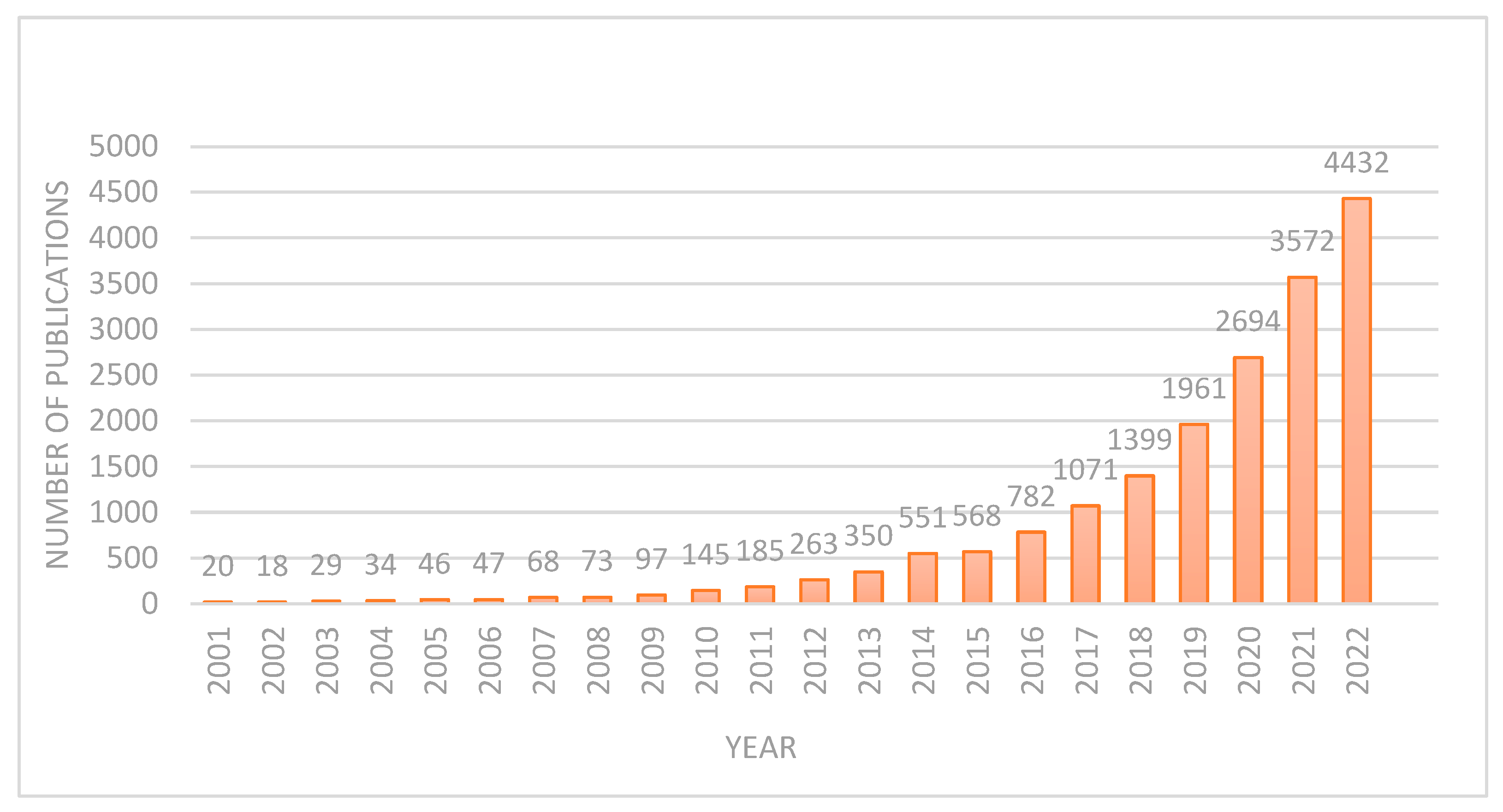Abstract
Brain tumors pose a significant challenge in medical research due to their associated morbidity and mortality. Magnetic Resonance Imaging (MRI) is the premier imaging technique for analyzing these tumors without invasive procedures. Recent years have witnessed remarkable progress in brain tumor detection, classification, and progression analysis using MRI data, largely fueled by advancements in deep learning (DL) models and the growing availability of comprehensive datasets. This article investigates the cutting-edge DL models applied to MRI data for brain tumor diagnosis and prognosis. The study also analyzes experimental results from the past two decades along with technical challenges encountered. The developed datasets for diagnosis and prognosis, efforts behind the regulatory framework, inconsistencies in benchmarking, and clinical translation are also highlighted. Finally, this article identifies long-term research trends and several promising avenues for future research in this critical area.
1. Introduction
The brain plays a vital role in our cognitive and emotional well-being, controlling functions like information processing, memory storage, and emotional regulation. Brain tumors, unfortunately, pose serious health threats, as indicated by increasing incidence rates [1]. According to U.S. cancer statistics for 2021 and 2023, the estimated brain tumor cases were 24,530 and 24,810, respectively, with projected mortality rates of 18,600 and 18,990 adults [2,3]. These tumors can lead to a wide range of neurological symptoms, from cognitive impairments and memory loss to motor dysfunction, sensory disturbances, and emotional changes. Brain anomalies can lead to a wide range of neurological symptoms, including cognitive impairments, memory loss, motor dysfunction, sensory disturbances, emotional and behavioral changes, seizures, communication disorders, psychiatric conditions, headaches, and neurodegenerative diseases [4,5,6].
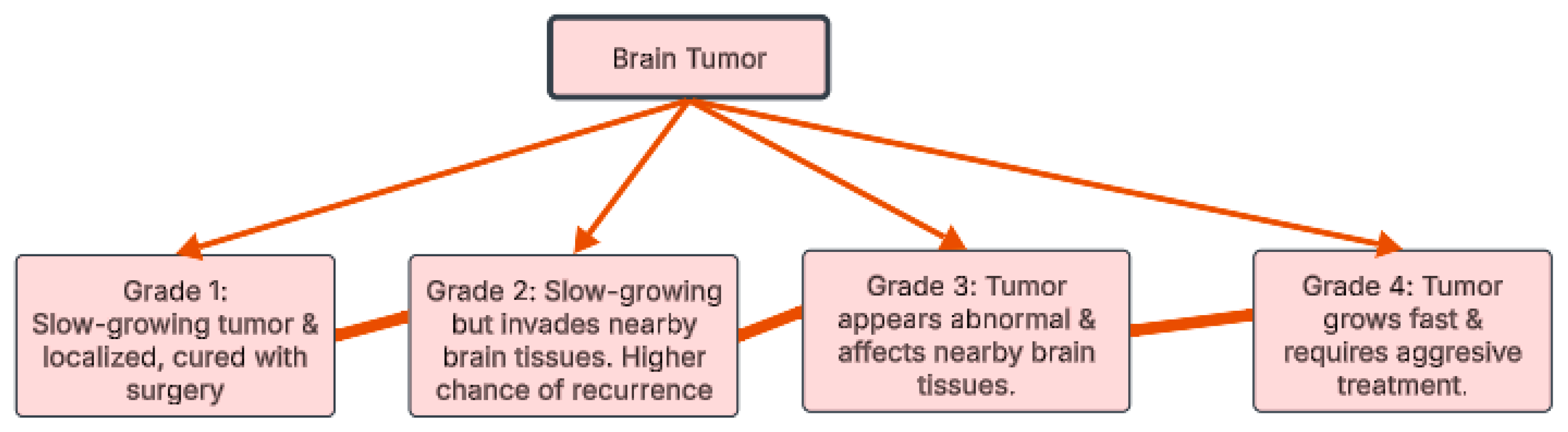
Over 120 types of brain tumors exist, all stemming from uncontrolled cell growth. These tumors are categorized, as illustrated in Figure 1, by grade (1–4) based on their aggressiveness [7]. Grades 1 and 2 are slower growing; Grade 1 tumors are benign, while Grade 2 may invade surrounding tissue. Grades 3 and 4 are malignant, with Grade 3 capable of spreading and Grade 4 known for rapid growth and frequent recurrence [8]. This grading system is crucial for predicting tumor behavior and determining the appropriate treatment strategy.
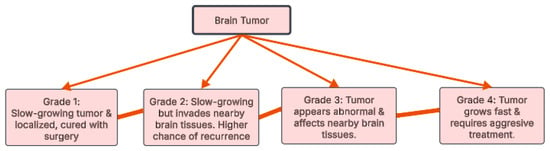
Figure 1.
Brain tumor grades.
The early detection of brain tumors enables timely intervention and treatment, significantly improving patient outcomes. Brain abnormalities in their initial stages are generally more susceptible to therapeutic approaches like radiation therapy, surgery, chemotherapy, and antibiotic regimens. Identifying these conditions early on also helps mitigate the risks associated with elevated intracranial pressure, seizures, and various neurological complications.
1.1. Why MRI for Brain Tumor Detection
MRI scans are considered the most effective method for brain cancer evaluation, offering several key advantages over other diagnostic techniques:
Superior soft tissue contrast: MRI excels at distinguishing between different brain tissues, enabling the detection of subtle abnormalities like tumors. It provides highly detailed images of the brain’s structure, revealing tumors that may go unnoticed with other imaging methods [7].
High-resolution imaging: MRI produces precise and detailed images, allowing physicians to locate tumors accurately, assess their size and shape, and evaluate their impact on surrounding tissues.
Radiation-free: Unlike CT scans, MRI does not use ionizing radiation, making it a safer option, particularly for children and patients requiring multiple scans.
Multi-view imaging: MRI captures images in multiple planes (axial, coronal, and sagittal), providing a comprehensive view of a tumor’s size, shape, location, and relationship to nearby structures. This is crucial for effective surgical planning and treatment decisions.
Enhanced tumor visibility: Contrast agents used in MRI can highlight even small abnormalities, improving tumor detection.
Functional imaging capabilities: Functional MRI (fMRI) can evaluate brain activity and map functional areas impacted by tumors.
Better detection of small or low-grade tumors: MRI is more sensitive than CT scans for identifying small or low-grade brain tumors.
