligent Computing Techniques (ICICT), Ghaziabad, India, 27–28 September 2019; pp. 1–6. [Google Scholar]
- Wang, G.; Sun, Y.; Wang, J. Automatic image-based plant disease severity estimation using deep learning. Comput. Intell. Neurosci. 2017, 2017, 2917536. [Google Scholar] [CrossRef] [Green Version]
- Amara, J.; Bouaziz, B.; Algergawy, A. A deep learning-based approach for banana leaf diseases classification. In Proceedings of the Datenbanksysteme für Business, Technologie und Web (BTW 2017)—Workshopband, Stuttgart, Germany, 6–10 March 2017. [Google Scholar]
- Statistics, F. Food and Agriculture Organization of the United Nations. Retrieved 2010, 3, 2012. [Google Scholar]
- Adeoye, I.; Aderibigbe, O.; Amao, I.; Egbekunle, F.; Bala, I. Tomato Products’market Potential and Consumer Preference In Ibadan, Nigeria. Sci. Pap. Manag. Econ. Eng. Agric. Rural Dev. 2017, 17, 9–15. [Google Scholar]
- Kaur, M.; Bhatia, R. Of An Improved Tomato Leaf Disease Detection And Classification Method. In Proceedings of the IEEE Conference on Information and Communication Technology, Allahabad, India, 6–8 December 2019; pp. 1–5. [Google Scholar]
- Rahman, M.A.; Islam, M.M.; Mahdee, G.S.; Kabir, M.W.U. Improved Segmentation Approach for Plant Disease Detection. In Proceedings of the 1st International Conference on Advances in Science, Engineering and Robotics Technology (ICASERT), Dhaka, Bangladesh, 3–5 May 2019; pp. 1–5. [Google Scholar]
- Fuentes, A.; Yoon, S.; Kim, S.C.; Park, D.S. A robust deep-learning-based detector for real-time tomato plant diseases and pests recognition. Sensors 2017, 17, 2022. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, M.; Singh, A.; Arjaria, S.; Sinha, A.; Gupta, S. ToLeD: Tomato leaf disease detection using convolution neural network. Procedia Comput. Sci. 2020, 167, 293–301. [Google Scholar] [CrossRef]
- Durmuş, H.; Güneş, E.O.; Kırcı, M. Disease detection on the leaves of the tomato plants by using deep learning. In Proceedings of the 6th International Conference on Agro-Geoinformatics, Fairfax, VA, USA, 7–10 August 2017; pp. 1–5. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, A.T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019; pp. 6105–6114. [Google Scholar]
- Tan, M.; Chen, B.; Pang, R.; Vasudevan, V.; Sandler, M.; Howard, A.; Le, Q.V. MnasnetPlatform-aware neural architecture search for mobile. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–21 June 2019; 2019; pp. 2820–2828. [Google Scholar]
- Howard, A.; Zhmoginov, A.; Chen, L.-C.; Sandler, M.; Zhu, M. Inverted Residuals and Linear Bottlenecks: Mobile Networks for Classification, Detection and Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Huang, Y.; Cheng, Y.; Bapna, A.; Firat, O.; Chen, M.X.; Chen, D.; HyoukJoong, L.; Jiquan, N.; Quoc, V.L.; Yonghui, W.; et al. Gpipe: Efficient training of giant neural networks using pipeline parallelism. arXiv 2018, arXiv:1811.06965. [Google Scholar]
- Lung-Segmentation-2d. Available online: https://github.com/imlab-uiip/lung-segmentation-2d#readme (accessed on 1 August 2020).
