Detecting and Classifying Pests in Crops Using Proximal Images and Machine Learning: A Review
Abstract
1. Introduction
- -
- It summarizes the progress achieved so far on the use of digital images and machine learning for an effective pest monitoring, thus providing a complete picture on the subject in a single source (Section 2).
- -
- It provides a detailed discussion on the main weaknesses and research gaps that still remain, with emphasis on technical aspects that discourage practical adoption (Section 3).
- -
- It proposes some possible directions for future research (Section 4).
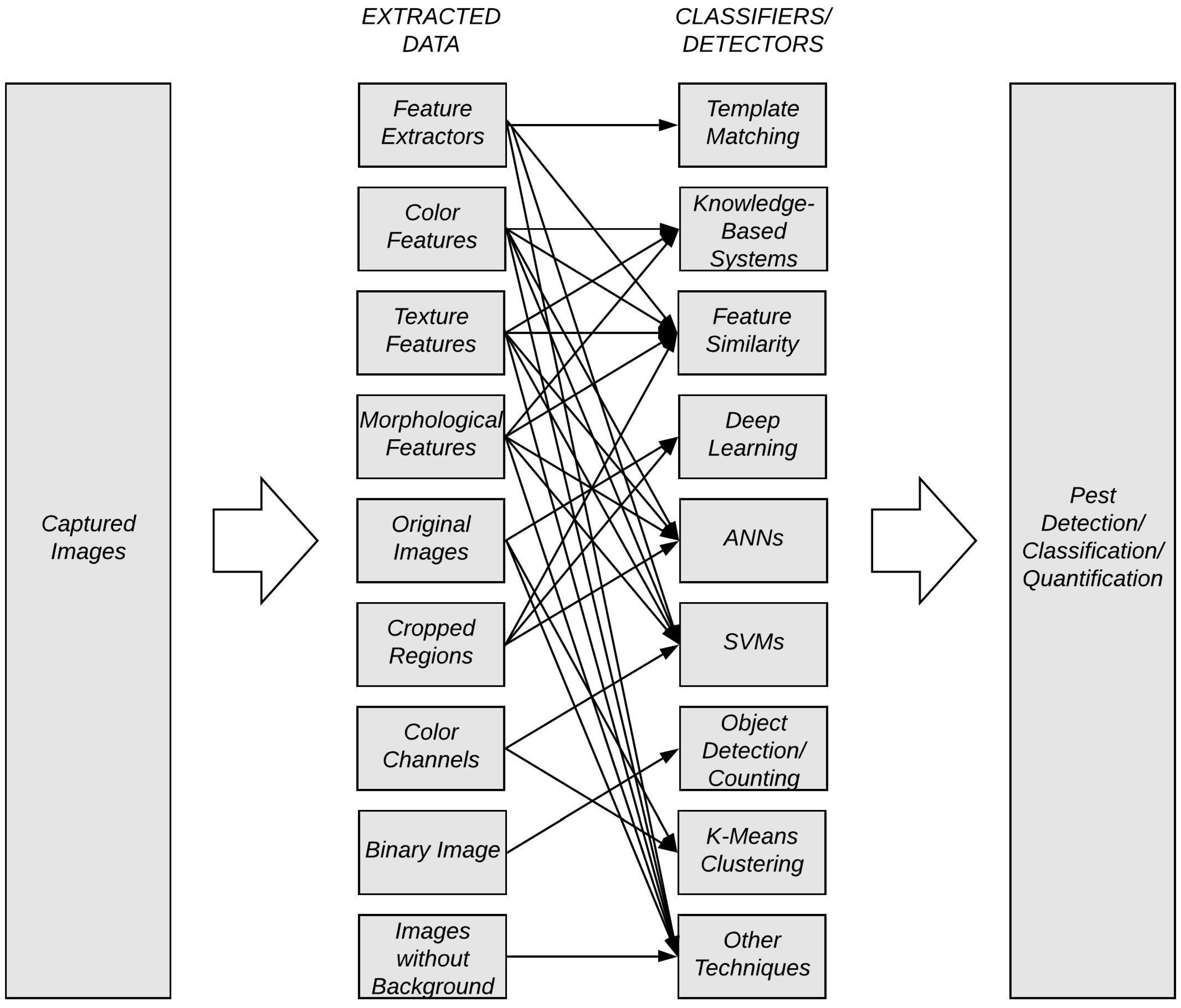
2. State of the Art of Pest Monitoring Using Digital Images and Machine Learning
2.1. Pest Detection Methods
2.2. Pest Classification Methods
3. Discussion
3.1. The Data Gap Problem
3.2. Difficulties Related to the Strategy Adopted—Images of Traps
3.3. Difficulties Related to the Strategy Adopted—Images in the Field
3.4. Difficulties Related to the Insects Themselves
3.5. Difficulties Related to the Imaging Equipment
3.6. Difficulties Related to Model Learning
4. Conclusions and Future Directions
Funding
Conflicts of Interest
References
- Oerke, E.C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Nalam, V.; Louis, J.; Shah, J. Plant defense against aphids, the pest extraordinaire. Plant Sci. 2019, 279, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Van Lenteren, J.; Bolckmans, K.; Köhl, J.; Ravensberg, W.; Urbaneja, A. Biological control using invertebrates and microorganisms: Plenty of new opportunities. BioControl 2017, 63, 39–59. [Google Scholar] [CrossRef]
- Yen, A.L.; Madge, D.G.; Berry, N.A.; Yen, J.D.L. Evaluating the effectiveness of five sampling methods for detection of the tomato potato psyllid, Bactericera cockerelli (Sulc) (Hemiptera: Psylloidea: Triozidae). Aust. J. Entomol. 2013, 52, 168–174. [Google Scholar] [CrossRef]
- Barbedo, J.G.A.; Castro, G.B. Influence of image quality on the identification of psyllids using convolutional neural networks. Biosyst. Eng. 2019, 182, 151–158. [Google Scholar] [CrossRef]
- Sun, Y.; Cheng, H.; Cheng, Q.; Zhou, H.; Li, M.; Fan, Y.; Shan, G.; Damerow, L.; Lammers, P.S.; Jones, S.B. A smart-vision algorithm for counting whiteflies and thrips on sticky traps using two-dimensional Fourier transform spectrum. Biosyst. Eng. 2017, 153, 82–88. [Google Scholar] [CrossRef]
- Huang, M.; Wan, X.; Zhang, M.; Zhu, Q. Detection of insect-damaged vegetable soybeans using hyperspectral transmittance image. J. Food Eng. 2013, 116, 45–49. [Google Scholar] [CrossRef]
- Ma, Y.; Huang, M.; Yang, B.; Zhu, Q. Automatic threshold method and optimal wavelength selection for insect-damaged vegetable soybean detection using hyperspectral images. Comput. Electron. Agric. 2014, 106, 102–110. [Google Scholar] [CrossRef]
- Clément, A.; Verfaille, T.; Lormel, C.; Jaloux, B. A new colour vision system to quantify automatically foliar discolouration caused by insect pests feeding on leaf cells. Biosyst. Eng. 2015, 133, 128–140. [Google Scholar] [CrossRef]
- Liu, H.; Lee, S.; Chahl, J. A review of recent sensing technologies to detect invertebrates on crops. Precis. Agric. 2017, 18, 635–666. [Google Scholar] [CrossRef]
