Abstract
Crown rot is one of the major stubble soil fungal diseases that bring significant yield loss to the cereal industry. The most effective crown rot management approach is removal of infected crop residue from fields and rotation of nonhost crops. However, disease screening is challenging as there are no clear visible symptoms on upper stems and leaves at early growth stages. The current manual screening method requires experts to observe the crown and roots of plants to detect disease, which is time-consuming, subjective, labor-intensive, and costly. As digital color imaging has the advantages of low cost and easy use, it has a high potential to be an economical solution for crown rot detection. In this research, a crown rot disease detection method was developed using a smartphone camera and machine learning technologies. Four common wheat varieties were grown in greenhouse conditions with a controlled environment, and all infected group plants were infected with crown rot without the presence of other plant diseases. We used a smartphone to take digital color images of the lower stems of plants. Using imaging processing techniques and a support vector machine algorithm, we successfully distinguished infected and healthy plants as early as 14 days after disease infection. The results provide a vital first step toward developing a digital color imaging phenotyping platform for crown rot detection to enable the management of crown rot disease effectively. As an easy-access phenotyping method, this method could provide support for researchers to develop an efficiency and economic disease screening method in field conditions.
1. Introduction
Crown rot is a stubble soil fungal disease primarily caused by Fusarium pseudograminearum and threatens cereal crop production worldwide. The Australian wheat industry suffered from economic losses of approximately AUD 80 million due to crown rot in 2009, and potential losses may reach AUD 434 million per year [1,2]. Crown rot contributes to failure of grain/seed establishment and causes prematurely senescing culms, which can cause up to 59% yield loss in wheat [3]. There is currently no effective biological or chemical control method that can effectively control crown rot disease, and there are no varieties resistant to it [4,5]. Currently, the most effective disease management solution is reducing disease inoculum levels in the field by growing nonhost rotation crops for at least 3 years [6]. Disease screening can identify the disease infection region and the severity of disease infection to help farmers adjust planting plans for minimizing the disease inoculum level in the field [7]. Therefore, disease detection is extremely important for the management of crown rot. However, it is difficult for farmers to detect crown rot at an early stage in the field as infected wheat plants do not show obvious symptoms on upper stems and leaves. At this stage, the symptoms are usually not obvious, because the major symptom of crown rot in early growth stages is brown discoloration of the stem close to the soil surface [8,9,10]. Furthermore, it can be challenging to screen the crop in a field by observing the lower stems of wheat in different growing regions many times, as it is labor-intensive, time-consuming, and costly.
Recently, computer vision technologies have shown great advantages in agricultural applications, from precision agriculture to imaging plant phenotyping [11,12]. In imaging-based crop phenotyping, research directions can be roughly grouped into two categories: hyperspectral imaging and digital color imaging [13]. Hyperspectral imaging includes both the visible light spectrum and the infrared range, and it can provide unique spectral characteristics of the target plant in the electromagnetic spectrum for understanding the growth status of the plant [14,15]. For example, near-infrared imaging can detect changes in reflectance at 680–730 nm in the epidermis and plant cell wall due to Fusarium infection [16,17]. This occurs when plants are invaded by fungi that secrete the toxic mycotoxin deoxynivalenol, which destroys the chloroplasts of the plants, thereby limiting the photosynthetic activity of the plants [18,19]. In addition, F. pseudograminearum can colonize a large part of a plant’s vascular system and limit the water transmission of plants [20]. This leads to water loss in infected plants detected by a change in 682–733 nm and 927–931 nm wavelengths after growing for 70 days [7]. Although the above studies used hyperspectral image technology to detect Fusarium-infected plants, the plants were close to maturity, which is not practical for the detection of the disease in early growth stages. In addition, hyperspectral cameras are expensive and hardly affordable for use by plant researchers or farmers. Therefore, using digital color imaging for the early detection of crown rot presents a viable alternative since digital color camera technology has the advantages of being low-cost, easy to access, and simple to operate [14,15]. Color imaging can successfully distinguish Fusarium head blight-infected plants from healthy plants in the early maturity stage of wheat by imaging wheat head [21]. However, this study is also limited by late detection of Fusarium disease. This research aims to develop a computer vision method using digital color images to detect crown rot disease in wheat at an early stage. We developed an algorithm to distinguish infected wheat from healthy wheat under greenhouse conditions. Crown rot is a disease that is greatly affected by environmental factors [22]. Compared with the complex environment in the field, the relatively controllable environment provided by the greenhouse can help researchers understand the development of crown rot disease and evaluate different wheat varieties’ typical phenotypic data and symptoms after being infected with disease at the seedlings stage [23,24,25]. We found that crown rot can be detected from the stems near the ground using the developed computer vision method after 14 days of infection. This research provides a basis for developing a ground-based robotic system that can detect crown rot disease at an early stage in a high-throughput, accurate, economic, and nondestructive manner.
2. Materials and Methods
2.1. Wheat Plant Growth and Disease Infection
Four common Australian wheat varieties were chosen based on their susceptibility to crown rot, including Aurora, which is durum wheat and most susceptible to crown rot; Emu Rock, which is susceptible to crown rot; and Yitpi and Trojan, which are moderately susceptible [26]. All wheat plants were planted in a greenhouse located in the Plant Accelerator® at the Waite Campus of the University of Adelaide (latitude and longitude are 34°58′16.4″ S and 138°38′23.5″ E, shown in Figure S1 in Supplementary Materials). The plants were grown in a soil substrate under two treatments of “controlled” and “infected” with 26 replicates, resulting in a total of 208 plants for analysis. All wheat was planted in a 150 mm common white plastic pot, where the growing medium was a 50:50 mixture of UC Davis mix [27] and cocopeat. To avoid spatial bias, a random block design was utilized based on the dae program, which is a package for the R statistical computing environment [28]. Moreover, plants were grown under 12 h of natural light, supplemented with light-emitting diode lights from 7:30 a.m. to 5:30 p.m., and grown at 22 °C (day) and 15 °C (night). All seeds were sown on 21 June 2021 and grew in the greenhouse until disease infection. Three seeds were sown in each pot to ensure an even germination rate, and two of the seedlings were removed after 14 days. The removal principle was to only keep one representative seedling per pot. The disease infection occurred at 14 days after sowing (defined as first day after infection—1 DAI), and the infection method was developed based on a previous study [29]. We placed 10 F. pseudograminearum-infected seeds (courtesy of colleagues at the South Australian Research and Development Institute) near seedlings and also used a PVC tube (30 mm diameter and 20 mm height) to reduce the movement of infected seeds. The plants were watered daily to set a target value at 30% of soil weight.
2.2. Manual Screening
In this experiment, a manual assessment was taken by recording the disease severity levels to understand the disease development at an early infection period. The manual assessment protocol was modified from a previous study [24], and the manual scores presented crown rot severity levels: 0 represents healthy plant, 1 represents the first leaf sheath showing necrotic lesions, 2 represents the first leaf sheath and the subcrown internode showing occurring brown discoloration symptoms, 3 represents partial second leaf sheath showing necrotic lesions, 4 represents brown discoloration symptoms on the second leaf sheath and the subnodes of the subcorona, and 5 and 6 describe the necrosis of the leaf sheath. The manual assessment was taken three times on 14, 21, and 28 DAI. A pathogen isolation for pathogen identification occurred on 43 DAI when a plant was randomly sampled from each treatment and variety for pathogen isolation. The plant’s crown was excised from the lower stems and placed in plates for pathogen growth for 5 days and then identified by observation under microscope [30].