Reduced artifacts: Unlike CT, MRI minimizes artifacts caused by bone, ensuring a clearer evaluation of brain structures.
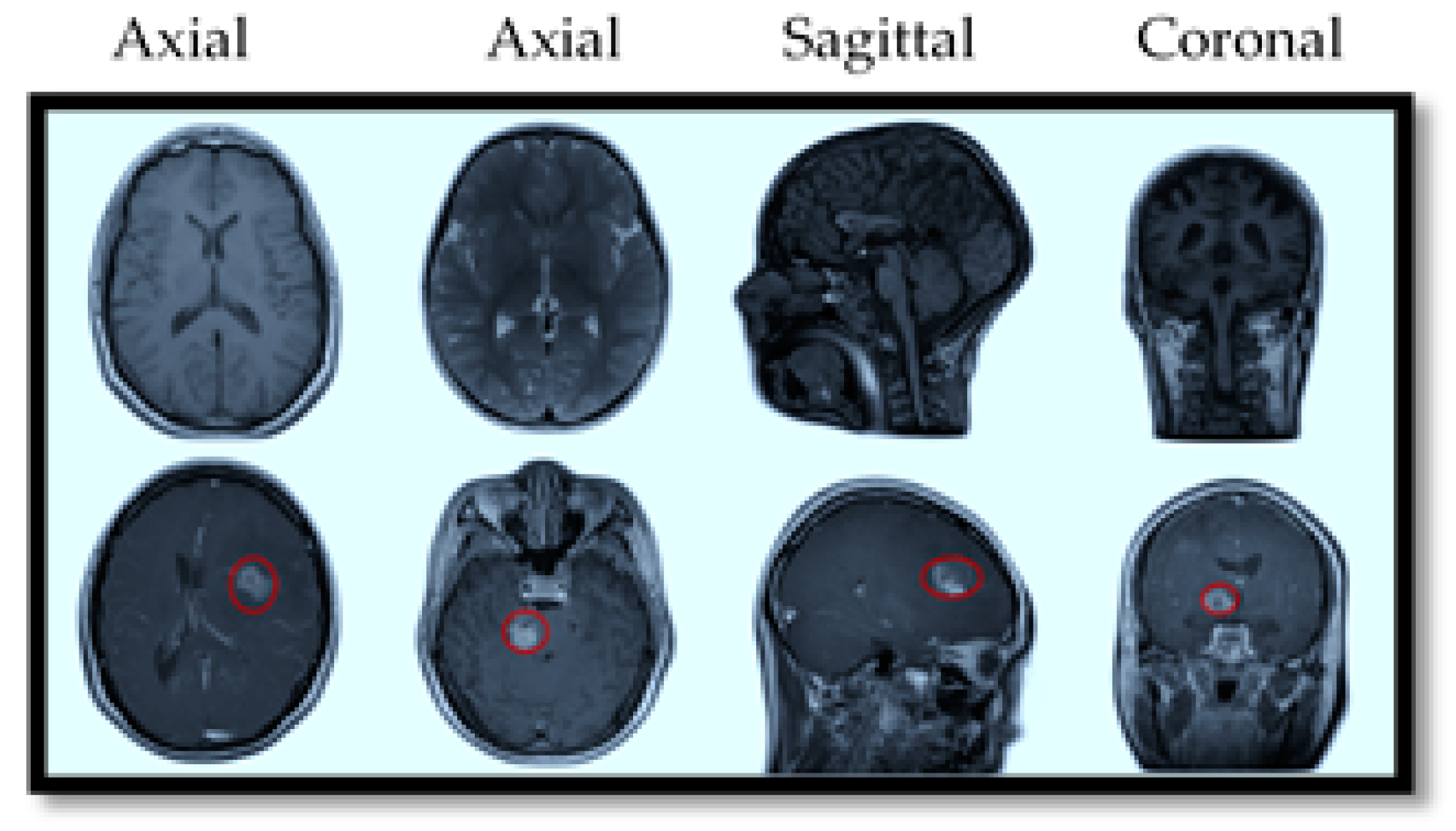
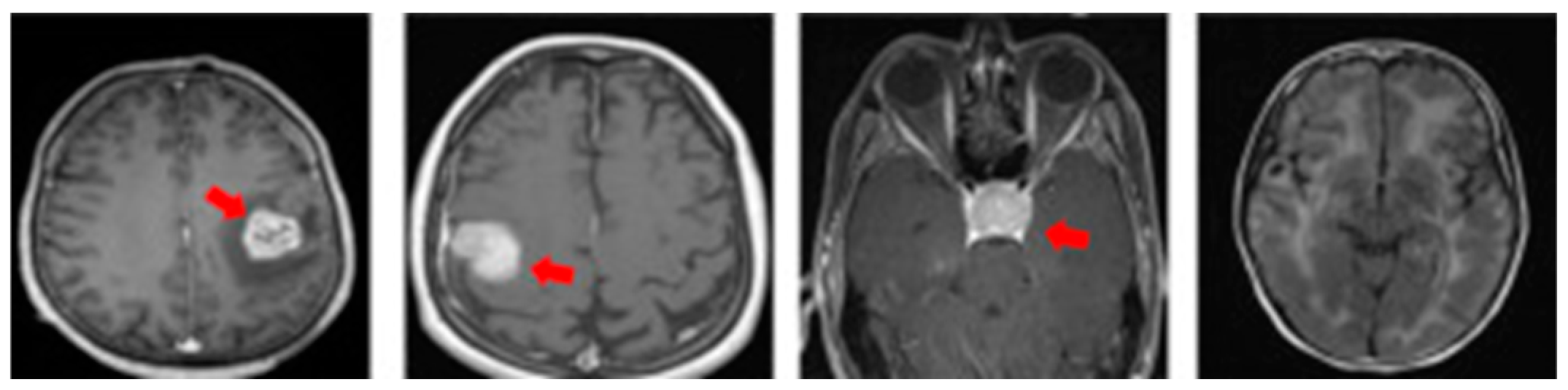
MRI also offers specialized imaging sequences that provide valuable insights into brain tumors, such as diffusion-weighted imaging (DWI), which helps differentiate tumor cells from healthy brain tissue; perfusion imaging, which assesses blood flow within the tumor to evaluate its aggressiveness; and magnetic resonance spectroscopy (MRS), which analyzes the tumor’s chemical composition to aid in diagnosis and treatment planning. For illustration purposes, Figure 2 provides a visual comparison of normal and tumorous brain MRI images with encircled areas, and Figure 3 illustrates the types of brain tumors in MRI images.
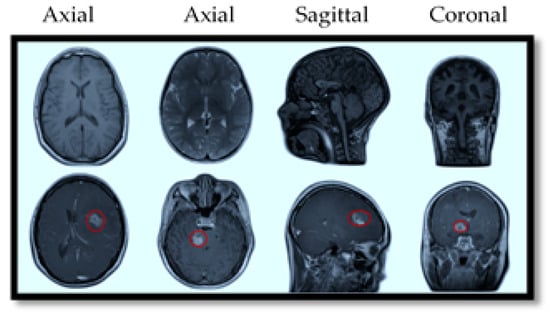
Figure 2.
MRI slices of human brain without tumor (top) and with tumor (bottom).
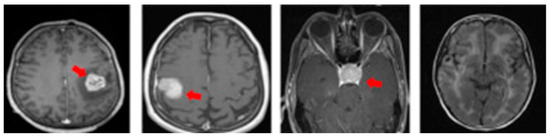
Figure 3.
MRI slices of brain tumor types (from left: glioma, meningioma, pituitary, normal).
1.2. Overview of Current Technology
Manual analysis of brain MRI images is often time-consuming and error-prone. Factors like human variability and fatigue can lead to inaccurate diagnoses. Subtle abnormalities in MRI scans are frequently missed or misinterpreted, highlighting the need for more reliable methods [9,10,11]. As technology advances, automated and computer-aided diagnostic techniques are becoming increasingly essential for improving the accuracy and reliability of brain tumor diagnosis. These methods can systematically analyze large datasets of MRI images, identifying subtle patterns that human observers may miss. By providing consistent and precise results, these techniques significantly enhance diagnostic outcomes in neuroimaging. Researchers are actively developing AI-based approaches to automate the detection of brain tumors from MRI scans. These automated systems offer the potential for more efficient and accurate diagnoses, ultimately improving patient care.
The objective of this research is to highlight research contributions conducted during the last two decades for brain MRI tumor detection and classification. The following are the main contributions of this paper:
A comparative analysis of prominent recent (the 2000–2025 period) single-view, multiple-view, and progression models is presented, detailing their limitations and outlining contemporary research trends.
A detailed section presents a comprehensive overview of datasets developed for single-view, multiple-view, and progression models, alongside a review of relevant benchmarking initiatives from 2000 to 2025.
An examination of technical challenges related to data, models, domain, benchmarking, computations, clinical applications, and practical implementation is provided. Additional discussion is presented on patient privacy, algorithmic bias, and the efforts behind developing ethical guidelines.
Special emphasis is given to future perspectives in the field, including integrated MRI sequences, federated learning, lightweight models, Explainable AI, regulatory successes, and market growth. Recent activities of funding agencies are also highlighted.
The paper is structured as follows. In Section 2, the background literature review is discussed from 2001–2010, 2010–2020, and beyond. Section 3 addresses datasets developed during that period for tumor diagnosis and prognosis, along with their specifications. The architectural models and comparative experimental results of related approaches are analyzed in Section 4. Section 5 discusses benchmarking and regulatory efforts for brain MRI tumor diagnosis. Section 6 discusses technical challenges, and Section 7 presents the future outlook related to optimized models, AI-powered diagnostic tools, Explainable AI, benchmarking, integration with clinical workflows, and market growth.
2. Research Review
DL applications in brain tumor diagnosis and prognosis have seen a surge in research over the past two decades. This work examines the evolution of DL methods for brain tumor diagnosis and prognosis, analyzing publications from 2001 to 2022. Figure 4 demonstrates a clear upward trend in publication volume, with a notable peak in 2022. This indicates a substantial increase in research activity, likely driven by the field’s growing market value and associated research and development investments. Before the 21st century, and especially before 2000, research on using DL for brain MRI tumor diagnosis was limited. This was largely due to the fact that the DL methods commonly used today, particularly convolutional neural networks (CNNs) for image analysis, only became practically feasible and widely adopted in the 2010s. As a result, DL was not a significant focus in brain MRI tumor diagnosis research before 2000. Below, the different aspects of brain MRI tumor diagnosis during 2001–2010 and 2011–2020 are discussed.