- Martineau, M.; Conte, D.; Raveaux, R.; Arnault, I.; Munier, D.; Venturini, G. A survey on image-based insect classification. Pattern Recognit. 2017, 65, 273–284. [Google Scholar] [CrossRef]
- Lehmann, J.R.K.; Nieberding, F.; Prinz, T.; Knoth, C. Analysis of Unmanned Aerial System-Based CIR Images in Forestry—A New Perspective to Monitor Pest Infestation Levels. Forests 2015, 6, 594–612. [Google Scholar] [CrossRef]
- Vanegas, F.; Bratanov, D.; Powell, K.; Weiss, J.; Gonzalez, F. A Novel Methodology for Improving Plant Pest Surveillance in Vineyards and Crops Using UAV-Based Hyperspectral and Spatial Data. Sensors 2018, 18, 260. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lee, S.; Chahl, J.S. A Multispectral 3-D Vision System for Invertebrate Detection on Crops. IEEE Sensors J. 2017, 17, 7502–7515. [Google Scholar] [CrossRef]
- Liu, H.; Chahl, J.S. A multispectral machine vision system for invertebrate detection on green leaves. Comput. Electron. Agric. 2018, 150, 279–288. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, T.; Qiu, Z.; Peng, J.; Zhang, C.; He, Y. Fast Detection of Striped Stem-Borer (Chilo suppressalis Walker) Infested Rice Seedling Based on Visible/Near-Infrared Hyperspectral Imaging System. Sensors 2017, 17, 2470. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, M.; Khoshtaghaza, M.; Minaei, S.; Jamshidi, B. Vision-based pest detection based on SVM classification method. Comput. Electron. Agric. 2017, 137, 52–58. [Google Scholar] [CrossRef]
- Al-doski, J.; Mansor, S.; Mohd Shafri, H.Z. Thermal imaging for pests detecting—A review. Int. J. Agric. For. Plant. 2016, 2, 10–30. [Google Scholar]
- Barbedo, J.G.A. Detection of nutrition deficiencies in plants using proximal images and machine learning: A review. Comput. Electron. Agric. 2019, 162, 482–492. [Google Scholar] [CrossRef]
- Neethirajan, S.; Karunakaran, C.; Jayas, D.; White, N. Detection techniques for stored-product insects in grain. Food Control 2007, 18, 157–162. [Google Scholar] [CrossRef]
- Haff, R.P.; Saranwong, S.; Thanapase, W.; Janhiran, A.; Kasemsumran, S.; Kawano, S. Automatic image analysis and spot classification for detection of fruit fly infestation in hyperspectral images of mangoes. Postharvest Biol. Technol. 2013, 86, 23–28. [Google Scholar] [CrossRef]
- Lu, R.; Ariana, D.P. Detection of fruit fly infestation in pickling cucumbers using a hyperspectral reflectance/transmittance imaging system. Postharvest Biol. Technol. 2013, 81, 44–50. [Google Scholar] [CrossRef]
- Shah, M.; Khan, A. Imaging techniques for the detection of stored product pests. Appl. Entomol. Zool. 2014, 49, 201–212. [Google Scholar] [CrossRef]
- Shen, Y.; Zhou, H.; Li, J.; Jian, F.; Jayas, D.S. Detection of stored-grain insects using deep learning. Comput. Electron. Agric. 2018, 145, 319–325. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, S.; Wang, Z.; Liu, Z.; Yang, F. Mobile smart device-based vegetable disease and insect pest recognition method. Intell. Autom. Soft Comput. 2013, 19, 263–273. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Using digital image processing for counting whiteflies on soybean leaves. J. Asia Pac. Entomol. 2014, 17, 685–694. [Google Scholar] [CrossRef]