2.3. Image Collection
The images were collected on 14 and 21 DAI using an iPhone XS (64 GB) between 1:00 p.m. and 3:00 p.m. The lighting was provided by a combination of LED light and natural indoor light. The photos were acquired at approximately 10 cm between the phone and wheat, focusing on the low stem regions, including the first node. All images were saved in JPEG (Joint Photographic Experts Group) format for future analysis. On 14 DAI, 60 images were randomly collected from the controlled and infected group. The disease levels of the manual assessment of the infected wheat were between 0 and 2. On 21 DAI, 60 images from the infected group were collected, and the disease levels of the infected wheat covered a large range from 0 to 6. As all of the plants in the controlled group had the same disease level of 0, only 30 images of the controlled group were collected.
2.4. Image Processing and Analysis
2.4.1. Background Segmentation
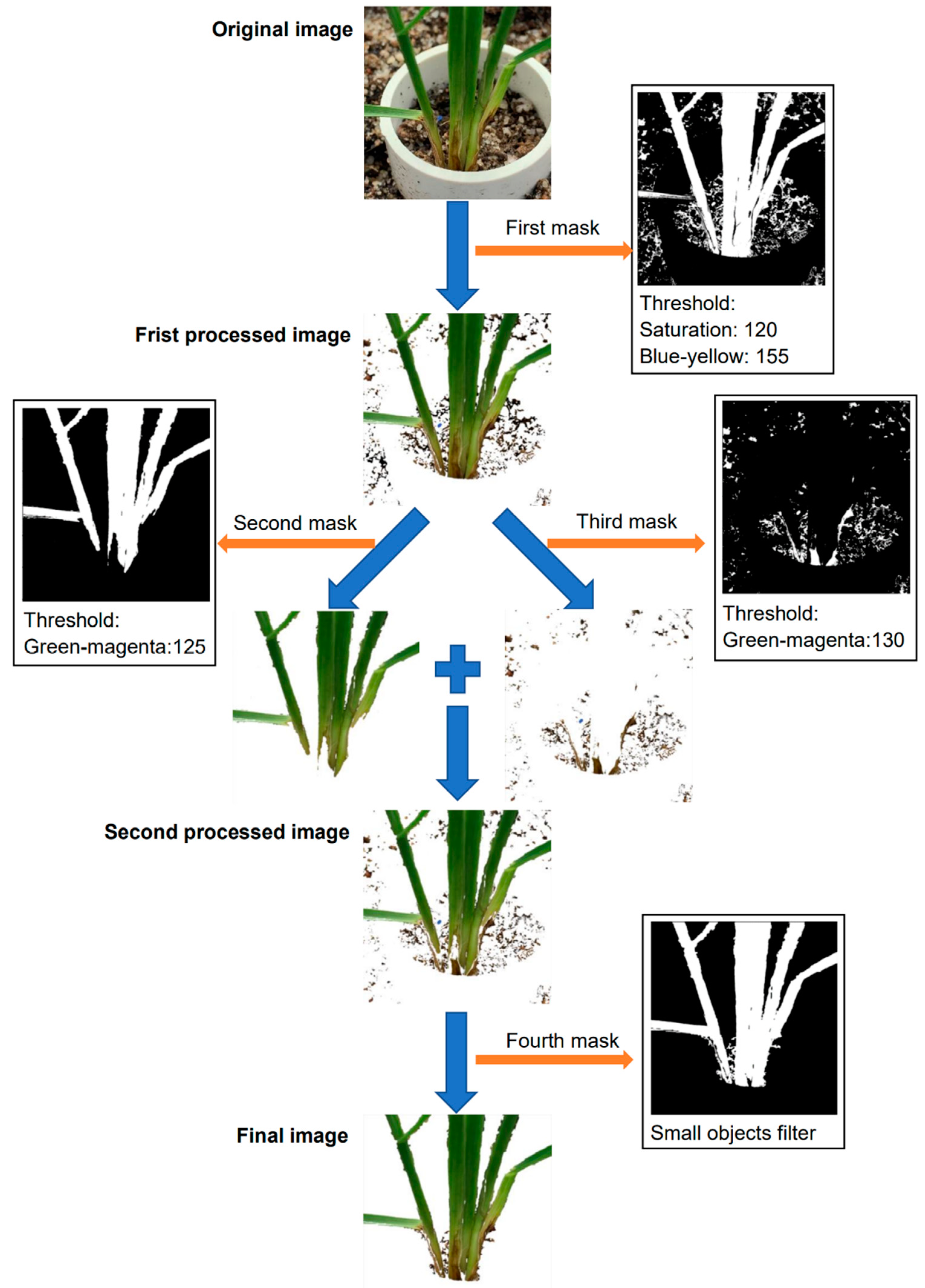
The image segmentation was conducted using Plant Computer Vision, which is a Python package [31]. In background segmentation, all of the original RGB (red, green, blue) images were converted into the HSV (hue, saturation, value) and LAB (lightness, greens and magentas channel, blues and yellows channel) color spaces, and then the color features were used to filter out backgrounds while maintaining regions of interest (ROIs). After an original image was converted into an HSV and LAB color space, four masks were applied to filter out unnecessary parts of the image in Figure 1. The purpose of the first mask is to remove most of the background pixels and maintain the plant. The grayscale images of saturation and blue-yellow were binarized and then combined to form the first mask, where the saturation threshold was 120 and the blue-yellow threshold was 155, which were determined by experiment. The first mask filtered out most pixels of the soil area and the PVC tube (Figure 1) but had a limitation to remove the cocopeat particles. As the healthy areas were close to the green color, and the infected areas were close to the brown color, two masks were developed to further extract the plant region, one for extracting the healthy areas and the other for the infected areas. Both of the masks used grayscale images of green-magenta, and the thresholds were set to 125 and 130 for the second and third masks, respectively. In the experiment, we found that if only one of the masks was applied, it was impossible to extract the complete part of the plant, including the brown discoloration region and healthy section of the plants (Figure 1). The final mask was to eliminate the small objects in the backgrounds, such as small soil particles. Notably, all the images were processed with the same method using the same masks and the same threshold values.
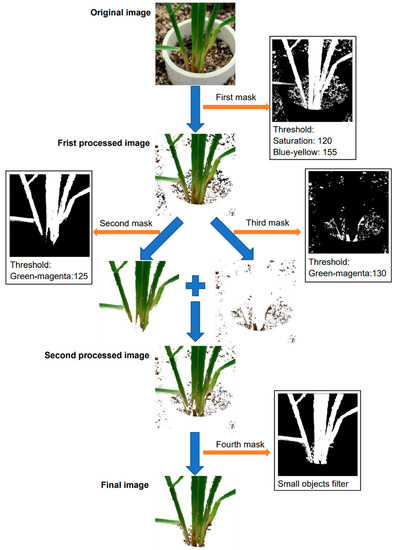
Figure 1.
Workflow for image segmentation process. Image segmentation occurred by applying four different masks in sequence to the original image to extract the ROI and produce the final image.