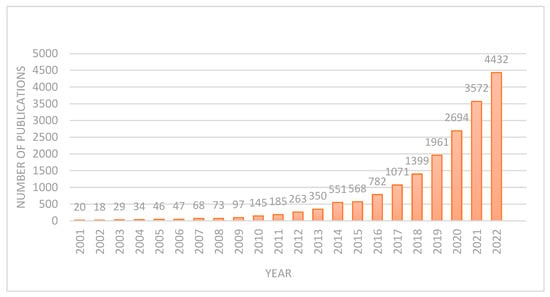
Figure 4.
Number of publications after 2000.
2001 and 2010: During this period, significant strides were made in brain tumor detection and classification using MRI images. Researchers focused on developing automated computer-aided diagnosis (CAD) systems to enhance the accuracy and efficiency of brain tumor diagnosis. Key areas of research included the extraction of features such as texture, shape, and intensity distributions; machine learning algorithms, including hybrid approaches; image processing techniques, such as histogram equalization, segmentation using region growing, edge detection employed for preprocessing, multimodal fusion, multichannel processing, and benchmark datasets and validation.
One notable development during this period was the introduction of a CAD system that combined conventional MRI and perfusion MRI for differential diagnosis of brain tumors [12]. This system involved several stages: region of interest (ROI) definition, feature extraction, feature selection, and classification. Researchers extracted various features, including tumor shape, intensity, and texture characteristics, and employed Support Vector Machines (SVMs) with recursive feature elimination for feature selection. This system was successfully applied to a diverse cohort of brain tumors, demonstrating its ability to differentiate between various tumor types, including metastases, meningiomas, and gliomas of different grades. In addition, researchers explored the use of various MRI techniques for brain tumor classification. MR spectroscopy and diffusion tensor imaging (DTI) were investigated for their potential to differentiate between primary and metastatic brain tumors. By combining multiple MRI modalities and employing advanced image processing and machine learning techniques, researchers significantly improved the accuracy and reliability of brain tumor classification. The following table (Table 1) lists a few notable studies with corresponding performance results.

Table 1.
Brain MRI tumor diagnosis models, datasets, and performance results (2001–2010).
Despite significant advancements in brain tumor detection and classification using brain MRI images between 2001 and 2010, challenges persisted, including variability in MRI scanner settings, patient-specific anatomical differences, and the computational cost of processing high-dimensional data. Notable limitations were small datasets leading to potential overfitting, binary classification, lower precision, etc. Researchers addressed these challenges by advocating more robust feature selection methods, improved model generalization, and the integration of domain knowledge into machine learning frameworks. The research during this period laid the foundation for subsequent advancements, particularly with the emergence of DL and high-performance computing. These contributions further propelled the automated brain tumor detection and classification field.
2011 and 2020: Between 2011 and 2020, research focused on machine learning algorithms for improving accuracy; DL-based methods, including multi-modal and transfer learning to improve accuracy and reduce reliance on manual feature extraction; hybrid methods, such as integrated segmentation and classification, ensemble classifiers, enhanced public and custom clinical datasets, and real-time processing [13,14]. These contributions collectively advanced the state-of-the-art in brain tumor classification, enabling reliable computer-aided diagnosis systems for radiologists.
The research study [15] presents a novel neural network (NN) approach for the automated classification of MR brain images into normal and abnormal categories. The method incorporates wavelet transform for effective feature extraction and principal component analysis (PCA) for dimensionality reduction, followed by a backpropagation algorithm optimized with the scaled conjugate gradient (SCG) algorithm for image classification. Evaluated on a dataset comprising 66 MR images (18 normal, 48 abnormal), the proposed method claims exceptional performance, achieving 100% classification accuracy on both training and test sets, requiring only 0.0451 s per image for classification.
The study [16] overcomes the limitations of manual analysis by employing an automated segmentation technique to extract tumor regions, followed by feature extraction using texture, shape, and boundary information. An ensemble classifier combining support vector machines, artificial neural networks, and k-nearest neighbors classifies tumors as benign or malignant. Evaluated on a dataset of 550 patients using leave-one-out cross-validation, the system achieved high accuracy (99.09%), with 100% sensitivity and 98.21% specificity, demonstrating its potential as a reliable tool to assist radiologists in accurate and efficient brain tumor diagnosis.
The research [17] introduces a novel brain tumor classification system that leverages a hybrid feature extraction approach in conjunction with a regularized extreme learning machine (RELM) classifier to achieve highly accurate tumor identification. The system commences with a preprocessing step involving min-max normalization to enhance image contrast, followed by the extraction of tumor features using a hybrid method. Subsequently, the RELM classifier is employed to effectively categorize tumor types. Rigorous evaluation of a newly released public brain image dataset demonstrates the superior performance of the proposed system, achieving a notable improvement in classification accuracy from 91.51% to 94.23% using the random holdout technique.
Another notable contribution involved developing an efficient multilevel segmentation method that combined optimal thresholding and watershed segmentation, followed by morphological operations to separate tumors from MRI images [18]. This approach, coupled with CNN feature extraction and Kernel Support Vector Machine classification, showed promising results in detecting and classifying tumors as cancerous or non-cancerous. The following table (Table 2) lists a few notable studies during this period with corresponding performance results.

Table 2.
Brain MRI tumor diagnosis models, datasets, and performance results (2010–2020).
Notable shifts noticed during this period were a shift to DL replacing traditional machine learning methods, the availability of larger datasets, and performance improvements in terms of accuracy and Dice score, etc. The research contributions significantly advanced the field of brain tumor detection and classification using MRI images, paving the way for more accurate and efficient diagnostic tools in clinical settings. Despite significant advancements, integrating research into clinical practice remained a persistent challenge. Efforts mostly centered on creating robust and interpretable models considered suitable for seamless integration into clinical workflows.
Research after 2020: During this period, the research related to brain MRI tumor detection and classification accelerated due to the maturity of DL-based architectures and the release of enhanced datasets.
The study in [19] introduces a significant advancement in unsupervised anomaly detection using a 3D deep autoencoder network. Trained on a dataset of 578 normal T2-weighted MRIs, this model surpasses previous methods like VAEs and GANs by 7%. However, it doesn’t account for the intricate structural differences within the human brain. Likewise, the research in [20] highlights the potential of automated machine learning for anomaly detection in medical imaging. While offering substantial advantages, it shares a common limitation with previous studies by focusing solely on detection without considering segmentation, which could provide a more comprehensive and informative analysis.
Shifting the focus to the second subcategory under anomaly detection, we encounter recent scientific studies exploring advanced methods to enhance our understanding and management of brain anomalies. Recent studies have explored advanced methods to detect and segment brain anomalies using CNNs [21]. While these models have been effective in identifying anomalies, they often struggle to differentiate tumors from other types of abnormalities. Similarly, transformer-based models [22] have shown promise in anomaly segmentation but still face challenges in accurately locating and classifying lesions. To overcome these limitations, we focus on state-of-the-art models for brain tumor detection. This category can be further divided into detection-only and segmentation-based approaches. Research in detection-only methods often prioritizes lightweight and efficient models [23] to enable deployment on various devices. However, accurate tumor boundary delineation is essential for clinical decision-making. Segmentation models, such as UNet, DeepLabv3, 3D-CNNs, ResNet50, DenseNet, and GANs [24,25], have shown promise in precisely identifying tumor boundaries, thereby improving the accuracy and reliability of brain tumor detection and analysis.
Vankdothu et al. [26] emphasize the challenges of early tumor detection and the need for advanced imaging techniques. While their proposed CNN-LSTM model claims an accuracy of 92%, the lack of supporting evidence raises doubts about its effectiveness. Additionally, the model lacks a robust segmentation strategy. Integrating MRI-based segmentation techniques could improve detection accuracy and understanding of complex tumor structures, as accurate segmentation is crucial for determining tumor location and size. The DeepSeg framework [27] proposes a generic UNet-based architecture for tumor segmentation. While it utilizes multiple DL models for feature extraction, it lacks automation and is outperformed by alternative methods in certain cases. The model’s limitations include a narrow focus on specific tumor boundaries, difficulties in handling diverse brain structures, and a lack of computational efficiency considerations. Integrating advanced MRI segmentation techniques could improve the model’s accuracy and adaptability.
Combining neuroimaging with histopathology may enhance glioblastoma management. The authors [28] investigate predictive models for high accuracy (AUC 0.902) for glioma-grade prediction. The results showed that FLAIR abnormality resection improved survival, while DWI best depicted tumor infiltration. Glioblastomas exhibited irregular shapes, margins, and enhancement, whereas metastases appeared round with clear edges and uniform contrast. Further studies with larger datasets are needed for validation.
Overall, this period marks a significant transition from proof-of-concept DL models to clinically viable tools that promise to revolutionize brain tumor diagnosis while addressing key challenges in standardization and validation. Based on the literature review, it was inferred that most of the approaches are tested on a single limited-size dataset; most of the works did not detect the type of the tumor (shown in Figure 3), and few research studies addressed multiple views of MRI images (shown in Figure 2) and exploited them for progression of tumor stages and thus helped the physician plan treatment accordingly. Despite this, benchmarking and regulatory efforts accelerated to bridge the clinical translation of these efforts.