- Cho, J.; Choi, J.; Qiao, M.; Ji, C.; Kim, H.; Uhm, K.B.; Chon, T.S. Automatic identification of whiteflies, aphids and thrips in greenhouse based on image analysis. Int. J. Math. Comput. Simul. 2007, 1, 46–53. [Google Scholar]
- Qiao, M.; Lim, J.; Ji, C.W.; Chung, B.K.; Kim, H.Y.; Uhm, K.B.; Myung, C.S.; Cho, J.; Chon, T.S. Density estimation of Bemisia tabaci (Hemiptera: Aleyrodidae) in a greenhouse using sticky traps in conjunction with an image processing system. J. Asia Pac. Entomol. 2008, 11, 25–29. [Google Scholar] [CrossRef]
- Maharlooei, M.; Sivarajan, S.; Bajwa, S.G.; Harmon, J.P.; Nowatzki, J. Detection of soybean aphids in a greenhouse using an image processing technique. Comput. Electron. Agric. 2017, 132, 63–70. [Google Scholar] [CrossRef]
- Al-Saqer, S.M.; Hassan, G.M. Red Palm Weevil (Rynchophorus Ferrugineous, Olivier) Recognition by Image Processing Techniques. Am. J. Agric. Biol. Sci. 2011, 6, 365–376. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldu, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Liakos, K.; Busato, P.; Moshou, D.; Pearson, S.; Bochtis, D. Machine Learning in Agriculture: A Review. Sensors 2018, 18, 2674. [Google Scholar] [CrossRef]
- Rustia, D.J.A.; Chao, J.J.; Chung, J.Y.; Lin, T.T. Crop Losses to Pests. In Proceedings of the 2019 ASABE Annual International Meeting, Boston, MA, USA, 7–10 July 2019. [Google Scholar] [CrossRef]
- Boissard, P.; Martin, V.; Moisan, S. A cognitive vision approach to early pest detection in greenhouse crops. Comput. Electron. Agric. 2008, 62, 81–93. [Google Scholar] [CrossRef]
- Cheng, X.; Zhang, Y.; Chen, Y.; Wu, Y.; Yue, Y. Pest identification via deep residual learning in complex background. Comput. Electron. Agric. 2017, 141, 351–356. [Google Scholar] [CrossRef]
- Dawei, W.; Limiao, D.; Jiangong, N.; Jiyue, G.; Hongfei, Z.; Zhongzhi, H. Recognition pest by image-based transfer learning. J. Sci. Food Agric. 2019, 99, 4524–4531. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Wang, Y.; Han, Z.; Yu, R. Research on insect pest image detection and recognition based on bio-inspired methods. Biosyst. Eng. 2018, 169, 139–148. [Google Scholar] [CrossRef]
- Dimililer, K.; Zarrouk, S. ICSPI: Intelligent Classification System of Pest Insects Based on Image Processing and Neural Arbitration. Appl. Eng. Agric. 2017, 33, 453–460. [Google Scholar] [CrossRef]
- Ding, W.; Taylor, G. Automatic moth detection from trap images for pest management. Comput. Electron. Agric. 2016, 123, 17–28. [Google Scholar] [CrossRef]
- Espinoza, K.; Valera, D.L.; Torres, J.A.; López, A.; Molina-Aiz, F.D. Combination of image processing and artificial neural networks as a novel approach for the identification of Bemisia tabaci and Frankliniella occidentalis on sticky traps in greenhouse agriculture. Comput. Electron. Agric. 2016, 127, 495–505. [Google Scholar] [CrossRef]
- Faithpraise, F.O.; Birch, P.; Young, R.; Obu, J.; Faithpraise, B.; Chatwin, C. Automatic plant pest detection and recognition using k-means clustering algorithm and correspondence filters. Int. J. Adv. Biotechnol. Res. 2013, 4, 189–199. [Google Scholar]