2.4.2. Disease Detection
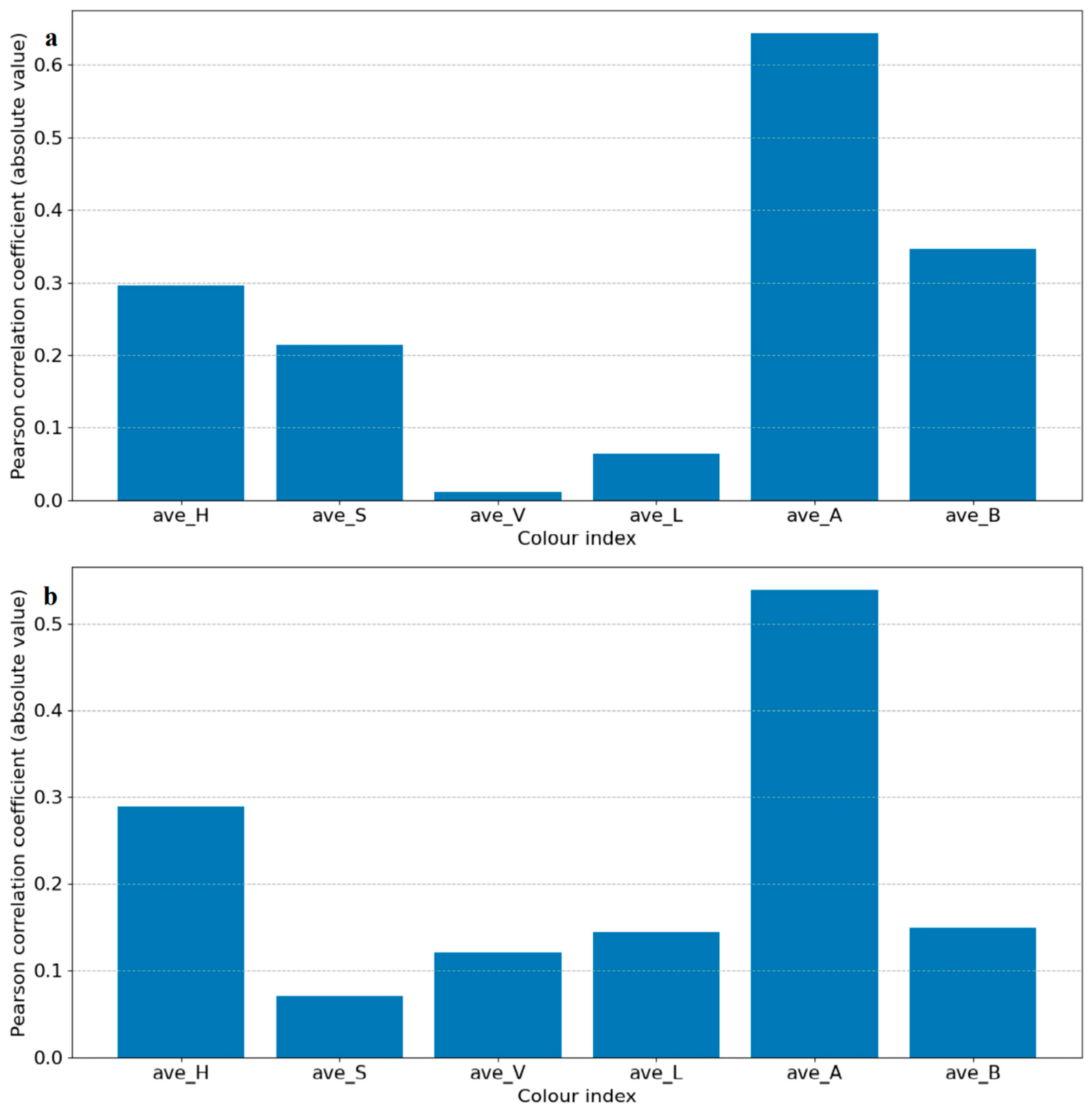
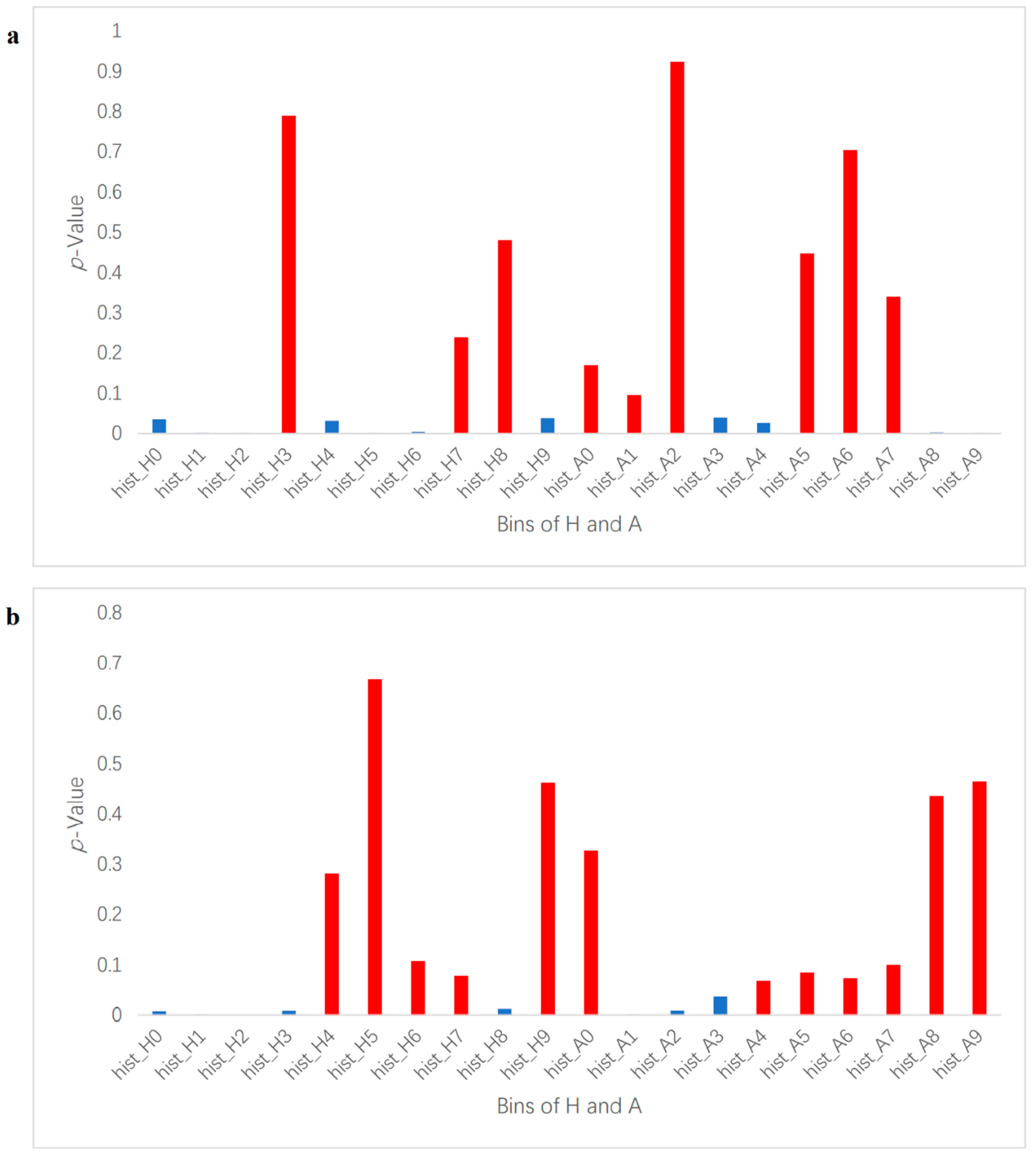
In this study, we used a machine learning method to perform binary classification of infected plants and controlled plants. If a plant was correctly classified into the corresponding class, then the detection was considered successful. Each segmented image was transformed from an RGB color space to an HSV and LAB color space, and then each image’s average values of H, S, V, L, A, and B were used to identify the degree of correlation between the color indices and the infection status (control and infected) by Pearson correlation coefficient analysis. Select color indexes whose absolute correlation values were the highest for further classification in HSV and LAB, respectively, and H in HSV and A in LAB were chosen in this case, as shown in Figure 2. To extract feature vectors, the pixels of ROI in the images of H and A were converted to normalized histograms of 10 bins with values in the range of 0–1 (examples shown in Figure S2). The histograms represent the distributions of the colors of the plants and were considered as the initial color feature vectors. The initial color feature vectors were further processed so that only the bins made significant contributions to further classification, which were kept to make it easier to establish a training model to complete the classification task. By conducting an analysis of variance of each bin of the histograms and the disease infection levels, only bins with p-values smaller than 0.05 were kept (Figure 2). It is worth noting that in this experiment, two groups of photos (14 DAI and 21 DAI) were used for analysis. The two different data collection dates represent different plant growth and infection stages, so bins whose p-values were smaller than 0.05 at different infection and growth stages were different. Therefore, the analysis of variance of histograms was divided into 14 DAI and 21 DAI dataset for analysis. The bins to be removed for 14 DAI were hist_h3, 7, 8 and hist_a1, 2, 5, 6, 7, while for 21 DAI hist_h4, 5, 6, 9 and hist_a0, 4, 5, 6, 7, 8, 9, as shown in red bars in Figure 3. The final feature vectors were input into a support vector machine (SVM) for binary classification, and the kernel method was radial basis function (RBF) kernel. Radial basis function kernel is based on the Gaussian function and is a monotonic function that finds the Euclidean distance from any point in the space to a certain center [32]. Its function is to deal with the example when the relationship between class labels and features is nonlinear [33]. We used accuracy and F1 score to evaluate the performance of the classification, as shown in Equations (1)–(4) [34,35].