3. Datasets
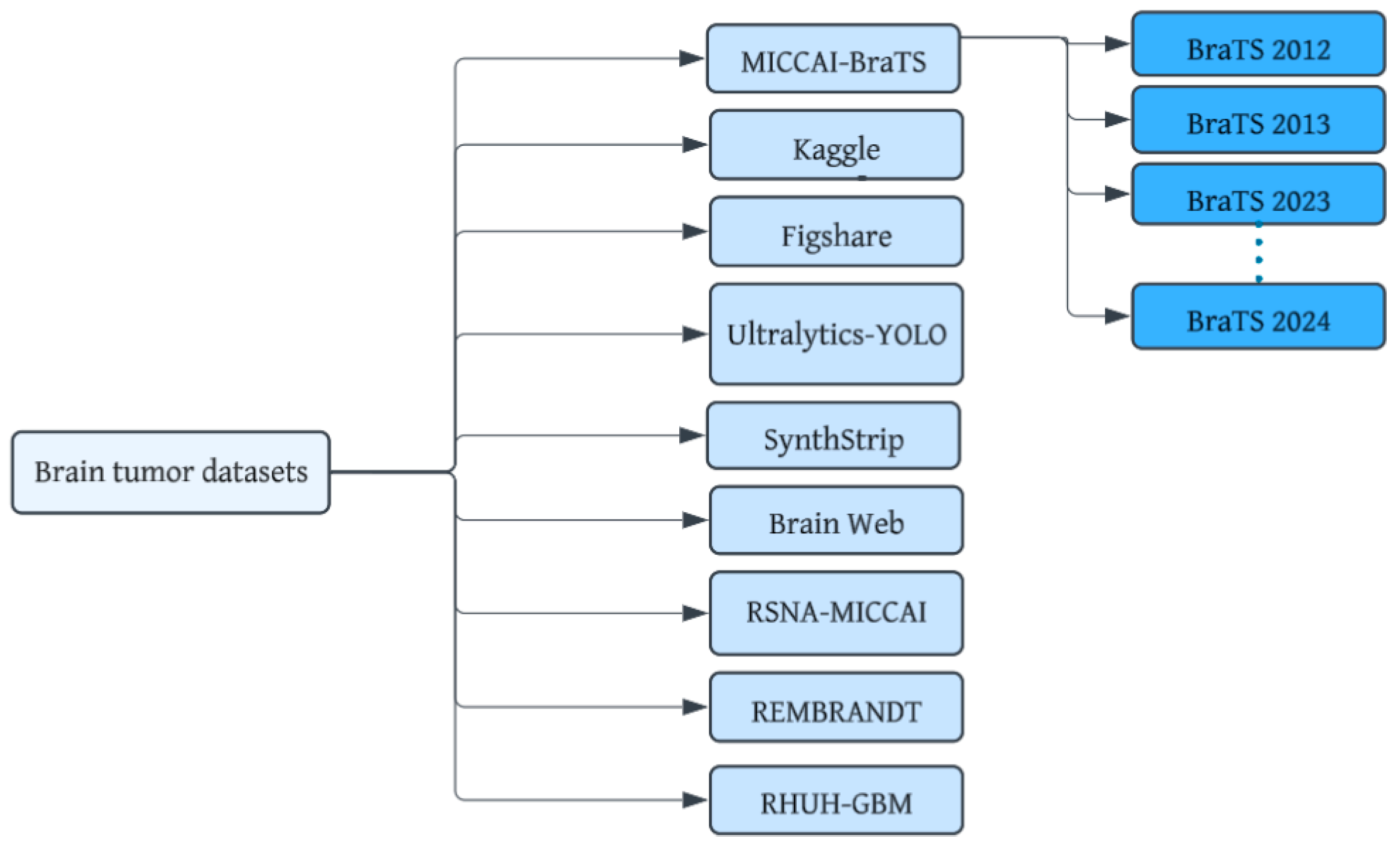
Brain tumor datasets are a cornerstone for bridging the gap between cutting-edge technology and real-world medical applications. These datasets provide the foundation for developing sophisticated machine-learning models capable of accurately recognizing complex patterns within brain images. Figure 5 shows datasets that have been developed for brain tumor diagnosis using MRI images. Important considerations include dataset size and diversity, image quality, annotation quality, data privacy, and ethics. The notable datasets are detailed below:
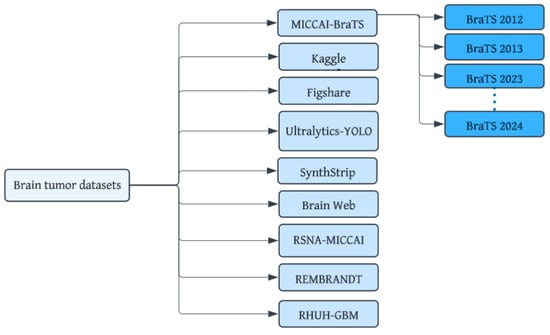
Figure 5.
Brain MRI tumor datasets.
Kaggle (SARTAJ + Br35H) dataset: This is a comprehensive open-access dataset [29] containing images with four classes (glioma, pituitary, meningioma, no tumor).
Figshare Brain Tumor MRI dataset: This dataset [30] categorizes MRI images into four types: glioma, meningioma, no tumor, and pituitary tumor.
Ultralytics YOLO: This is a larger dataset [31] containing 1116 structured format images in the directory structure for YOLO models.
BraTS (Brain Tumor Segmentation Challenge) dataset: This annually updated dataset [32] offers MRI images with detailed annotations for tumor segmentation, making it a popular choice for researchers. The dataset includes four structural MRI modalities: T1-weighted, post-contrast T1-weighted, T2-weighted, and T2-FLAIR volumes.
TCGA (The Cancer Genome Atlas) dataset: A comprehensive resource, TCGA includes a vast collection of clinical and imaging data [33], including MRI scans together with manual FLAIR abnormality segmentation masks.
RSNA-MICCAI Brain Tumor Radiogenomic Classification Challenge Dataset: This dataset with associated clinical and genomic data was specifically designed for brain tumor classification as a part of the 10th anniversary of the BraTs challenge [34].
View-specific datasets: The neurofeedback skull-stripped (NFBS) Repository [35], the SynthStrip dataset [36], and the Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2016 Challenge Dataset (MSSEG) [37] may be used for training, validation, and testing of T1, T2-weighted, FLAIR, and PD MRI sequences.
REMBRANDT dataset: The REMBRANDT dataset [38] comprises data from 874 glioma specimens. It includes pre-surgical magnetic resonance multi-sequence images (from 130 patients) linked to the clinical data in the larger REMBRANDT collection.
Brain Cancer for Tumor Recurrence Dataset: This dataset [39] is developed from 47 patients (21 males, 26 females) and includes T1 MPRAGE with gadolinium contrast. This includes MRI scans that show the progression of brain tumors over time (pre-operative and post-operative MRI scans).
Brain-Tumor-Progression dataset: This dataset [40] is a collection of MRI data from 20 glioblastoma patients. It includes pre- and post-chemo-radiation therapy scans acquired within 90 days of treatment initiation and at the time of tumor progression. The dataset encompasses a range of MRI sequences, including T1-weighted, FLAIR, T2-weighted, ADC, and perfusion images.
RHUH-GBM dataset: This dataset [41] comprises 600 MRI series acquired at three time points: preoperatively, early postoperatively, and during follow-up. It includes T1-weighted, T2-weighted, FLAIR, T1-contrast-enhanced, and ADC map sequences. Additionally, expert-validated segmentations of tumor sub-regions are available for all three time points.
Multi-center, multi-origin brain tumor MRI (MOTUM) dataset: This dataset [42] is collected from 67 patients with various types of brain tumors and includes FLAIR, T1-weighted, contrast-enhanced T1-weighted, and T2-weighted sequences. This dataset offers data for assessing disease status and progression for multi-origin brain tumors.

Table 3.
Specifications of MRI datasets (with no tumor progression).

Table 4.
Specifications of view-specific MRI datasets.

Table 5.
Specifications of MRI datasets (with tumor progression).
4. Architecture Models
In this section, various single-view and multiple-view brain MRI tumor models are discussed, along with experimental results.
4.1. Single-View Brain MRI Tumor Models
Brain tumor detection and classification methods primarily utilize CNNs as their backbone. While sharing architectural similarities, these methods diverge in depth, complexity, and specific components. Hybrid approaches combining vision transformers and recurrent units have emerged, offering improved feature extraction and relationship identification. Transfer learning with pre-trained models like EfficientNets, ResNets, MobileNets, or VGG variants is commonly employed to leverage prior knowledge. Newer architectures integrate object detection and segmentation, while multi-task models aim to simultaneously detect, classify, and localize tumors. Attention mechanisms are incorporated to focus on relevant features, and neural architecture search automates the design of optimal network structures. Data augmentation, addressing class imbalance, and various optimization techniques are commonly used to improve accuracy and efficiency in these tasks. Below, well-known architectural models are briefly discussed to highlight the significance of such architectures:
BCM-CNN: The research [43] introduces a state-of-the-art 3D CNN model that combines the benefits of sine, cosine, and grey wolf optimization algorithms. By utilizing the pre-trained Inception-ResNetV2 model, the BCM-CNN effectively extracts relevant features from brain MRI images. When evaluated on the challenging BRaTS 2021 Task 1 dataset, the model achieved an impressive accuracy of 99.98%.
AlexNet-based CNN: The study [44] introduces a hybrid DL architecture that integrates the strengths of CNNs and recurrent neural networks (RNNs). Specifically, the model combines AlexNet and Gated Recurrent Units (GRU) to extract both spatial and temporal features from brain tumor images. The proposed model demonstrates impressive performance, achieving 97% accuracy, 97.63% precision, 96.78% recall, and a 97.25% F1-score.
GoogleNet-based model: The study [45] proposes a novel approach for brain tumor classification using a modified GoogleNet architecture. The researchers fine-tuned the last three fully connected layers of the pre-trained GoogleNet model on a dataset of 3064 T1w (Figshare) MRI images. By combining GoogleNet with SVM, the model significantly improved classification accuracy, reaching approximately 98% for distinguishing between glioma, meningioma, and pituitary tumors.
VGG19 with SVM: The research [46] introduces a novel approach for brain tumor classification that integrates the power of CNNs and support vector machines (SVMs). The model utilizes the pre-trained VGG19 architecture to extract high-level features from MRI images. Subsequently, SVM classifiers are employed to accurately classify different types of brain tumors. The model demonstrates superior performance, achieving an accuracy of 95.68% on the Brats and Sartaj datasets.