- Han, R.; He, Y.; Liu, F. Feasibility Study on a Portable Field Pest Classification System Design Based on DSP and 3G Wireless Communication Technology. Sensors 2012, 12, 3118–3130. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Dong, S.; Zhang, S.; Xie, C.; Wang, H. AF-RCNN: An anchor-free convolutional neural network for multi-categories agricultural pest detection. Comput. Electron. Agric. 2020, 174, 105522. [Google Scholar] [CrossRef]
- Li, Y.; Xia, C.; Lee, J. Detection of small-sized insect pest in greenhouses based on multifractal analysis. Optik Int. J. Light Electron Opt. 2015, 126, 2138–2143. [Google Scholar] [CrossRef]
- Liu, T.; Chen, W.; Wu, W.; Sun, C.; Guo, W.; Zhu, X. Detection of aphids in wheat fields using a computer vision technique. Biosyst. Eng. 2016, 141, 82–93. [Google Scholar] [CrossRef]
- Liu, Z.; Gao, J.; Yang, G.; Zhang, H.; He, Y. Localization and Classification of Paddy Field Pests using a Saliency Map and Deep Convolutional Neural Network. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Liu, L.; Wang, R.; Xie, C.; Yang, P.; Wang, F.; Sudirman, S.; Liu, W. PestNet: An End-to-End Deep Learning Approach for Large-Scale Multi-Class Pest Detection and Classification. IEEE Access 2019, 7, 45301–45312. [Google Scholar] [CrossRef]
- Roldán-Serrato, K.L.; Escalante-Estrada, J.; Rodríguez-González, M. Automatic pest detection on bean and potato crops by applying neural classifiers. Eng. Agric. Environ. Food 2018, 11, 245–255. [Google Scholar] [CrossRef]
- Solis-Sánchez, L.O.; Castañeda-Miranda, R.; García-Escalante, J.J.; Torres-Pacheco, I.; Guevara-González, R.G.; Castañeda-Miranda, C.L.; Alaniz-Lumbreras, P.D. Scale invariant feature approach for insect monitoring. Comput. Electron. Agric. 2011, 75, 92–99. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, X.; Yuan, M.; Ren, L.; Wang, J.; Chen, Z. Automatic in-trap pest detection using deep learning for pheromone-based Dendroctonus valens monitoring. Biosyst. Eng. 2018, 176, 140–150. [Google Scholar] [CrossRef]
- Vakilian, K.A.; Massah, J. Performance evaluation of a machine vision system for insect pests identification of field crops using artificial neural networks. Arch. Phytopathol. Plant Prot. 2013, 46, 1262–1269. [Google Scholar] [CrossRef]
- Venugoban, K.; Ramanan, A. Image Classification of Paddy Field Insect Pests Using Gradient-Based Features. Int. J. Mach. Learn. Comput. 2014, 4, 1–5. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, K.; Liu, Z.; Wang, X.; Pan, S. A Cognitive Vision Method for Insect Pest Image Segmentation. IFAC Pap. Online 2018, 51, 85–89. [Google Scholar] [CrossRef]
- Wang, F.; Wang, R.; Xie, C.; Yang, P.; Liu, L. Fusing multi-scale context-aware information representation for automatic in-field pest detection and recognition. Comput. Electron. Agric. 2020, 169, 105222. [Google Scholar] [CrossRef]
- Wen, C.; Guyer, D.E.; Li, W. Local feature-based identification and classification for orchard insects. Biosyst. Eng. 2009, 104, 299–307. [Google Scholar] [CrossRef]
- Wen, C.; Guyer, D. Image-based orchard insect automated identification and classification method. Comput. Electron. Agric. 2012, 89, 110–115. [Google Scholar] [CrossRef]