where TP is true positive, TN is true negative, FP is false positive, and FN is false negative [34,36]. F1 score and accuracy are both essential metrics for evaluating and determining the statistical analysis of binary classification results (Mishra & Kumar, 2020). Cross-validation is a resampling method that can train and validate a model iteratively by using different portions of the data [37,38]. In machine learning, it is used to estimate how the model (built by training sets) is expected to perform in general when used to make predictions on data not used during the training (validation sets) in a limited sample size [38,39]. In this experiment, five-fold cross-validation with three repetitions was performed for data analysis, and we used the weighted average values of the classification metrics of each validation to represent the results of the cross-validation. This experiment performed two sets of analyses according to the different varieties. One set analyzed four varieties, and the other set analyzed only three types of bread wheat (Trojan, Emu Rock, and Yitpi). The reason was that the lower stem region of the durum wheat variety (Aurora) expresses the purple color naturally, which is different from other three bread wheat varieties with the green color on the lower stems.
Accuracy = TP + TN/TP + FP + FN + TN
Precision = TP/TP + FP
Recall = TP/TP + FN
F1 Score = 2 × (Recall × Precision)/(Recall + Precision)
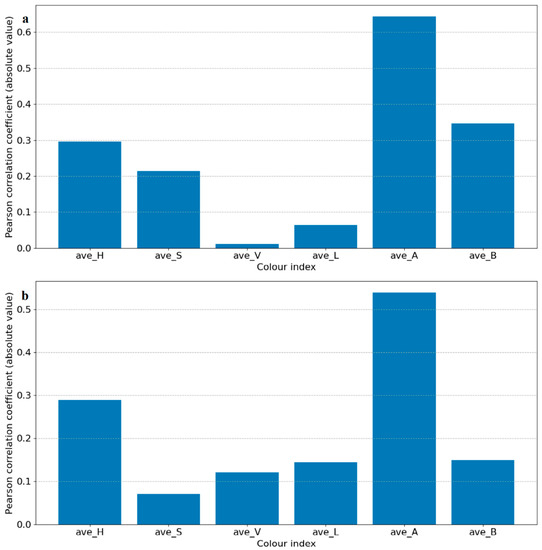
Figure 2.
Results of Pearson correlation t analysis of each color index. It depicts the correlation of each color index in the HSV and LAB color spaces. The correlation coefficients are in absolute values for convenient observation. (a) is for 14 DAI and (b) is for 21 DAI.
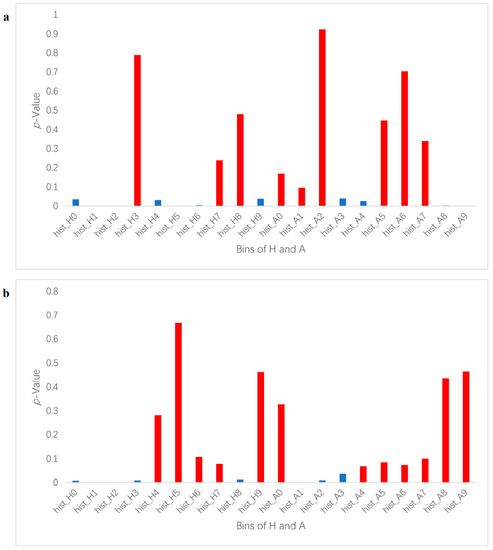
Figure 3.
p-Value of histogram bins of H and A. It depicts p-value results for each histogram bin in the ROI. Hist_H represents H (hue) and hist_A represents A (greens and magentas channel). (a) is for 14 DAI and (b) is for 21 DAI. The red bars mark the bins whose p-values are over 0.05 and removed; the blue bins were kept as feature vectors for binary classification.
3. Results
3.1. Manual Assessment Result
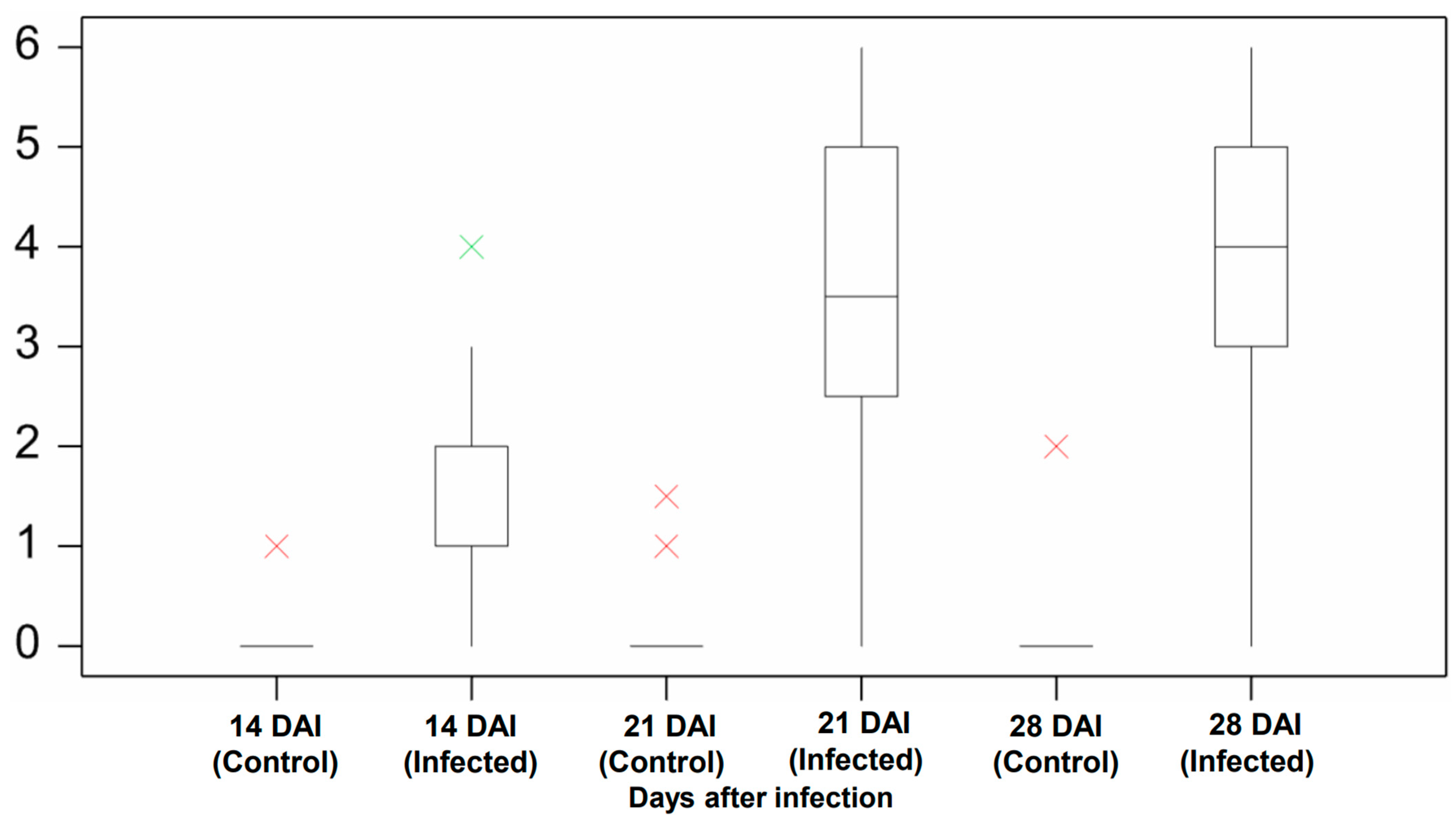
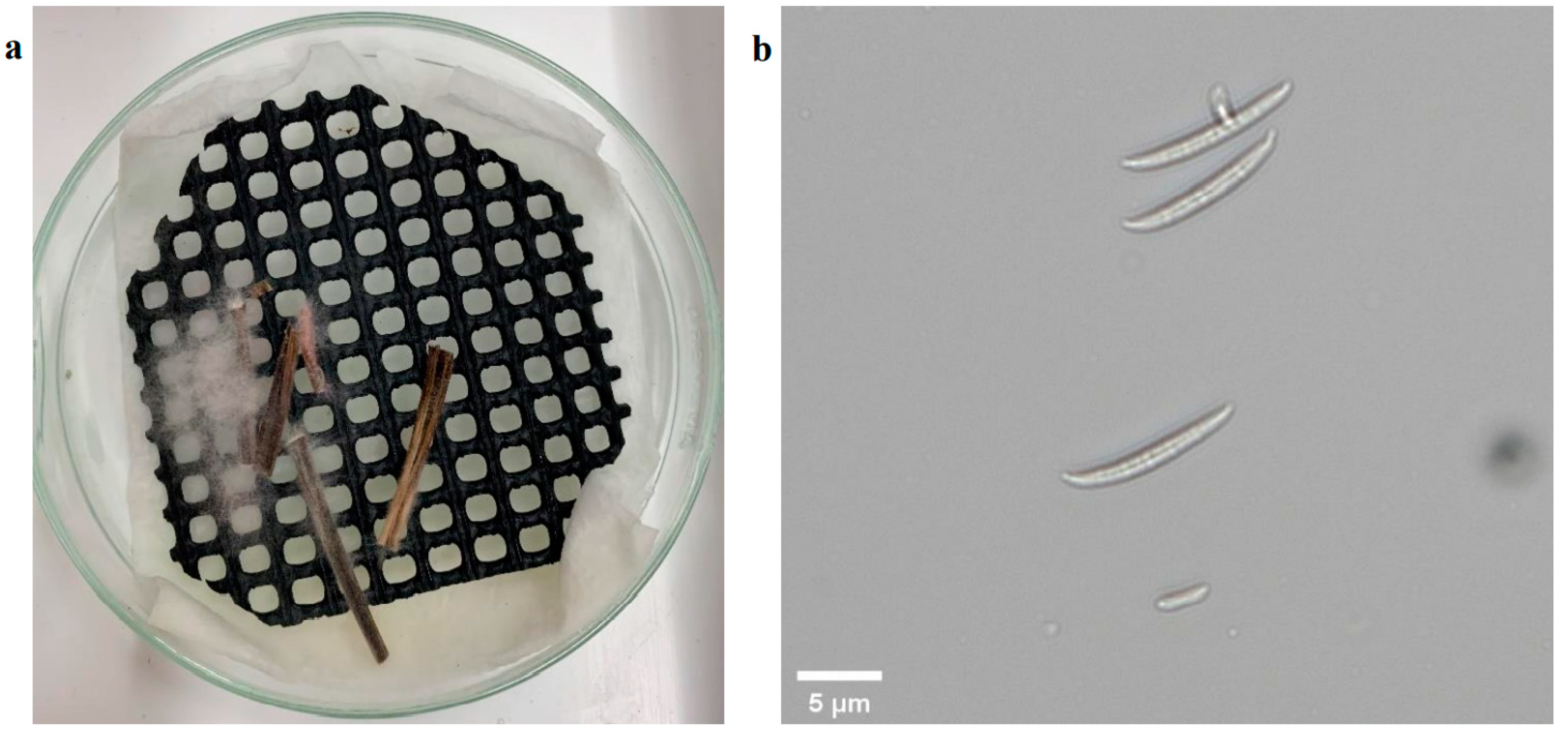