VGG16 and VGG19 with ELM: The authors in [47] present a multimodal DL framework for accurate brain tumor classification. The model utilizes VGG16 and VGG19 pre-trained CNNs to extract relevant features from MRI images. Subsequently, an Extreme Learning Machine (ELM) classifier, enhanced with a correntropy-based feature selection strategy, is employed for the final classification task. The model demonstrates superior performance on the BraTS2015, BraTS2017, and BraTS2018 datasets, achieving accuracy rates of 97.8%, 96.9%, and 92.5%, respectively.
EfficientNets: The study [48] proposes a novel DL approach that utilizes transfer learning to detect brain tumors. The model incorporates six pre-trained architectures: VGG16, ResNet50, MobileNetV2, DenseNet201, EfficientNetB3, and InceptionV3. To enhance performance, the models are fine-tuned using Adam and AdaMax optimizers. The proposed approach demonstrates high accuracy, ranging from 96.34% to 98.20%, while requiring minimal computational resources. Another research [49] introduces a hybrid DL approach for accurate brain tumor classification. The model utilizes EfficientNets, a state-of-the-art CNN architecture, for feature extraction. Grad-CAM visualization is employed to understand the model’s decision-making process. The model demonstrates superior performance on the CE-MRI Figshare dataset, achieving an impressive accuracy of 99.06%, precision of 98.73%, recall of 99.13%, and F1-score of 98.79%.
YOLO NAS: The research [50] investigates the application of the YOLO NAS (Neural Architecture Search) DL model for accurate brain tumor detection and classification in MRI images. The model’s performance is enhanced by a segmentation process utilizing a deep neural network with a pre-trained EfficientNet decoder and a U-Net encoder. The dataset, consisting of 2570 training images and 630 validation/testing images, was used to train and evaluate the model. The results demonstrate exceptional performance, with 99.7% accuracy, a 99.2% F1-score, and other key metrics exceeding 98%.
Hybrid ViT-GRU: The authors [51] introduce a hybrid DL model that combines the strengths of Vision Transformers (ViT) and GRU for effective brain tumor detection and classification. To improve model transparency, Explainable AI techniques, such as attention maps, SHAP, and LIME, are integrated. The model was evaluated on the BrTMHD-2023 and brain tumor Kaggle datasets, achieving remarkable results, including a 98.97% F1-score and 96.08% accuracy, respectively.
MobileNetv3: The study [52] investigates the potential of the MobileNetv3 architecture for improving brain tumor diagnosis accuracy. The model was trained and validated on the Kaggle dataset, with image enhancement techniques applied to balance the dataset. A five-fold cross-validation strategy was employed to ensure robust performance. The proposed approach, which integrates the DenseNet201 architecture with Principal Component Analysis (PCA) and Support Vector Machines (SVM), demonstrated exceptional performance. It achieved 100% accuracy, recall, and precision on dataset 1 and 98% accuracy on dataset 2.
UNet: The architecture comprises two key pathways: a contracting path (encoder) and an expansive path (decoder), creating a distinctive U-shape [53,54]. This design enables the network to effectively grasp local and global information within the image, making it well-suited for the accurate segmentation of tumors. Several variations of the U-Net have been developed, including the 3D U-Net [55], which is designed to capture spatial context across multiple image slices, hybrid approaches such as VGG16-U-Net [56] that leverage the feature extraction capabilities of pre-trained networks like VGG16, and YOLO-U-Net [57], among others.
Hybrid models: The study [58] introduces a cutting-edge approach to brain tumor classification, leveraging DL and advanced optimization techniques. The framework involves modifying pre-trained neural networks, utilizing a quantum theory-based Marine Predator Optimization algorithm for feature selection, and employing Bayesian optimization for hyperparameter tuning. By fusing features through a serial-based approach, the proposed framework achieved remarkable performance on an augmented Figshare dataset, with an accuracy of 99.80%, sensitivity of 99.83%, precision of 99.83%, and a low false negative rate of 17%.
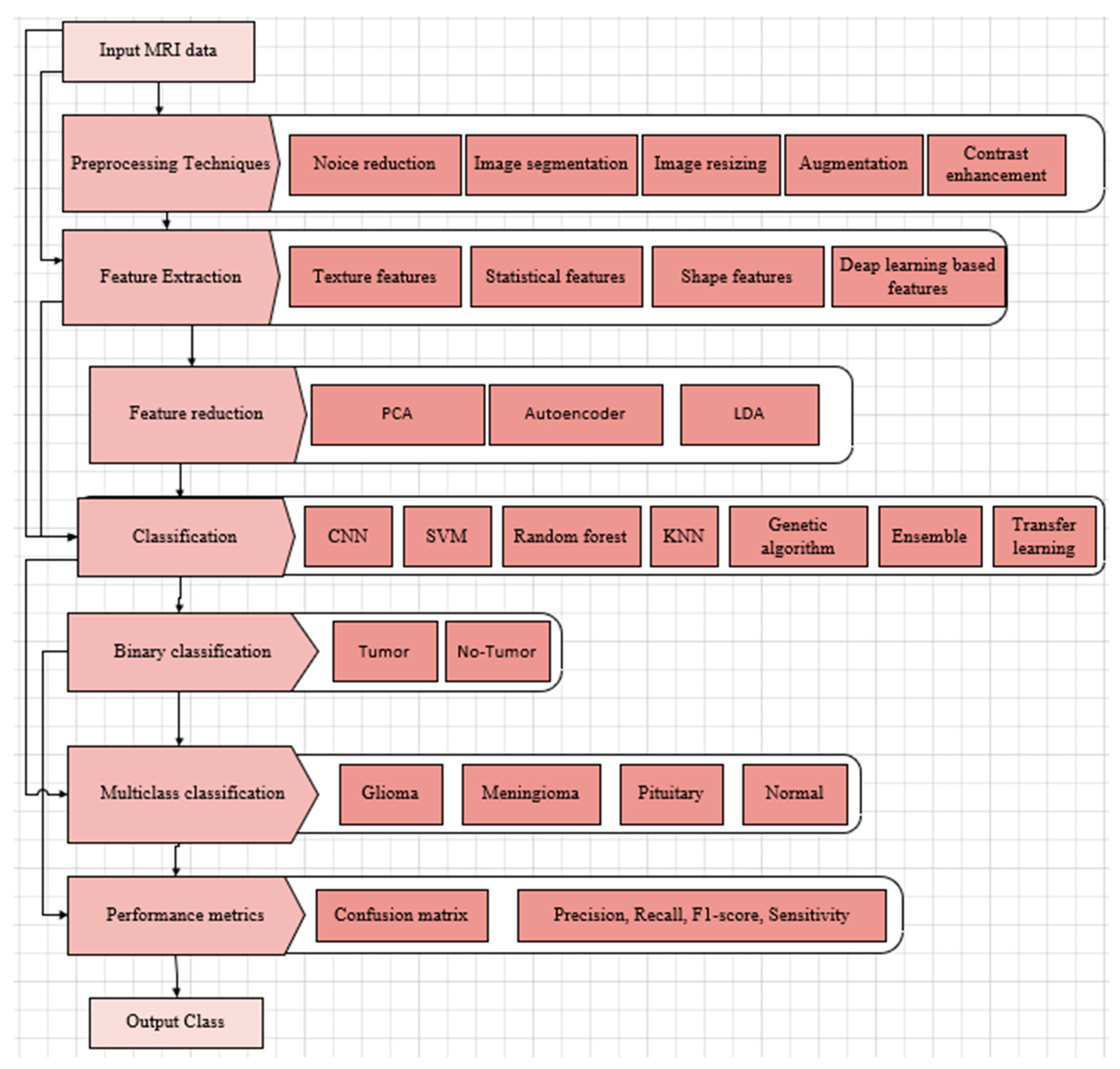
Based on the discussion in this section, two important aspects of the architectural models discussed in this section can be outlined. The first aspect is the clinical application, shown in Table 6, where each row shows which of the applications (classification, detection, segmentation) is targeted by each model. The other aspect is the process diagram presented in Figure 6. The process begins with an MRI brain tumor dataset (shown in Table 3) that enters image pre-processing, including noise reduction, resizing, contrast enhancement, and normalization.

Table 6.
Models mapped to clinical applications.
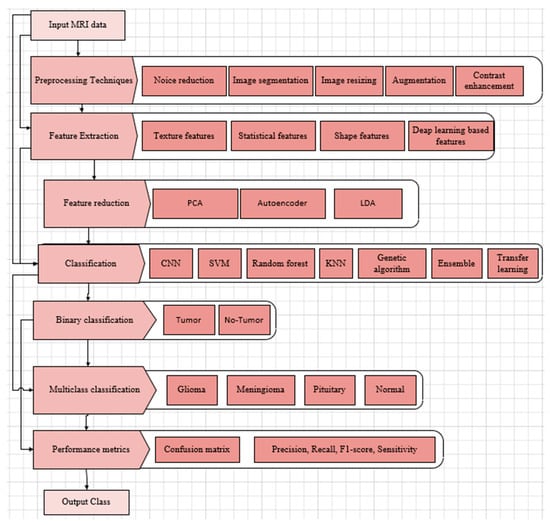
Figure 6.
DL-based brain tumor detection and classification process.
Accurate segmentation of brain tumors in MRI scans is essential for effective diagnosis, treatment planning, and survival prediction. A variety of techniques are used to distinguish tumor regions from healthy brain tissue. Automatic segmentation methods strive to fully automate this process, utilizing approaches such as thresholding (which separates tissues based on intensity), region-based techniques (grouping pixels with similar properties), edge-based methods (detecting boundaries), and clustering algorithms (grouping voxels by feature similarity). Modern methods are designed to capture unique pathological features. Recent DL models have advanced the field by focusing computational attention on tumor regions, achieving high Dice scores for both whole tumor and tumor core segmentation. However, challenges persist due to variations in tumor size, shape, location, and appearance, along with image artifacts and ambiguous tumor boundaries. The ongoing improvements in segmentation highlight the field’s progress toward reliable, automated clinical tools while also emphasizing the need for larger datasets and better generalization across diverse imaging conditions and tumor types.