- Wen, C.; Wu, D.; Hu, H.; Pan, W. Pose estimation-dependent identification method for field moth images using deep learning architecture. Biosyst. Eng. 2015, 136, 117–128. [Google Scholar] [CrossRef]
- Xia, C.; Lee, J.M.; Li, Y.; Chung, B.K.; Chon, T.S. In situ detection of small-size insect pests sampled on traps using multifractal analysis. Opt. Eng. 2012, 51. [Google Scholar] [CrossRef]
- Xia, C.; Chon, T.S.; Ren, Z.; Lee, J.M. Automatic identification and counting of small size pests in greenhouse conditions with low computational cost. Ecol. Inform. 2015, 29, 139–146. [Google Scholar] [CrossRef]
- Xia, D.; Chen, P.; Wang, B.; Zhang, J.; Xie, C. Insect Detection and Classification Based on an Improved Convolutional Neural Network. Sensors 2018, 18, 4169. [Google Scholar] [CrossRef]
- Yao, Q.; Lv, J.; Liu, Q.J.; Diao, G.Q.; Yang, B.J.; Chen, H.M.; Tang, J. An Insect Imaging System to Automate Rice Light-Trap Pest Identification. J. Integr. Agric. 2012, 11, 978–985. [Google Scholar] [CrossRef]
- Yao, Q.; Liu, Q.; Dietterich, T.G.; Todorovic, S.; Lin, J.; Diao, G.; Yang, B.; Tang, J. Segmentation of touching insects based on optical flow and NCuts. Biosyst. Eng. 2013, 114, 67–77. [Google Scholar] [CrossRef]
- Yao, Q.; Xian, D.X.; Liu, Q.J.; Yang, B.J.; Diao, G.Q.; Tang, J. Automated Counting of Rice Planthoppers in Paddy Fields Based on Image Processing. J. Integr. Agric. 2014, 13, 1736–1745. [Google Scholar] [CrossRef]
- Yue, Y.; Cheng, X.; Zhang, D.; Wu, Y.; Zhao, Y.; Chen, Y.; Fan, G.; Zhang, Y. Deep recursive super resolution network with Laplacian Pyramid for better agricultural pest surveillance and detection. Comput. Electron. Agric. 2018, 150, 26–32. [Google Scholar] [CrossRef]
- Zhong, Y.; Gao, J.; Lei, Q.; Zhou, Y. A Vision-Based Counting and Recognition System for Flying Insects in Intelligent Agriculture. Sensors 2018, 18, 1489. [Google Scholar] [CrossRef] [PubMed]
- Barbedo, J.G.A. Factors influencing the use of deep learning for plant disease recognition. Biosyst. Eng. 2018, 172, 84–91. [Google Scholar] [CrossRef]
- Arel, I.; Rose, D.C.; Karnowski, T.P. Deep Machine Learning—A New Frontier in Artificial Intelligence Research [Research Frontier]. IEEE Comput. Intell. Mag. 2010, 5, 13–18. [Google Scholar] [CrossRef]
- Lake, B.M.; Salakhutdinov, R.; Tenenbaum, J.B. Human-level concept learning through probabilistic program induction. Science 2015, 350, 1332–1338. [Google Scholar] [CrossRef]
- Hu, G.; Peng, X.; Yang, Y.; Hospedales, T.M.; Verbeek, J. Frankenstein: Learning Deep Face Representations Using Small Data. IEEE Trans. Image Process. 2018, 27, 293–303. [Google Scholar] [CrossRef]
- Barth, R.; Hemming, J.; Henten, E.V. Optimising realism of synthetic images using cycle generative adversarial networks for improved part segmentation. Comput. Electron. Agric. 2020, 173. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. A review on the main challenges in automatic plant disease identification based on visible range images. Biosyst. Eng. 2016, 144, 52–60. [Google Scholar] [CrossRef]
- Sugiyama, M.; Nakajima, S.; Kashima, H.; Bünau, P.v.; Kawanabe, M. Direct Importance Estimation with Model Selection and Its Application to Covariate Shift Adaptation. In Proceedings of the 20th International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 3–6 December 2007; pp. 1433–1440. [Google Scholar]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef]