The manual assessment was conducted at three time points to measure the pathogen’s impact on plants during the early development period. The plants in the control group did not show symptoms of infection except for two outliers (Figure 4). These two outliers were from the Aurora variety and likely resulted from the natural purple and slight brown color stem (Figure 5). For the infection group, most plants started to present infection symptoms on 14 DAI, which confirmed that the plants were successfully infected. As expected, the symptoms in the infected group at this stage were minor, as most of their severity levels were between 0 and 2. As shown in Figure 6a, only the first leaf sheath showed discoloration at the wheat’s lower stem region on 14 DAI. On 21 DAI, the symptoms in the infected plants were obvious, and the severity levels of the infected group covered a range from 3.0 to 5.0, and the median level was approximately 3.5. There was more brown discoloration at the wheat’s lower stem region at this stage, as shown in Figure 6b. On 28 DAI, the mean value for infection level in the infected group increased slightly to approximately 4.0, and the range was between 3.0 and 5.0. In general, there was a significant increase from 14 to 21 DAI but a slight change from 21 to 28 DAI. From the pathogen isolation, the infection source was determined as F. pseudograminearum (Figure 7b), and there were pink and white mycelia around the plant tissue (Figure 7a), which are diagnostic of the pathogen by comparison with a previous study [30].
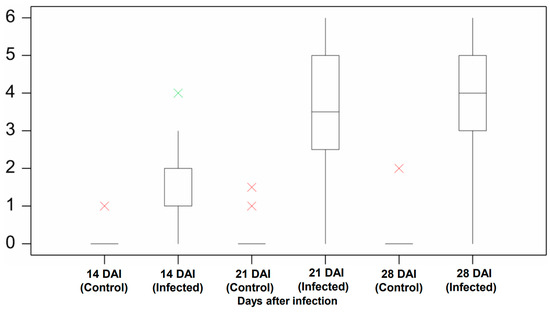
Figure 4.
Manual screening result of the control and infected group. Compares the severity level between the control and infected group at 14, 21, and 28 DAI. The boxplot shows the disease severity level in the median, lower, and upper quartile value with interval, as well as the minimum and maximum value. The green cross is the outlier, and the red cross is the extreme outlier. Analysis was undertaken in Genstat (VSN International).

Figure 5.
Healthy Aurora (a) and bread wheat (b). Shows that the lower stem and leaf sheath of healthy Aurora is brown and purple, while the bread wheat has a normal green color lower stem and leaf sheath.

Figure 6.
Infected plants on 14 DAI and 21 DAI. These images compare the brown discoloration symptoms of infection wheat between 14 DAI (a) and 21 DAI (b).
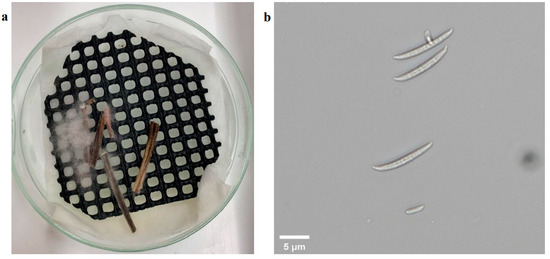
Figure 7.
Pathogen isolation result. Depicts fungi cultivated on plates in the image (a) and fungi observed under 20× microscope in the image (b).