Data augmentation is employed to expand the dataset and ensure a balanced representation of different tumor types. In some models, it may be desirable to have features extracted and reduced. The augmented dataset is partitioned into training and testing sets (typically 80% and 20%, respectively). DL models are then selected, modified if needed, and then trained on the training set, with hyperparameters optimized for optimal performance. In some instances, additional optimization is performed to refine the feature set with an additional feature fusion mechanism in case multiple models are trained. Next, a classifier is employed—either binary to detect the presence of a brain tumor or multiclass to identify specific tumor types. The trained model is then evaluated on the held-out test set using a set of performance metrics.
Comparative Performance of Single-View MRI Models
The performance of single-view brain MRI tumor models can be judged by examining the evaluation of well-known models found in the literature. Table 7 lists several single-view brain MRI tumor models, along with datasets used and estimated performance metrics. Table 7 shows that the DL methods used demonstrate superior performance for the detection and classification of brain tumors involving metrics such as accuracy, precision, recall, and F1-score. These methods achieved high accuracy rates, generally exceeding 93%.

Table 7.
Comparative performance of recent single-view brain MRI brain tumor models.
Despite impressive performance metrics, many models face critical limitations that hinder their real-world clinical application. Three major challenges—data bias, validation practices, and model generalizability—highlight a significant disconnect between laboratory success and practical utility. Data bias remains a foundational issue, beginning with pronounced class imbalances in training datasets. Moreover, variations in imaging protocols introduce further complications. Models trained on single-institution MRI data frequently experience performance drops when applied to external datasets. Validation practices also reveal systemic flaws. Many models are evaluated using internal datasets, which tend to overstate readiness for clinical deployment. External validation remains inconsistent, with test sets often lacking diversity in scanner types and patient demographics, undermining confidence in model robustness. Generalizability is the ultimate determinant of clinical viability. Single-plane models are particularly vulnerable to view dependency, with performance declining when processing coronal or sagittal MRI slices—views commonly used in clinical settings. Real-world imaging artifacts, such as motion blur, further degrade performance, especially in tumor segmentation tasks. Additionally, clinicians face trust barriers, citing a mismatch between AI-generated tumor boundaries and radiologist annotations, along with “black box” decision-making processes. In response, the field is shifting toward multi-center trials, emphasizing diverse patient cohorts and standardized imaging protocols to bridge the gap between controlled research settings and real-world clinical environments.
4.2. Multiple-View Brain MRI Tumor Models
MRI scans are typically obtained in three anatomical planes—axial, coronal, and sagittal—each offering distinct views of brain structures and abnormalities, illustrated in Figure 2. Utilizing data specific to these planes in brain tumor detection and classification models allows for a comprehensive understanding of tumor characteristics. These MRI view-specific models capitalize on the strengths inherent to each anatomical plane, enhancing diagnostic precision and accuracy. Figure 3 shows these multiple-view MRI slices and brain tumor classifications.
Meningiomas, typically benign but potentially problematic, originate near the protective membranes of the brain and spinal cord [59]. Gliomas, arising from glial cells, are the deadliest brain tumors and constitute about one-third of all cases [60]. Pituitary tumors are generally benign growths within the pituitary gland [61]. While accurate diagnosis is vital for determining related treatment, traditional biopsy methods face challenges due to their invasive nature, time consumption, and potential for non-representative sampling [62,63]. Moreover, histopathological grading based on biopsies is limited by intratumor variability and subjective interpretation among pathologists [64], complicating the diagnostic process and restricting treatment options.
The axial (transverse) view offers a horizontal cross-section of the brain, dividing it into upper (superior) and lower (inferior) sections. This view is extensively used in clinical practice because it provides a detailed visualization of key brain regions, such as the ventricles, corpus callosum, and basal ganglia. Axial images are predominant in clinical datasets because of their widespread use in diagnostic procedures. DL models such as GoogLeNet, InceptionV3, DenseNet201, AlexNet, and ResNet50 [43,65] are trained specifically on axial slices to analyze spatial relationships and capture critical features across slices. The models can also be used to capture spatial dependencies across slices.
The coronal view offers a frontal cross-section of the brain, dividing it into anterior and posterior regions. This orientation is especially advantageous for examining the brainstem, thalamus, and temporal lobes. It is particularly effective in identifying tumors located in midline structures, such as pituitary adenomas and gliomas in the temporal lobe. Additionally, the coronal perspective is critical for assessing brain symmetry, which helps detect mass effects and midline shifts caused by tumors. Coronal datasets require meticulous preparation since they are less commonly used on their own than axial views. Convolutional neural network (CNN) architectures like ResNet-50, AlexNet, VGGNet, and MobileNet-v2 [43,66,67] can be fine-tuned to process coronal slices effectively. Incorporating attention mechanisms can enhance the model’s ability to focus on midline structures. Moreover, pre-trained models developed for axial views can be further trained on coronal images to take advantage of shared features between the two perspectives.
The sagittal view presents a side-oriented cross-section of the brain, dividing it into the left and right hemispheres. This perspective is particularly important for examining midline structures and understanding overall brain morphology. Sagittal images are crucial for evaluating tumors in areas like the corpus callosum and brainstem. Additionally, they help detect structural abnormalities such as ventricular compression or displacement of the cerebellum caused by tumors. Although sagittal datasets are often smaller, they contain distinctive features essential for diagnosing specific tumor types. To analyze spatial patterns effectively across sagittal slices, models [43,67,68,69] often employ sequence-based architectures, such as RNNs or transformer models, which can capture the sequential dependencies within the data.
While single-view models demonstrate efficacy for specific tasks, integrating information from axial, coronal, and sagittal MRI views offers a more comprehensive understanding of tumor characteristics, with approaches ranging from feature fusion using separate DL models for enhanced classification to employing attention layers for dynamic weighting of view contributions, utilizing 3D input volumes to capture spatial relationships across planes, and combining predictions from view-specific models through voting or averaging mechanisms, ultimately leading to more robust and accurate brain tumor detection and classification.
Multi-view modeling in brain MRI analysis faces challenges such as data imbalance due to the predominance of axial views in datasets, necessitating augmentation techniques for balanced training while also demanding significant computational resources, particularly for 3D CNNs. The architecture primarily remains the same, as shown in Figure 6. Accurate model training demands consistent alignment of anatomical structures across all views. Despite these challenges, integrating axial, coronal, and sagittal views provides comprehensive diagnostic insights. The axial view offers broad information, and coronal and sagittal views contribute critical details about midline structures and tumor morphology. When effectively implemented, multi-view modeling can enhance diagnostic accuracy. However, future research must address issues like data imbalance, computational efficiency, generalization, and the impact of subtle variations in slice thickness and orientation. This research is crucial to fully realize the potential of view-specific and multi-view models in clinical AI applications.
Comparative Performance of Multiple-View Brain MRI Tumor Models
The performance of multiple-view MRI brain tumor models can be assessed by analyzing the performance of multiple-view MRI brain tumor models found in the literature. Table 8 provides a list of various multiple-view MRI brain tumor models, along with the datasets (Table 3 and Table 4), and the corresponding performance metrics. As shown in Table 8, multiple-view brain tumor MRI models exhibit superior performance in detecting and classifying brain tumors, achieving high accuracy rates—often exceeding 98%—based on metrics such as accuracy, precision, recall, specificity, and F1-score.

Table 8.
Comparative performance of recent view-specific methods for brain MRI tumor.
Despite achieving high accuracy, multi-view brain tumor MRI models face significant hurdles in clinical translation due to data bias stemming from imbalanced, demographically narrow datasets and institution-specific scanner protocols, which often lead to performance degradation on external data. Validation practices frequently suffer from overly optimistic internal testing and cross-validation leakage, undermining model generalizability across different MRI views and making them vulnerable to real-world imaging artifacts. Clinician adoption is further hindered by the “black-box” nature of model-generated tumor boundaries, which limits trust. Although multi-center trials and Explainable AI offer promising paths forward, progress remains constrained by the limited availability of annotated data and ongoing patient privacy concerns, highlighting the urgent need for diverse, representative datasets and standardized imaging protocols to enable broader clinical integration.