- Smilkov, D.; Thorat, N.; Kim, B.; Viégas, F.; Wattenberg, M. Smoothgrad: Removing noise by adding noise. arXiv 2017, arXiv:1706.03825. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Chattopadhay, A.; Sarkar, A.; Howlader, P.; Balasubramanian, V.N. Grad-cam++: Generalized gradient-based visual explanations for deep convolutional networks. In Proceedings of the IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; pp. 839–847. [Google Scholar]
- Wang, H.; Wang, Z.; Du, M.; Yang, F.; Zhang, Z.; Ding, S.; Mardziel, P.; Hu, X. Score-CAM: Score-weighted visual explanations for convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 14–19 June 2020; pp. 24–25. [Google Scholar]
- Chaerani, R.; Voorrips, R.E. Tomato early blight (Alternaria solani): The pathogen, genetics, and breeding for resistance. J. Gen. Plant Pathol. 2006, 72, 335–347. [Google Scholar] [CrossRef]
- Wu, Q.; Chen, Y.; Meng, J. DCGAN-based data augmentation for tomato leaf disease identification. IEEE Access 2020, 8, 98716–98728. [Google Scholar] [CrossRef]
- Cabral, R.N.; Marouelli, W.A.; Lage, D.A.; Café-Filho, A.C. Septoria leaf spot in organic tomatoes under diverse irrigation systems and water management strategies. Hortic. Bras. 2013, 31, 392–400. [Google Scholar] [CrossRef] [Green Version]
- Café-Filho, A.C.; Lopes, C.A.; Rossato, M. Management of plant disease epidemics with irrigation practices. In Irrigation in Agroecosystems; Books on Demand: Norderstedt, Germany, 2019; p. 123. [Google Scholar]
- Zou, J.; Rodriguez-Zas, S.; Aldea, M.; Li, M.; Zhu, J.; Gonzalez, D.O.; Lila, O.V.; DeLucia, E.; Steven, J.C. Expression profiling soybean response to Pseudomonas syringae reveals new defense-related genes and rapid HR-specific downregulation of photosynthesis. Mol. Plant. Microbe Interact. 2005, 18, 1161–1174. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Chen, T.; Zhang, Z.; Li, B.; Tian, S. Roles of Aquaporins in Plant-Pathogen Interaction. Plants 2020, 9, 1134. [Google Scholar] [CrossRef] [PubMed]
- Schlub, R.; Smith, L.; Datnoff, L.; Pernezny, K. An overview of target spot of tomato caused by Corynespora cassiicola. In Proceedings of the II International Symposium on Tomato Diseases 808, Kusadasi, Turkey, 8 October 2007; pp. 25–28. [Google Scholar]
- Zhu, J.; Zhang, L.; Li, H.; Gao, Y.; Mu, W.; Liu, F. Development of a LAMP method for detecting the N75S mutant in SDHI-resistant Corynespora cassiicol. Anal. Biochem. 2020, 597, 113687. [Google Scholar] [CrossRef] [PubMed]
- Pernezny, K.; Stoffella, P.; Collins, J.; Carroll, A.; Beaney, A. Control of target spot of tomato with fungicides, systemic acquired resistance activators, and a biocontrol agent. Plant. Prot. Sci. Prague 2002, 38, 81–88. [Google Scholar] [CrossRef] [Green Version]
- Abdulridha, J.; Ampatzidis, Y.; Kakarla, S.C.; Roberts, P. Detection of target spot and bacterial spot diseases in tomato using UAV-based and benchtop-based hyperspectral imaging techniques. Precis. Agric. 2020, 21, 955–978. [Google Scholar] [CrossRef]
- De Jong, C.F.; Takken, F.L.; Cai, X.; de Wit, P.J.; Joosten, M.H. Attenuation of Cf-mediated defense responses at elevated temperatures correlates with a decrease in elicitor-binding sites. Mol. Plant. Microbe Interact. 2002, 15, 1040–1049. [Google Scholar] [CrossRef] [Green Version]