3.2. Results of Disease Detection
We adopted the standard proposed by a previous study [40] in which F1 values above 0.75 were considered as acceptable and above 0.80 as satisfactory. The F1 scores for most datasets were higher than 0.80, and only the F1 scores in four varieties on 14 DAI were 0.73 (Table 1). The F1 scores on 14 DAI were lower than those on 21 DAI regardless of whether there were four or three varieties. The three-variety dataset showed a higher F1 score than the four-variety dataset on 14 DAI; the F1 score in the four-variety dataset was 0.730, while that in the three-variety dataset was 0.830. A similar trend was shown on 21 DAI as the four-variety dataset had a 0.830 F1 score, and the three-variety dataset had a 0.890 F1 score. The accuracy at 14 DAI was higher than on 21 DAI, 0.739 on 14 DAI compared with 0.723 on 21 DAI in four varieties, and 0.827 on 14 DAI in comparison with 0.821 on 21 DAI in three varieties. This likely occurred because the accuracy measurement did not take into account class imbalance as on 21 DAI, there were fewer images taken in controlled plants than in infected plants. A few of misclassification errors in controlled plants could dramatically reduce the accuracy.

Table 1.
F1 and accuracy metrics for disease detection. Depicts the F1 score and accuracy of SVM classification between the control and infection groups on 14 and 21 DAI. There were 60 wheat plants imaged (29 plants in control and 31 in infected) on 14 DAI, and 120 wheat plants were imaged (30 plants in control and 90 in infected) on 21 DAI. In the variety section, the four-variety dataset refers to Aurora, Trojan, Emu Rock, and Yitpi, and the three-verity dataset refers to Trojan, Emu Rock, and Yitpi.
4. Discussion
The manual scoring results indicated that on 14 DAI, most of the infected wheat showed moderate symptoms with levels between 0 and 2. Although the symptoms of the infected plants on 14 DAI were not obvious, we still obtained an acceptable classification result (F1 score was over 0.75) by using computer vision and machine learning technologies. When the infection progressed to 21 DAI, the symptoms of the infected plants were obvious, and levels increased to 3 to 5, meaning that the second and third leaf sheaths showed brown coloration. With the worsening symptoms of the infection, the F1 scores on 21 DAI increased significantly, reaching the satisfactory level (over 0.80). These results are representative of the infection process of crown rot on cereal plants in other experimental systems [22,24,41,42]. When the plants come into contact with the source of infection, F. pseudograminearum enters the leaf sheath tissue through the stomata of the leaves. However, there is no significant increase in fungal biomass and observable symptoms on the plant during approximately the first couples of weeks of the disease process [43]. In this experiment, the results of the 21 DAI and 28 DAI were similar, and there was no significant change from 21 DAI to 28 DAI shown in Figure 4, so it can be considered that the symptoms of infected plants at 21 DAI are relatively obvious and stable. Therefore, most of wheat plants showed mild symptoms of brown discoloration on 14 DAI when the crown rot disease had only just begun to move from the roots to the crowns for colonization [20,44]. As the infection progressed, the F1 scores increased on 21 DAI, likely indicating that the colonization in the lower stem had begun. Furthermore, the manual scoring results on 28 DAI also showed that there was no significant change from 21 to 28 DAI, which indicated that the brown discoloration symptom development of the infected plant became stable after 21 DAI.
Although the experiment was conducted in a greenhouse condition and the images were collected by a handheld device, the technology has a high potential to be applied to autonomous mobile devices. One of the challenges of the proposed technology is that the imaging needs to be adjusted to focus on the lower part of the wheat; however, several studies have verified that a ground-based robotic system can enter the inter-row in the field to identify and photograph different parts of plants [45,46]. For example, a field-tested robotic harvesting system can precisely identify the lower stem location of iceberg lettuce and complete harvesting [47]. The key challenge in the field condition comes from the segmentation of a single plant from the image. Compared with the laboratory environment, a single field image usually contains multiple plant information, and the background colors are more complex [48]. Therefore, when this method is applied to the field conditions, it is necessary to cooperate with a more robust plant segmentation method to ensure that the ROIs of individual plants can be separated from the original images with sufficient accuracy.
In this experiment, we observed that Aurora had unique color features around the stems compared with the other varieties. Most of the lower stem and leaf sheaths of Aurora used in this experiment were purple (Figure 7a). However, the lower stems of the other three types of bread wheat were all green (Figure 7b). The underlying reason for the purple leaf base is that this wheat variety has anthocyanin pigmentation, and anthocyanin pigmentation is closely related to the genetic changes in the wheat breeding process, but it does not affect plant growth [49,50]. When comparing the analysis of all four varieties with the three varieties without Aurora, the F1 score significantly improved when removing Aurora. This result indicates that the performances of the computer vision and machine learning algorithms are affected by the varieties if they have different color features. Although Aurora reduced the F1 score due to the purple stem, F1 scores on 21 DAI of the two datasets were still higher than 0.8. In addition, the results of Aurora also support that this experiment is not limited to specific wheat varieties. This is because when wheat is infected with crown rot, although different genotypes may show a different symptom severity due to a different disease resistance, the initial and early symptom is relatively concentrated in brown discoloration at the lower stem, such as dark necrotic lesions at the crown and stem base [51]. Moreover, the method used in this experiment also has certain potential in the detection of other plant diseases that cause discoloration at the lower stem, such as common root rot in wheat [52] and blackleg in canola [53]. This is because these diseases have a high probability of showing early infection in discoloration at the lower part of plants. By adjusting the thresholds in the segmentation method in this experiment, there is a great potential to segment plants with infected areas from the original images. Therefore, the methodology worked in this study can be indicated to be robust enough to deal with coloration differences for crown rot disease detection. Although a well-controlled greenhouse environment can provide a consistent environment for wheats to present relatively stable symptoms after crown rot infection, it is supportive to train the basic model. However, in the natural environment, the possibility of plant coloration differences will be very high, and different diseases and stress may cause plants to show coloration differences, even if the plant infection is not crown rot. For example, nitrogen deficiency or rust will cause the plant to show yellow leaves and stem [54]. Increasing the number of training samples could train more robust models for disease detection; however, this would be challenging when a large number of varieties have to be involved. Using more powerful imaging technologies, such as hyperspectral imaging technologies, to detect crown rot disease and understand disease development would be a further research direction [13].
5. Conclusions
Crown rot disease seriously affects wheat yield, and it requires an effective disease detection method to help control the disease spread and reduce the reinfection rate. However, crown rot does not show obvious symptoms on the leaves and upper stem in the early stage of infection, and early screening usually requires observation of the crown section, which is time-consuming and expensive and inevitably subjective. This study shows that by using color images of the lower stems of the plants combined with proper computer vision and machine learning technologies, infected plants can be detected after 14 DAI with satisfactory F1 scores above 0.75. With the days of infection increasing, the F1 score increased by over 0.80 on 21 DAI. The limitations of this research and further study are as follows:
- This study was conducted in a greenhouse where the environment was well controlled. However, there will be many factors that will affect crown rot disease development and spread in the field; hence, the time of infection start would not be as clear as in the greenhouse. Furthermore, unstable solar illumination and shadow in the field could also affect the colorful imaging collection process.
- In this study, all plants were only infected with crown rot and grown under a well-controlled greenhouse environment. However, when plants grow in the field condition, they may be infected with several different diseases. A necessary further research is to determine whether the current method will be suitable to screen crown rot in the field environment with the potential presence of other plant diseases.
- In this study, the images were collected using a smartphone. In a future work of field applications, a ground-based robotic system could be developed to collect the images of the lower stems of the plants in a more efficient way.