4.3. Brain MRI Tumor Progression Models
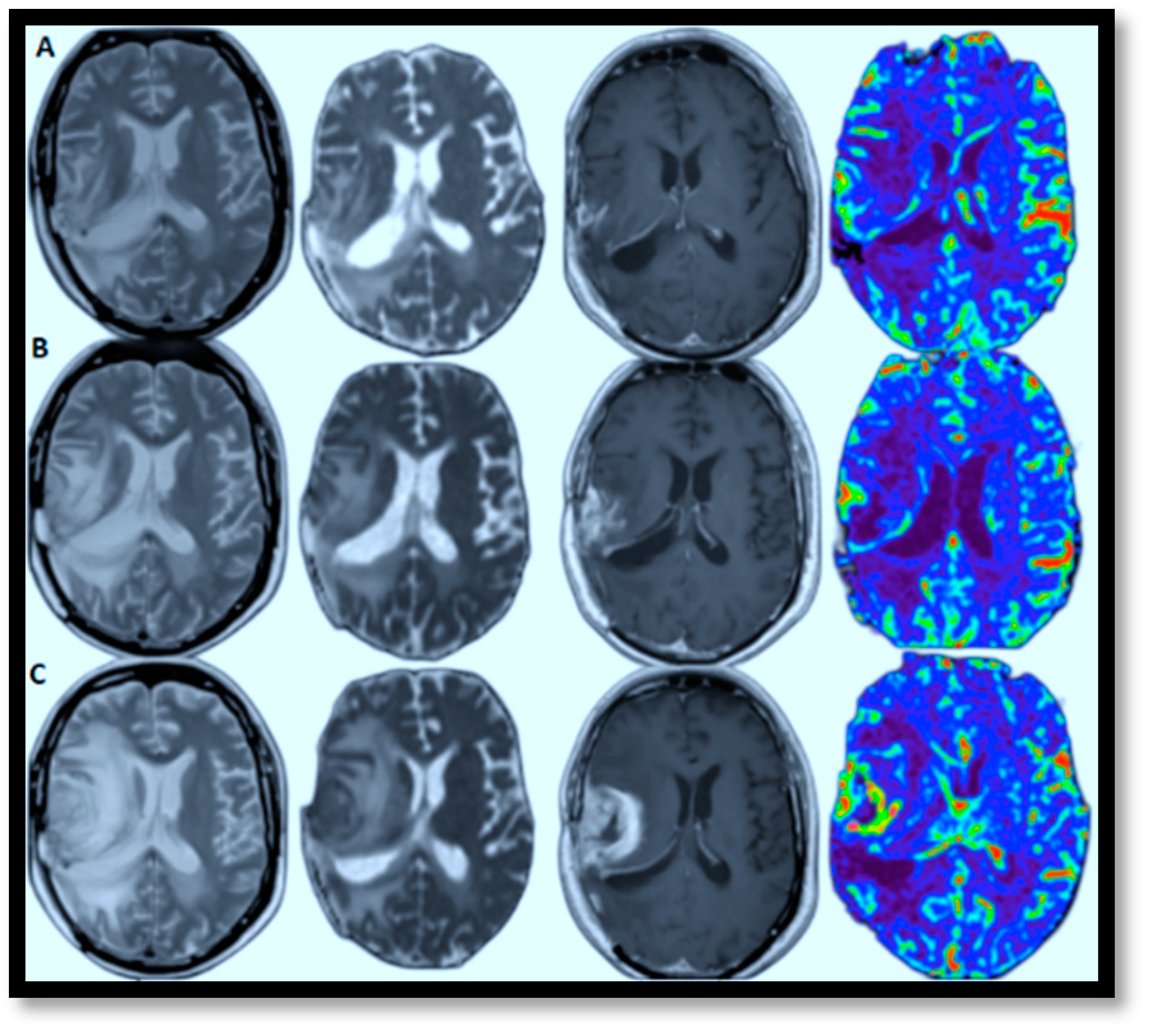
AI-powered predictive analytics enables proactive and personalized interventions by forecasting disease progression. In brain MRI brain tumor progression modeling, AI methods excel because they can identify complex patterns and relationships within high-dimensional data. These approaches effectively process large datasets, capture non-linear relationships, and learn features directly from raw data, which is crucial for medical imaging. View-specific brain MRI models exemplify this, demonstrating automated feature extraction with reliable accuracy and scalable architecture. This facilitates personalized medicine by identifying critical regions driving disease progression. Figure 7A–C illustrates this by showcasing glioblastoma progression through (T2, ADC map, T1-enhanced, and CBV) MRI scans taken at the 2nd, 4t, and 6th months, highlighting relapses and guiding the development of tailored treatment plans [70]. The color in the rightmost image signifies the volume of blood in a given amount of brain tissue. Below, a few specific models are discussed that have successfully tried to model brain MRI tumor progression with reasonable accuracy.
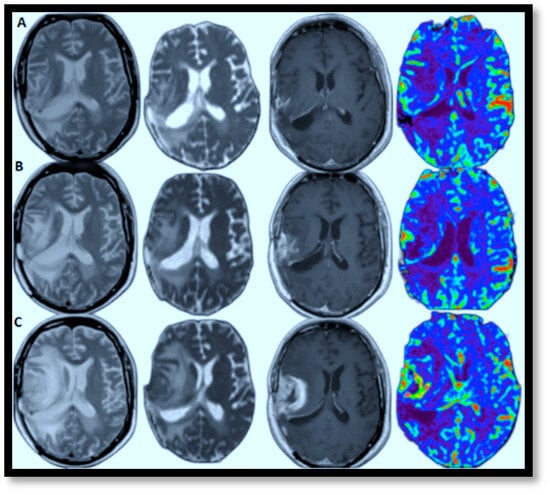
Figure 7.
Relapsing of Glioblastoma (lesion edema degree aggravated from left to right: T2, ADC map, T1-enhanced, and CBV-MRIs performed in the 2nd, 6th, and 8th months, respectively).
The study [71] introduces a new DL method for analyzing glioblastoma multiforme (GBM) tumors. By developing a model that estimates how quickly tumor cells spread (diffusivity) and multiply (proliferation rate), the researchers can predict how the tumor will grow over time. The model was tested on both simulated and real patient data, successfully generating complete growth trajectories for all five GBM patients in the study. Importantly, the model not only predicts tumor growth but also provides an assessment of the reliability of its predictions. This research demonstrates a significant step forward in using DL to understand brain tumors through DWI. These findings could lead to more accurate and individualized treatment plans for GBM patients.
The research [72] introduces a novel method for brain tumor segmentation that combines tumor growth modeling with DL. The approach leverages the Lattice Boltzmann Method (LBM) to extract intensity features from initial MRI scans, enhancing segmentation accuracy, and a Modified Sunflower Optimization (MSFO) algorithm for optimization. Furthermore, the method incorporates texture features such as fractal and multi-fractal Brownian motion (mBm). The extracted features are then fed into a full-resolution convolutional network (FrCN) for final segmentation. Evaluated on three benchmark datasets (BRATS 2020, 2019, and 2018), the method demonstrated high accuracy, achieving 97%, 95.56%, and 95.23%, respectively.
The research [73] introduces an Enhanced Fuzzy Segmentation Framework (EFSF) designed to extract white matter from MRI scans. Recognizing the critical role in diagnosing neurological disorders, EFSF builds upon the traditional Fuzzy C-means (FCM) clustering technique and refines the derivation of fuzzy membership functions and prototype values, leading to enhanced segmentation accuracy. The white matter region is identified as the area with the highest prototype value. When evaluated on a dataset of 100 MR images, EFSF demonstrated a Dice Similarity Index of 0.8051 ± 0.0577, indicating strong agreement with reference segmentations. These results suggest that EFSF offers a promising solution for white matter segmentation in MR images, potentially improving the assessment of white matter atrophy across various neurological disorders.
The research [74] investigates both linear and nonlinear models for simulating brain tumor growth through numerical methods. Utilizing the Crank-Nicolson scheme, a finite difference approach, the study performed simulations to examine tumor characteristics such as peak concentration and the total count of cancerous cells. Although specific outcomes are not detailed, the work assesses the effectiveness of linear versus nonlinear models in forecasting tumor development patterns. This contributes to computational oncology by enhancing the understanding of how different mathematical frameworks can represent the intricate progression of brain tumors, potentially influencing clinical decision-making in treatment and prognosis.
The study [75] develops a mathematical model to explore the intricate interplay between key components of the immune system (dendritic cells and cytotoxic T-cells) and distinct cancer cell populations (cancer stem cells and non-stem cancer cells). The researchers employed a system of ordinary differential equations to simulate the impact of immunotherapy, specifically dendritic cell vaccines and T-cell adoptive therapy, on tumor growth, both in the presence and absence of chemotherapy. The model successfully replicated several experimental observations in the scientific literature, including the temporal dynamics of tumor size in in vivo studies. Notably, the model revealed a crucial finding: chemotherapy can inadvertently increase tumor growth, while immunotherapy targeting cancer stem cells can effectively reduce tumorigenicity.
The inherent complexity of GBM—characterized by its heterogeneous enhancement, irregular and infiltrative growth patterns, and considerable variability among patients—complicates accurate progression detection. Although imaging technologies have advanced significantly, reliably identifying and differentiating true tumor growth from treatment-related effects, such as pseudo-progression or radiation necrosis, remains a clinical obstacle. The review [76] focuses on existing criteria for assessing tumor progression and highlights the difficulties these methods face in achieving precise detection and consistent application in clinical practice.
Comparative Performance of Brain MRI Tumor Progression Models
Analyzing tumor progression with MRI scans is challenging due to inconsistencies in scan intervals and image quality. The scarcity of annotated data limits the model’s ability to accurately learn tumor growth patterns. The complex nature of tumor evolution, influenced by treatment and biological factors, adds to the difficulty. Furthermore, the high computational demands of these models make them less practical for immediate use in clinical settings.
The effectiveness of current MRI brain tumor progression models can be evaluated by reviewing recent studies in the literature. Table 9 presents a compilation of various MRI brain tumor progression models, detailing the datasets (Table 3 and Table 5) and their corresponding performance metrics. As illustrated in Table 9, these models demonstrate exceptional accuracy in predicting tumor progression, often exceeding 93% based on metrics such as accuracy, precision, recall, specificity, F1-score, Dice score index (DSI), and area under the curve. Despite impressive accuracy, the models face substantial limitations in real-world clinical use. Data bias poses a major challenge, as most models are trained on imbalanced datasets that underrepresent rare tumor types and diverse patient groups.

Table 9.
Comparative performance of brain tumor progression methods.
A lack of standardization across imaging protocols, scanner vendors, and annotation methods further undermines model reliability, often leading to significant performance declines. Internal evaluations may report near-perfect accuracy, yet external tests expose sharp drops in performance, such as drastically reduced Dice scores for metastases detection. Generalizability remains a key barrier to clinical adoption, as models struggle to maintain accuracy across various MRI views, scanner types, and common real-world artifacts. Without improved robustness and adaptability, the broader clinical impact of these models remains highly constrained.
5. Benchmarking and Regulatory Efforts
In this section, frequently used performance metric benchmarks and regulatory efforts of professional and government bodies are mentioned to apprise the translation pace of academic and commercial research into the clinical environment.