- Ben-David, S.; Blitzer, J.; Crammer, K.; Kulesza, A.; Pereira, F.; Vaughan, J.W. A theory of learning from different domains. Mach. Learn. 2010, 79, 151–175. [Google Scholar] [CrossRef]
- Bock, C.H.; Poole, G.H.; Parker, P.E.; Gottwald, T.R. Plant Disease Severity Estimated Visually, by Digital Photography and Image Analysis, and by Hyperspectral Imaging. Crit. Rev. Plant Sci. 2010, 29, 59–107. [Google Scholar] [CrossRef]
- Bekker, A.J.; Goldberger, J. Training Deep Neural-Networks Based on Unreliable Labels. In Proceedings of the 2016 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Shanghai, China, 20–25 March 2016; pp. 2682–2686. [Google Scholar] [CrossRef]
- Irwin, A. Citizen Science: A Study of People, Expertise and Sustainable Development, 1st ed.; Routledge: Abingdon-on-Thames, UK, 2002. [Google Scholar]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3. [Google Scholar] [CrossRef] [PubMed]
- Albani, D.; IJsselmuiden, J.; Haken, R.; Trianni, V. Monitoring and Mapping with Robot Swarms for Agricultural Applications. In Proceedings of the 2017 14th IEEE International Conference on Advanced Video and Signal Based Surveillance (AVSS), Lecce, Italy, 29 August–1 September 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Shi, W.; Cao, J.; Zhang, Q.; Li, Y.; Xu, L. Edge Computing: Vision and Challenges. IEEE Internet Things J. 2016, 3, 637–646. [Google Scholar] [CrossRef]
| Ref. | Problem | Image | Pest | Input | Classifier/Detector | Acc. |
|---|---|---|---|---|---|---|
| [30] | Detection | Trap | Red Palm Weevil | Zernike Moments and Region Properties | Template matching | 0.88–0.97 |
| [26] | Detection | Controlled | Whiteflies | Binary image | Connected objects counting | 0.68–0.95 |
| [5] | Detection | Trap | Psyllids | Image patches | CNN | 0.69–0.92 |
| [34] | Detection | Controlled | Whitefly | Spatial, color and texture features | Knowledge-based system | 0.70–0.97 |
| [35] | Classification | Field | 10 species | Original images | Deep residual learning | 0.98 |
| [27] | Classification | Trap | Whiteflies, aphids and thrips | Size, shape and color features | Feature similarity | 0.59–1.00 |
| [36] | Classification | Field | 10 species | Resized images | AlexNet (CNN) | 0.94 |
| [37] | Classification | Field | 10 species | SIFT, NNSC, LCP features | SVM | 0.85 |
| [38] | Classification | Field | 8 species | Segmented images | ANN | 0.93 |
| [39] | Detection | Trap | Moth | Patches from original image | CNN | 0.93 |
| [17] | Detection | Field | Thrips | HSI Color Channels | SVM | 0.98 |
| [40] | Detection | Trap | Western flower thrips, whitefly | Patches from original image | Multi-layer ANN | 0.92–0.96 |
| [41] | Classification | Field | 10 species | L*a*b* images | K-Means Clustering | N/A |
| [42] | Classification | Trap | 6 species | Morphological and color features | ANN | 0.82 |
| [43] | Detection | Trap | 24 species | Original Images | CNN + AFRPN | 0.55–0.99 |
| [44] | Detection | Field | Whiteflies | Image with background removed | Multifractal analysis | 0.88 |