- Calleja-Cabrera, J.; Boter, M.; Oñate-Sánchez, L.; Pernas, M. Root growth adaptation to climate change in crops. Front. Plant. Sci. 2020, 11, 544. [Google Scholar] [CrossRef]
- Louws, F.; Wilson, M.; Campbell, H.; Cuppels, D.; Jones, J.; Shoemaker, P.; Sahin, F.; Miller, S. A Field control of bacterial spot and bacterial speck of tomato using a plant activator. Plant. Dis. 2001, 85, 481–488. [Google Scholar] [CrossRef] [Green Version]
- Qiao, K.; Liu, Q.; Huang, Y.; Xia, Y.; Zhang, S. Management of bacterial spot of tomato caused by copper-resistant Xanthomonas perforans using a small molecule compound carvacrol. Crop. Prot. 2020, 132, 105114. [Google Scholar] [CrossRef]
- Nowicki, M.; Foolad, M.R.; Nowakowska, M.; Kozik, E.U. Potato and tomato late blight caused by Phytophthora infestans: An overview of pathology and resistance breeding. Plant. Dis. 2012, 96, 4–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buziashvili, A.; Cherednichenko, L.; Kropyvko, S.; Yemets, A. Transgenic tomato lines expressing human lactoferrin show increased resistance to bacterial and fungal pathogens. Biocatal. Agric. Biotechnol. 2020, 25, 101602. [Google Scholar] [CrossRef]
- Glick, E.; Levy, Y.; Gafni, Y. The viral etiology of tomato yellow leaf curl disease–a review. Plant. Prot. Sci. 2009, 45, 81–97. [Google Scholar] [CrossRef] [Green Version]
- Dhaliwal, M.; Jindal, S.; Sharma, A.; Prasanna, H. Tomato yellow leaf curl virus disease of tomato and its management through resistance breeding: A review. J. Hortic. Sci. Biotechnol. 2020, 95, 425–444. [Google Scholar] [CrossRef]
- Ghanim, M.; Czosnek, H. Tomato yellow leaf curl geminivirus (TYLCV-Is) is transmitted among whiteflies (Bemisia tabaci) in a sex-related manner. J. Virol. 2000, 74, 4738–4745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghanim, M.; Morin, S.; Zeidan, M.; Czosnek, H. Evidence for transovarial transmission of tomato yellow leaf curl virus by its vector, the whiteflyBemisia tabaci. Virology 1998, 240, 295–303. [Google Scholar] [CrossRef] [Green Version]
- He, Y.-Z.; Wang, Y.-M.; Yin, T.-Y.; Fiallo-Olivé, E.; Liu, Y.-Q.; Hanley-Bowdoin, L.; Xiao-Wei, W. A plant DNA virus replicates in the salivary glands of its insect vector via recruitment of host DNA synthesis machinery. Proc. Natl. Acad. Sci. USA 2020, 117, 16928–16937. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Jo, Y.; Cho, W.K.; Yu, J.; Tran, P.-T.; Salaipeth, L.; Hae-Ryun, K.; Hong-Soo, C.; Kook-Hyung, K. Identification of Viruses and Viroids Infecting Tomato and Pepper Plants in Vietnam by Metatranscriptomics. Int. J. Mol. Sci. 2020, 21, 7565. [Google Scholar] [CrossRef]
- Broadbent, L. Epidemiology and control of tomato mosaic virus. Annu. Rev. Phytopathol. 1976, 14, 75–96. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, S.; Shen, J.; Wu, Z.; Du, Z.; Gao, F. The phylogeographic history of tomato mosaic virus in Eurasia. Virology 2021, 554, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- SpMohanty/PlantVillage-Dataset. Available online: https://github.com/spMohanty/PlantVillage-Dataset (accessed on 24 January 2021).
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Daniel, R. Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 2019, 53, 197–207. [Google Scholar] [CrossRef]
- Tm, P.; Pranathi, A.; SaiAshritha, K.; Chittaragi, N.B.; Koolagudi, S.G. Tomato leaf disease detection using convolutional neural networks. In Proceedings of the 2018 Eleventh International Conference on Contemporary Computing (IC3), Surat, India, 7–8 February 2020; pp. 1–5. [Google Scholar]