- The performances of the proposed method are affected by the color features of the plants. Even with Aurora included, the results were acceptable, showing that the proposed method is a promising economical method for assessing crown rot. However, it would be challenging if a large number of varieties of different color features have to be involved. In the future, using hyperspectral imaging of shoot to understand the disease development process and how fungi affect plant growth is a new research direction.
- In this study, we only tested an SVM; other machine learning algorithms could perform differently on different data [11]. Therefore, it is worthy to study other machine learning techniques [55] for digital imaging analysis.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agriengineering4010010/s1, Figure S1. The location of greenhouse in this experiment. Shows the greenhouse’s location of the research conducted. The latitude and longitude are 34°58′16.4″ S and 138°38′23.5″ E. Figure S2. Example for histogram bins. Depicts examples for histogram bins for pixels of ROI in one image for hue (H) and greens and magentas channel (A). Hist_h represents H and hist_a represents A. Figure 2a,b was 14 DAI and Figure 2c,d was 21 DAI.
Author Contributions
Writing—original draft, Y.X.; writing—review and editing, H.L. and D.P.; project administration, H.L. and D.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Yitpi Foundation, grant ID: 0006009868.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available at https://doi.org/10.25909/19137308.v1.
Acknowledgments
We would like to thank Lidia Mischis, Fiona Norrish, Nicole Bond, and other technical staff in the Plant Accelerator for excellent technical assistance. We would like to thank Marg Evans and Mark Butt (South Australian Research and Development Institute) for crown rot infection advice. We would like to thank Jason Able and Andrew Mathew to provide the seed materials in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kazan, K.; Gardiner, D.M. Fusarium crown rot caused by Fusarium pseudograminearum in cereal crops: Recent progress and future prospects. Mol. Plant Pathol. 2018, 19, 1547–1562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, G.M.; Brennan, J.P. Estimating disease losses to the Australian wheat industry. Australas. Plant Pathol. 2009, 38, 558–570. [Google Scholar] [CrossRef]
- Hüberli, D.; Gajda, K.; Connor, M.; Van Burgel, A. Choosing the Best Yielding Wheat and Barley Variety under High Crown Rot. 2017. Available online: https://www.agric.wa.gov.au/barley/grdc-research-updates-2017-choosing-best-yielding-wheat-and-barley-variety-under-high-crown#:~:text=In%20the%20barley%20trials%2C%20La,59%25%20yield%20to%20the%20disease (accessed on 10 June 2021).
- Moya-Elizondo, E.A.; Jacobsen, B.J. Integrated management of Fusarium crown rot of wheat using fungicide seed treatment, cultivar resistance, and induction of systemic acquired resistance (SAR). Biol. Control 2016, 92, 153–163. [Google Scholar] [CrossRef]
- Simpfendorfer, S. Evaluation of the Seed Treatment Rancona Dimension as a Standalone Option for Managing Crown Rot in Wheat–2015; DAN00175; Department of Primary Industries NSW, NSW DPI Northern Grains Research Results: Tamworth, Australia, 2016; pp. 121–124.
- Backhouse, D. Modelling the behaviour of crown rot in wheat caused by Fusarium pseudograminearum. Australas. Plant Pathol. 2014, 43, 15–23. [Google Scholar] [CrossRef]
- Bauriegel, E.; Giebel, A.; Herppich, W. Hyperspectral and chlorophyll fluorescence imaging to analyse the impact of Fusarium culmorum on the photosynthetic integrity of infected wheat ears. Sensors 2011, 11, 3765–3779. [Google Scholar] [CrossRef] [Green Version]
- Erginbas-Orakci, G.; Poole, G.; Nicol, J.M.; Paulitz, T.; Dababat, A.A.; Campbell, K. Assessment of inoculation methods to identify resistance to Fusarium crown rot in wheat. J. Plant Dis. Prot. 2016, 123, 19–27. [Google Scholar] [CrossRef]
- Saad, A.; Macdonald, B.; Martin, A.; Knight, N.L.; Percy, C. Comparison of disease severity caused by four soil-borne pathogens in winter cereal seedlings. Crop Pasture Sci. 2021, 72, 325–334. [Google Scholar] [CrossRef]
- Shikur Gebremariam, E.; Sharma-Poudyal, D.; Paulitz, T.C.; Erginbas-Orakci, G.; Karakaya, A.; Dababat, A.A. Identity and pathogenicity of Fusarium species associated with crown rot on wheat (Triticum spp.) in Turkey. Eur. J. Plant Pathol. 2018, 150, 387–399. [Google Scholar] [CrossRef]
- Liu, H.; Chahl, J.S. Proximal detecting invertebrate pests on crops using a deep residual convolutional neural network trained by virtual images. Artif. Intell. Agric. 2021, 5, 13–23. [Google Scholar] [CrossRef]
- Liu, H.; Lee, S.H.; Saunders, C. Development of a Machine Vision System for Weed Detection during Both of Off-Season and in-Season in Broadacre No-Tillage Cropping Lands. Am. J. Agric. Biol. Sci. 2014, 9, 174–193. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Plett, D.; Liu, H. The promise of hyperspectral imaging for the early detection of crown rot in Wheat. AgriEngineering 2021, 3, 924–941. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.; Huang, D. A review of imaging techniques for plant phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef] [PubMed]
- Mutka, A.M.; Bart, R.S. Image-based phenotyping of plant disease symptoms. Front. Plant Sci. 2015, 5, 734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauriegel, E.; Giebel, A.; Geyer, M.; Schmidt, U.; Herppich, W.B. Early detection of Fusarium infection in wheat using hyper-spectral imaging. Comput. Electron. Agric. 2011, 75, 304–312. [Google Scholar] [CrossRef]
- Dammer, K.-H.; Möller, B.; Rodemann, B.; Heppner, D. Detection of head blight (Fusarium ssp.) in winter wheat by color and multispectral image analyses. Crop Prot. 2011, 30, 420–428. [Google Scholar] [CrossRef]
- Bushnell, W.; Perkins-Veazie, P.; Russo, V.; Collins, J.; Seeland, T. Effects of deoxynivalenol on content of chloroplast pigments in barley leaf tissues. Phytopathology 2010, 100, 33–41. [Google Scholar] [CrossRef] [Green Version]
- Nogués, S.; Cotxarrera, L.; Alegre, L.; Trillas, M.I. Limitations to photosynthesis in tomato leaves induced by Fusarium wilt. New Phytol. 2002, 154, 461–470. [Google Scholar] [CrossRef]
- Knight, N.L.; Macdonald, B.; Percy, C.; Sutherland, M.W. Disease responses of hexaploid spring wheat (Triticum aestivum) culms exhibiting premature senescence (dead heads) associated with Fusarium pseudograminearum crown rot. Eur. J. Plant Pathol. 2021, 159, 191–202. [Google Scholar] [CrossRef]
- Qiu, R.; Yang, C.; Moghimi, A.; Zhang, M.; Steffenson, B.J.; Hirsch, C.D. Detection of Fusarium Head Blight in wheat using a deep neural network and color imaging. Remote Sens. 2019, 11, 2658. [Google Scholar] [CrossRef] [Green Version]
- Alahmad, S.; Simpfendorfer, S.; Bentley, A.R.; Hickey, L.T. Crown rot of wheat in Australia: Fusarium pseudograminearum taxonomy, population biology and disease management. Australas. Plant Pathol. 2018, 47, 285–299. [Google Scholar] [CrossRef]
- Collard, B.; Grams, R.; Bovill, W.; Percy, C.; Jolley, R.; Lehmensiek, A.; Wildermuth, G.; Sutherland, M. Development of molecular markers for crown rot resistance in wheat: Mapping of QTLs for seedling resistance in a ‘2-49’בJanz’population. Plant Breed. 2005, 124, 532–537. [Google Scholar] [CrossRef] [Green Version]
- Shi, S.D.; Zhao, J.C.; Pu, L.F.; Sun, D.J.; Han, D.J.; Li, C.L.; Feng, X.J.; Fan, D.S.; Hu, X.P. Identification of new sources of resistance to crown rot and Fusarium head blight in Wheat. Plant Dis. 2020, 104, 1979–1985. [Google Scholar] [CrossRef] [PubMed]
- Erginbas-Orakci, G.; Sehgal, D.; Sohail, Q.; Ogbonnaya, F.; Dreisigacker, S.; Pariyar, S.R.; Dababat, A.A. Identification of novel quantitative trait loci linked to crown rot resistance in spring wheat. Int. J. Mol. Sci. 2018, 19, 2666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallwork, H.; Zwer, P. Cereal Variety Disease Guide. 2016. Available online: https://pir.sa.gov.au/__data/assets/pdf_file/0011/356429/Cereal_Variety_Disease_Guide_Feb_2020.pdf (accessed on 3 June 2021).