| [45] | Detection | Field | Aphids | HOG Features | SVM | 0.87 |
| [46] | Classification | Field | 12 species | Cropped regions | CNN | 0.83–0.95 |
| [10] | Survey | N/A | N/A | N/A | N/A | N/A |
| [47] | Classification | Trap | 16 butterfly species | Original images | CNN + RPN + PSSM | 0.75 |
| [29] | Detection | Controlled | Aphids | Binary image | Object counting | 0.81–0.96 |
| [11] | Survey | N/A | N/A | N/A | N/A | N/A |
| [28] | Detection | Trap | Whiteflies | Binary image | Object counting | 0.81–0.99 |
| [48] | Detection | Field | Mexican Bean Beetle, Colorado Potato Beetle | Original images | RSC and LRA | 0.76–0.89 |
| [33] | Classification | Trap | Flies, gnats, moth flies, thrips and whiteflies | Original images | Tiny YOLO v3 + multistage CNN | 0.92–0.94 |
| [49] | Classification | Trap | 6 species | LOSS V2 + SIFT | Feature Similarity | 0.96–0.99 |
| [50] | Detection | Trap | Red turpentine beetle | Original images | Downsized RetinaNet | 0.75 |
| [51] | Detection | Controlled | Beet armyworm | Morphological and texture features | ANN | 0.89 |
| [52] | Classification | Varied | 20 insect species | SURF + HOG features | SVM | 0.90 |
| [25] | Detection | Field | Whiteflies | Thresholding + morphology | Connected objects counting | 0.82–0.97 |
| [53] | Detection | Field | Whiteflies | Original images | K-means clustering | 0.92–0.98 |
| [54] | Detection | Field | 3 species | Original images | CNN + Decision Net | 0.62–0.91 |
| [55] | Classification | Trap | 5 species | SIFT features | MLSLC, KNNC, PDLC, PCALC, NMC, SVM | 0.77–0.89 |
| [56] | Classification | Trap | 8 species | Several features | MLSLC, KNNC, NMC, NDLC, DT | 0.60–0.87 |
| [57] | Classification | Trap | 9 moth species | 154 features | IpSDAE | 0.94 |
| [58] | Detection | Trap | Whiteflies | Original images | Multifractal analysis | 0.96 |
| [59] | Classification | Trap | Whiteflies, aphids, thrips | Segmented insects | Mahalanobis distance | 0.70–0.91 |
| [60] | Classification | Field | 24 species | Resized images | VGG19 (CNN) | 0.89 |
| [61] | Classification | Trap | 4 species of Lepidoptera | 156 color, shape and texture features | SVM | 0.97 |
| [62] | Detection | Trap | Several | Original images | Normalized Cuts, watershed, K-means | 0.96 |
| [63] | Detection | Field | Rice Planthoppers | Haar and HOG features | Adaboost and SVM | 0.85 |
| [64] | Detection | Field | Atractomorpha, Erthesina, Pieris | Original images | DSRNLP | 0.70–0.92 |
| [65] | Classification | Trap | 6 species | Original images, variety of features | YOLO, SVM | 0.90–0.92 |
| Issue | Source of the Issue | Potential Solutions |
|---|---|---|
| Image distortions caused by the trap structure | Trap | Enforce image capture protocols |
| Object configuration in the trap changes over time | Trap | Increase frequency of image capture |
| The poses in which insects are glued vary | Trap | Capture images as soon as insects land |
| Overlapping insects in the traps | Trap | Image processing techniques for separating clusters; statistical corrections |
| Presence of dust, debris and other insects | Trap | Trap and lure should be designed to minimize the presence of spurious objects |