- Zhang, K.; Wu, Q.; Liu, A.; Meng, X. Can deep learning identify tomato leaf disease? Adv. Multimed. 2018, 2018, 6710865. [Google Scholar] [CrossRef] [Green Version]
- Dookie, M.; Ali, O.; Ramsubhag, A.; Jayaraman, J. Flowering gene regulation in tomato plants treated with brown seaweed extracts. Sci. Hortic. 2021, 276, 109715. [Google Scholar] [CrossRef]








| Stage | Operator | Resolution | Channels | Layers |
|---|---|---|---|---|
| 1 | Conv3 × 3 | 224 × 224 | 32 | 1 |
| 2 | MBConv 1, k3 × 3 | 112 × 112 | 16 | 1 |
| 3 | MBConv 6, k3 × 3 | 112 × 112 | 24 | 2 |
| 4 | MBConv 6, k5 × 5 | 56 × 56 | 40 | 2 |
| 5 | MBConv 6, k3 × 3 | 28 × 28 | 80 | 3 |
| 6 | MBConv 6, k5 × 5 | 14 × 14 | 112 | 3 |
| 7 | MBConv 6, k5 × 5 | 14 × 14 | 192 | 4 |
| 8 | MBConv 6, k3 × 3 | 7 × 7 | 320 | 1 |
| 9 | Conv1 × 1 & Pooling & FC | 7 × 7 | 1280 | 1 |
| Class | Healthy | Fungi | Bacteria | Mold | Virus | Mite |
|---|---|---|---|---|---|---|
| Sub Class | Healthy (1591) | Early blight (1000) | Bacterial spot (2127) | Late bright mold (1910) | Tomato Yellow Leaf Curl Virus (5357) | Two-spotted spider mite (1676) |
| Septoria leaf spot (1771) | ||||||
| Tomato Mosaic Virus (373) | ||||||
| Target spot (1404) | ||||||
| Leaf mold (952) | ||||||
| Tomato (18,161) | ||||||
| Dataset | Number of Tomato Leaf Images and Their Corresponding Mask | Train Set Count/Fold | Validation Set Count/Fold | Test Set Count/Fold |
|---|---|---|---|---|
| Plant Village tomato leaf images | 18161 | 13076 | 1453 | 3632 |
| Classification | Types | Total No. of Images/Class | For Both Segmented and Unsegmented Experiment | ||
|---|---|---|---|---|---|
| Train Set Count/Fold | Validation Set Count/Fold | Test Set Count/Fold | |||
| Binary-class | Healthy | 1591 | 1147 × 10 = 11470 | 127 | 317 |
| Unhealthy (9 diseases) | 16,570 | 11930 | 1326 | 3314 | |
| Six-class | Healthy | 1591 | 1147 × 3 = 3441 | 127 | 317 |
| Fungi | 5127 | 3692 | 410 | 1025 | |
| Bacteria | 2127 | 1532 × 2 = 3064 | 170 | 425 | |
| Mold | 1910 | 1375 × 3 = 4125 | 153 | 382 | |
| Virus | 5730 | 4126 | 458 | 1146 | |
| Mite | 1676 | 1207 × 3 = 3621 | 134 | 335 | |
| Ten-class | Healthy | 1591 | 1147 × 3 = 3441 | 127 | 317 |
| Early Blight | 1000 | 720 × 5 = 3600 | 80 | 200 | |
| Septoria Leaf Spot | 1771 | 1275 × 3 = 3825 | 142 | 354 | |
| Target Spot | 1404 | 1011 × 3 = 3033 | 112 | 281 | |
| Leaf Mold | 952 | 686 × 5 = 3430 | 76 | 190 | |
| Bacterial Spot | 2127 | 1532 × 2 = 3064 | 170 | 425 | |
| Late Bright Mold | 1910 | 1375 × 3 = 4125 | 153 | 382 | |
| Tomato Yellow Leaf Curl Virus | 5357 | 3857 | 429 | 1071 | |
| Tomato Mosaic Virus | 373 | 268 × 13 = 3484 | 30 | 75 | |
| Parameters | Segmentation Model | Classification Model |
|---|---|---|
| Batch size | 16 | 16 |
| Learning rate | 0.001 | 0.001 |
| Epochs | 50 | 15 |
| Epochs patience | 8 | 6 |
| Stopping criteria | 8 | 5 |
| Loss function | NLL/BCE/MSE | BCE |
| Optimizer | ADAM | ADAM |
| Loss Function | Network | Test Loss | Test Accuracy | IoU | Dice | Inference Time T (s) |
|---|---|---|---|---|---|---|
| NLL loss | Unet | 0.0168 | 97.25 | 96.83 | 97.11 | 14.05 |
| BCE loss | Unet | 0.0162 | 97.32 | 96.9 | 97.02 | 13.89 |
| MSE loss | Unet | 0.0134 | 97.52 | 97.25 | 97.35 | 13.66 |
| NLL loss | Modified Unet | 0.0076 | 98.66 | 98.5 | 98.73 | 12.12 |