- University of California. Growing Media. Available online: https://cagardenweb.ucanr.edu//Houseplants/Growing_Media/ (accessed on 10 June 2021).
- Brien, C. Dae: Functions Useful in the Design and ANOVA of Experiments. Available online: https://cran.r-project.org/package=dae (accessed on 9 October 2021).
- Agustí-Brisach, C.; Raya-Ortega, M.C.; Trapero, C.; Roca, L.F.; Luque, F.; López-Moral, A.; Fuentes, M.; Trapero, A. First report of Fusarium pseudograminearum causing crown rot of wheat in Europe. Plant Dis. 2018, 102, 1670. [Google Scholar] [CrossRef]
- Su, J.; Zhao, J.; Zhao, S.; Shang, X.; Pang, S.; Chen, S.; Liu, D.; Kang, Z.; Wang, X. Genetic determinants of wheat resistance to common root rot (spot blotch) and Fusarium crown rot. bioRxiv 2020. [Google Scholar] [CrossRef]
- Fahlgren, N.; Feldman, M.; Gehan, M.A.; Wilson, M.S.; Shyu, C.; Bryant, D.W.; Hill, S.T.; McEntee, C.J.; Warnasooriya, S.N.; Kumar, I. A versatile phenotyping system and analytics platform reveals diverse temporal responses to water availability in Setaria. Mol. Plant 2015, 8, 1520–1535. [Google Scholar] [CrossRef] [Green Version]
- Omrani, E.; Khoshnevisan, B.; Shamshirband, S.; Saboohi, H.; Anuar, N.B.; Nasir, M.H.N.M. Potential of radial basis function-based support vector regression for apple disease detection. Measurement 2014, 55, 512–519. [Google Scholar] [CrossRef]
- Scholkopf, B.; Sung, K.-K.; Burges, C.J.; Girosi, F.; Niyogi, P.; Poggio, T.; Vapnik, V. Comparing support vector machines with Gaussian kernels to radial basis function classifiers. IEEE Trans. Signal Process. 1997, 45, 2758–2765. [Google Scholar] [CrossRef] [Green Version]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Stehman, S.V. Selecting and interpreting measures of thematic classification accuracy. Remote Sens. Environ. 1997, 62, 77–89. [Google Scholar] [CrossRef]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berrar, D. Cross-Validation. 2019. Available online: https://www.researchgate.net/publication/324701535_Cross-Validation (accessed on 7 October 2021).
- Refaeilzadeh, P.; Tang, L.; Liu, H. Cross-validation. Encycl. Database Syst. 2009, 5, 532–538. [Google Scholar]
- Browne, M.W. Cross-validation methods. J. Math. Psychol. 2000, 44, 108–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, B.K.; Kumar, R. Natural Language Processing in Artificial Intelligence; CRC Press: Boca Raton, FL, USA, 2020. [Google Scholar]
- Hagerty, C.H.; Irvine, T.; Rivedal, H.M.; Yin, C.; Kroese, D.R. Diagnostic Guide: Fusarium Crown Rot of Winter Wheat. Plant Health Prog. 2021, 22, 176–181. [Google Scholar] [CrossRef]
- Rahman, M.M.; Davies, P.; Bansal, U.; Pasam, R.; Hayden, M.; Trethowan, R. Relationship between resistance and tolerance of crown rot in bread wheat. Field Crops Res. 2021, 265, 108106. [Google Scholar] [CrossRef]
- Stephens, A.E.; Gardiner, D.M.; White, R.G.; Munn, A.L.; Manners, J.M. Phases of infection and gene expression of Fusarium graminearum during crown rot disease of wheat. Mol. Plant-Microbe Interact. 2008, 21, 1571–1581. [Google Scholar] [CrossRef] [Green Version]
- Knight, N.L.; Sutherland, M.W. Histopathological assessment of Fusarium pseudograminearum colonization of cereal culms during crown rot infections. Plant Dis. 2016, 100, 252–259. [Google Scholar] [CrossRef] [Green Version]
- Mueller-Sim, T.; Jenkins, M.; Abel, J.; Kantor, G. The Robotanist: A ground-based agricultural robot for high-throughput crop phenotyping. In Proceedings of the 2017 IEEE International Conference on Robotics and Automation (ICRA), Singapore, 29 May–3 June 2017; pp. 3634–3639. [Google Scholar]
- Oliveira, L.F.; Moreira, A.P.; Silva, M.F. Advances in agriculture robotics: A state-of-the-art review and challenges ahead. Robotics 2021, 10, 52. [Google Scholar] [CrossRef]
- Birrell, S.; Hughes, J.; Cai, J.Y.; Iida, F. A field-tested robotic harvesting system for iceberg lettuce. J. Field Robot. 2020, 37, 225–245. [Google Scholar] [CrossRef] [Green Version]
- Kelly, D.; Vatsa, A.; Mayham, W.; Ngô, L.; Thompson, A.; Kazic, T. An opinion on imaging challenges in phenotyping field crops. Mach. Vis. Appl. 2016, 27, 681–694. [Google Scholar] [CrossRef] [Green Version]
- Melz, G.; Thiele, V. Chromosome locations of genes controlling ‘purple leaf base’ in rye and wheat. Euphytica 1990, 49, 155–159. [Google Scholar] [CrossRef]
- Zhao, S.; Xi, X.; Zong, Y.; Li, S.; Li, Y.; Cao, D.; Liu, B. Overexpression of ThMYC4E Enhances Anthocyanin Biosynthesis in Common Wheat. Int. J. Mol. Sci. 2019, 21, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Liu, C.; Chakraborty, S.; Manners, J.M.; Kazan, K. A simple method for the assessment of crown rot disease severity in wheat seedlings inoculated with Fusarium pseudograminearum. J. Phytopathol. 2008, 156, 751–754. [Google Scholar] [CrossRef]
- Moore, K.; Manning, B.; Simpfendorfer, S.; Verrell, A. Root and Crown Diseases of Wheat and Barley in Northern NSW; NSW Department of Primary Industries: Orange, NSW, USA, 2005.
- Markell, S.G.; del Rio, L.; Halley, S. Blackleg of Canola. 2008. Available online: https://library.ndsu.edu/ir/bitstream/handle/10365/5289/pp1367.pdf?sequence=1 (accessed on 10 January 2022).
- Devadas, R.; Lamb, D.; Backhouse, D.; Simpfendorfer, S. Sequential application of hyperspectral indices for delineation of stripe rust infection and nitrogen deficiency in wheat. Precis. Agric. 2015, 16, 477–491. [Google Scholar] [CrossRef] [Green Version]
- Jahan, N.; Flores, P.; Liu, Z.; Friskop, A.; Mathew, J.; Zhang, Z. Detecting and distinguishing wheat diseases using image processing and machine learning algorithms. In Proceedings of the ASABE 2020 Annual International Meeting, St. Joseph, MI, USA, 10 September 2020. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).