| Delay between the beginning of infestation and the moment the trap has enough samples | Trap | Capture images directly in the field environment |
| Non-airborne pests are missed | Trap | Capture images directly in the field environment |
| Traps have too many problems associated | Trap | Capture images directly in the field environment |
| Images captured in the field have a high degree of variability | Condition variety in the field | Employ techniques for illumination correction; build more comprehensive datasets |
| Contrast between specimens of interest and their surroundings change | Condition variety in the field | Build more comprehensive datasets; enforce image capture protocols |
| Angle of image capture cause specimens to appear distorted | Condition variety in the field | Build more comprehensive datasets; enforce image capture protocols |
| Pest occlusion by leaves and other structures | Condition variety in the field | Use of autonomous robots for peering into occluded areas; statistical corrections |
| Failure to detect specimens at early stages of development | Insect’s intrinsic characteristics | Development of more sensitive algorithms; use of more sophisticated sensors |
| Other objects in the scene cause confusion | Uncontrolled environment | Use more sophisticated detection algorithms; build more comprehensive datasets |
| Visual similarities between the pest of interest and other species | Insect’s intrinsic characteristics | Use more sophisticated detection algorithms; build more comprehensive datasets |
| Different cameras produce images with different characteristics | Imaging equipment | Build more comprehensive datasets; enforce image capture protocols |
| More spatial resolution does not necessarily translate into better results | Imaging equipment | Specific experiments need to be designed to investigate the ideal resolution |
| Data overfitting | Model learning | Apply regularization techniques and other methods for reducing overfitting |
| Class imbalance | Model learning | Oversample smaller classes and/or undersample the larger ones |
| Covariate shift issues | Model learning | Apply domain adaptation techniques; build more comprehensive datasets |
| Inconsistencies in the reference annotations | Model learning | Image annotation performed redundantly by several people |
| Most datasets are not comprehensive enough | Model learning | Data sharing; citizen science; use of social networks concepts |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbedo, J.G.A. Detecting and Classifying Pests in Crops Using Proximal Images and Machine Learning: A Review. AI 2020, 1, 312-328. https://doi.org/10.3390/ai1020021
Barbedo JGA. Detecting and Classifying Pests in Crops Using Proximal Images and Machine Learning: A Review. AI. 2020; 1(2):312-328. https://doi.org/10.3390/ai1020021
Chicago/Turabian StyleBarbedo, Jayme Garcia Arnal. 2020. "Detecting and Classifying Pests in Crops Using Proximal Images and Machine Learning: A Review" AI 1, no. 2: 312-328. https://doi.org/10.3390/ai1020021
APA StyleBarbedo, J. G. A. (2020). Detecting and Classifying Pests in Crops Using Proximal Images and Machine Learning: A Review. AI, 1(2), 312-328. https://doi.org/10.3390/ai1020021