| BCE loss | Modified Unet | 0.016 | 97.12 | 96.82 | 97.1 | 12.04 |
| MSE loss | Modified Unet | 0.089 | 98.19 | 98.25 | 98.43 | 11.76 |
| Classification Scheme | Models | Result with 95% CI | |||||
|---|---|---|---|---|---|---|---|
| Overall | Weighted | ||||||
| Accuracy | Precision | Sensitivity | F1-Score | Specificity | Inference Time (T) | ||
| 2 Class | EfficientNet-b0 | 99.74 ± 0.07 | 99.75 ± 0.07 | 99.73 ± 0.08 | 99.73 ± 0.08 | 99.75 ± 0.07 | 19.32 |
| EfficientNet-b4 | 99.82 ± 0.06 | 99.83 ± 0.06 | 99.82 ± 0.06 | 99.82 ± 0.06 | 98.74 ± 0.16 | 34.25 | |
| EfficientNet-b7 | 99.95 ± 0.03 | 99.94 ± 0.03 | 99.95 ± 0.03 | 99.95 ± 0.03 | 99.77 ± 0.07 | 44.12 | |
| 6 Class | EfficientNet-b0 | 97.34 ± 0.23 | 97.38 ± 0.23 | 97.34 ± 0.23 | 97.33 ± 0.23 | 99.47 ± 0.11 | 20.45 |
| EfficientNet-b4 | 98.49 ± 0.18 | 98.51 ± 0.18 | 98.49 ± 0.18 | 98.49 ± 0.18 | 99.73 ± 0.08 | 38.02 | |
| EfficientNet-b7 | 99.12 ± 0.14 | 99.1 ± 0.14 | 99.11± 0.14 | 99.1 ± 0.14 | 99.81 ± 0.06 | 45.18 | |
| 10 Class | EfficientNet-b0 | 99.71 ± 0.08 | 98.69 ± 0.17 | 98.68 ± 0.17 | 98.68 ± 0.17 | 99.87 ± 0.05 | 22.16 |
| EfficientNet-b4 | 99.89 ± 0.05 | 99.45 ± 0.11 | 99.44 ± 0.11 | 99.4 ± 0.11 | 99.94 ± 0.04 | 41.24 | |
| EfficientNet-b7 | 99.84 ± 0.06 | 99.15 ± 0.13 | 99.13 ± 0.14 | 99.13 ± 0.14 | 99.92 ± 0.04 | 51.23 | |
| Paper | Classification | Dataset | Accuracy | Precision | Recall | F1-Score | Results |
|---|---|---|---|---|---|---|---|
| Mohit et al. [23] | Ten-class | Plant Village | 91% | 90% | 92% | 91% | Non-Segmented |
| P. Tm et al. [63] | Ten-class | Plant Village | 94% | 94.81% | 94.78% | 94.8% | Segmented |
| Keke et al. [64] | Two-class | Own dataset | 95% | - | - | - | Non-segmented |
| Madhavi et al. [65] | Two-class | Own dataset | 85% | - | 84% | - | Non-Segmented -- |
| Proposed study | Two-class | Plant Village | 99.95% | 99.94% | 99.95% | 99.95% | Segmented |
| Six-class | Plant Village | 99.12% | 99.10% | 99.11% | 99.10% | Segmented | |
| Ten-class | Plant Village | 99.89% | 99.45% | 99.44% | 99.4% | Segmented |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chowdhury, M.E.H.; Rahman, T.; Khandakar, A.; Ayari, M.A.; Khan, A.U.; Khan, M.S.; Al-Emadi, N.; Reaz, M.B.I.; Islam, M.T.; Ali, S.H.M. Automatic and Reliable Leaf Disease Detection Using Deep Learning Techniques. AgriEngineering 2021, 3, 294-312. https://doi.org/10.3390/agriengineering3020020
Chowdhury MEH, Rahman T, Khandakar A, Ayari MA, Khan AU, Khan MS, Al-Emadi N, Reaz MBI, Islam MT, Ali SHM. Automatic and Reliable Leaf Disease Detection Using Deep Learning Techniques. AgriEngineering. 2021; 3(2):294-312. https://doi.org/10.3390/agriengineering3020020
Chicago/Turabian StyleChowdhury, Muhammad E. H., Tawsifur Rahman, Amith Khandakar, Mohamed Arselene Ayari, Aftab Ullah Khan, Muhammad Salman Khan, Nasser Al-Emadi, Mamun Bin Ibne Reaz, Mohammad Tariqul Islam, and Sawal Hamid Md Ali. 2021. "Automatic and Reliable Leaf Disease Detection Using Deep Learning Techniques" AgriEngineering 3, no. 2: 294-312. https://doi.org/10.3390/agriengineering3020020
APA StyleChowdhury, M. E. H., Rahman, T., Khandakar, A., Ayari, M. A., Khan, A. U., Khan, M. S., Al-Emadi, N., Reaz, M. B. I., Islam, M. T., & Ali, S. H. M. (2021). Automatic and Reliable Leaf Disease Detection Using Deep Learning Techniques. AgriEngineering, 3(2), 294-312. https://doi.org/10.3390/agriengineering3020020