5.1. Performance Metric Benchmarking
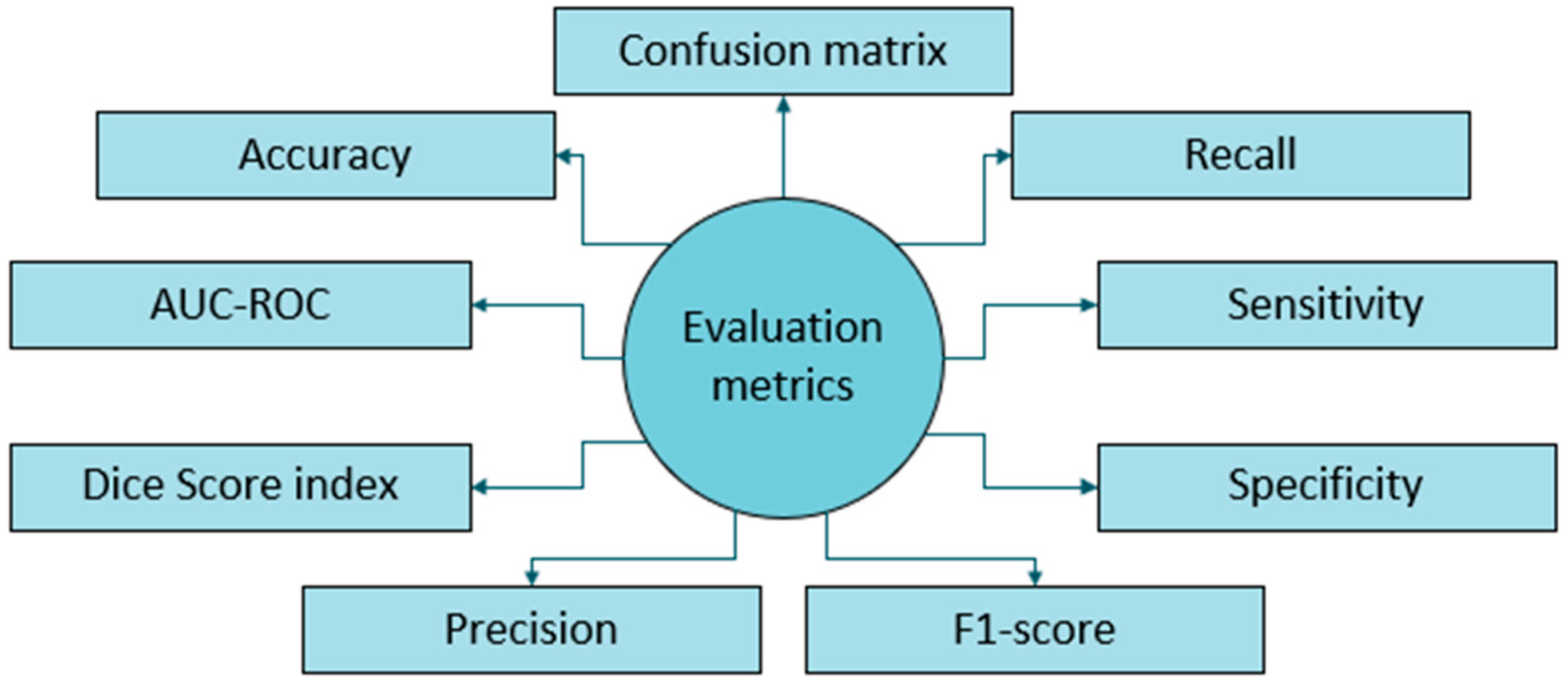
The performance metric benchmarking frequently found in literature includes accuracy, recall, sensitivity, specificity, F1-score, precision, Dice score index, AUC-ROC, and plotting a confusion matrix that shows true and false detections, illustrated in Figure 8. Several factors have hindered the establishment of uniform performance benchmarks for DL-based brain tumor detection and progression analysis. The factors include:
Figure 8.
Evaluation metrics employed for brain tumor MRI.
- Dataset Variability: Inconsistent tumor class distributions, heterogeneous data augmentation methodologies, and divergent validation cohorts across studies lead to significant confounding variables [80]. This variability can bias performance metrics, skew robustness assessments, and limit external validity, hindering the ability to make meaningful comparisons of algorithmic efficacy.
- Model Diversity: A wide range of DL architectures is being explored, preventing the adoption of a single benchmark applicable to all models.
- Task Specificity: Differences between detection and segmentation tasks create substantial challenges since each targets a different clinical application. Performance benchmarks optimized for detection accuracy become irrelevant for assessing models designed for precise boundary delineation [81]. Likewise, annotation complexity disparities impact dataset creation costs and model error interpretation.
- Evaluation Metrics: Studies employ various performance metrics, including accuracy, precision, recall, F1-score, Dice coefficient, etc., which can lead to inconsistencies in evaluation.
- Imaging Modalities: While MRI is the predominant imaging technique, some studies incorporate other modalities, further complicating standardization efforts.
- Rapid Advancements: The field is evolving rapidly, with new models and techniques constantly emerging, making it challenging to maintain consistent benchmarks.
- Lack of Standardization: No universally accepted framework exists for reporting results or conducting evaluations, leading to inconsistencies in performance assessment.
Despite these challenges, recent efforts to standardize performance metrics for DL-based MRI brain tumor diagnosis are gaining momentum. Studies are increasingly adopting a more consistent set of evaluation metrics, including accuracy for classification tasks, sensitivity, precision, recall, and F1-score for comprehensive model assessment, Dice coefficient and IOU for segmentation tasks, and AUC for evaluating overall model discrimination ability.
Several initiatives are working to establish standardized metrics for reporting DL-based MRI brain tumor diagnoses, aiming to enhance consistency, transparency, and reproducibility in research and clinical applications. One significant effort is the Radiomics Quality Score (RQS) [82] to evaluate the quality of radiomics studies, including DL applications, and the Quantitative Imaging Network (QIN) initiative aimed at improving the stability and reproducibility of radiomic features [83]. The RQS assesses key aspects such as image acquisition, preprocessing, feature extraction, and model validation. Another important initiative is the Image Biomarker Standardization Initiative (IBSI), which seeks to standardize terminology and methodologies in image biomarker research for reporting DL-based image analysis results. Additionally, the MICCAI Society has been organizing challenges and workshops to advance standardization in DL-based medical image analysis. These events facilitate the evaluation and comparison of different methods by providing common datasets and evaluation metrics, fostering collaboration within the research community.
5.2. Brain Tumor Diagnosis Challenge Competitions
DL-based brain tumor diagnosis has greatly benefited from the rise of challenge competitions. These events accelerate research, encourage collaboration, and establish benchmarks for cutting-edge methods. Participants develop and submit algorithms for tasks like tumor segmentation, classification, or detection, often using provided datasets of annotated medical images. Below, the notable challenges are discussed:
- BraTS Challenge
First held as the MICCAI 2012 [84], BraTS challenge competitions focus on segmenting brain tumors from multi-modal MRI scans, creating a standardized evaluation framework. It has also, in some years, included patient survival prediction based on imaging data. Participants delineate tumor sub-regions (e.g., enhancing tumor, necrotic core, edema) using multi-institutional MRI scans (T1, T1c, T2, FLAIR) with expert annotations. Evaluation metrics included the Dice score, Hausdorff distance, sensitivity, and specificity. BraTS has expanded to include a pediatric dataset, a BraTS-Africa dataset (for adult-type diffuse glioma in underrepresented patients), and, in 2023, challenges for segmenting brain metastases and meningiomas [85]. BraTS has significantly propelled brain tumor segmentation research, providing a standard dataset used to benchmark numerous DL models (e.g., U-Net, nnU-Net).
- b.
- RSNA-ASNR-MICCAI Brain Tumor AI Challenge
In 2021, the RSNA, ASNR, and MICCAI partnered to launch their first challenge, which focused on using AI to classify brain tumor types from MRI scans. The challenge dataset comprised 2040 diffuse glioma cases contributed by 37 institutions globally. Leveraging the foundation laid by the BraTS challenges (running since 2012), the dataset included pre- and post-contrast MRI scans with manual expert annotations. Model performance was evaluated using accuracy, F1-score, and AUC-ROC metrics.
- c.
- Fets-MICCAI Challenge on Brain Tumor Segmentation
The 2022 MICCAI Federated Tumor Segmentation (FeTS) challenge tackled the privacy issues surrounding medical data sharing by focusing on federated learning for brain tumor segmentation. Using distributed MRI datasets from various institutions, FeTS evaluated performance with the Dice score and Hausdorff distance. This challenge showcased the promise of federated learning in medical imaging [86], enabling decentralized learning across hospitals while protecting data privacy.
- d.
- ATLAS (Anatomical Tracings of Lesions After Stroke) Challenge
A 2018 MICCAI challenge, while primarily focused on stroke lesions, also incorporated tasks relevant to brain tumor segmentation and analysis. This challenge provided manually annotated brain lesion data from stroke patients, with performance evaluated using metrics such as the Dice similarity coefficient (DSC), Hausdorff distance, and precision/recall.
- e.
- CPM-RadPath Challenge
In 2020, the Children’s Brain Tumor Network (CBTN) and the Pacific Pediatric Neuro-Oncology Consortium (PNOC) collaborated on a challenge centered on integrating radiology and pathology data for diagnosing pediatric brain tumors. The dataset comprised multi-modal data, including MRI scans and histopathology images, with performance measured using accuracy and the Dice score.
- f.
- UPenn-GBM Challenge
A 2021 challenge organized by the University of Pennsylvania concentrated on glioblastoma multiforme (GBM) segmentation and survival prediction. Using a dataset of GBM patient MRI scans with corresponding survival data, the challenge evaluated segmentation performance with the Dice score and survival prediction with the concordance index. This challenge contributed to a better understanding of GBM imaging biomarkers and their relationship to patient outcomes.
- g.
- Kaggle Competitions
Several competitions, like the RSNA-MCCAI Brain Tumor Radiogenomic Classification challenge, are hosted on platforms like Kaggle. This particular challenge focused on predicting MGMT promoter methylation status in glioblastoma patients using MRI scans, with the AUC-ROC as the primary evaluation metric. These competitions foster the development of models capable of non-invasive genetic prediction [87].
- h.
- TCIA Challenges
The Cancer Imaging Archive (TCIA) hosts numerous cancer imaging challenges, including those focused on brain tumor diagnosis. These challenges cover tasks like tumor lesion segmentation, classification, and diagnostic predictions using publicly available imaging data [88]. For example, the autoPET 2022 challenge aimed to improve automated tumor lesion segmentation in PET/CT scans. It provided a large dataset of 1014 studies from 900 patients, emphasizing accurate and rapid lesion segmentation while minimizing false positives.
5.3. Regulatory Efforts
Recent years have witnessed significant advancements in regulatory efforts surrounding MRI-based brain tumor diagnosis. These efforts have focused on standardizing imaging protocols, improving access to diagnostic services, and enhancing early detection capabilities. A key initiative in this area is the development of a standardized Brain Tumor Imaging Protocol (BTIP) for multicenter studies. The BTIP aims to create consistency in MRI protocols across different clinical centers, enabling the use of comparable imaging data in clinical trials. Endorsed by the Response Assessment in Neuro-Oncology (RANO) working group, the BTIP directly addresses priorities identified at a 2014 workshop. This standardization is crucial for improving the reliability of radiographic endpoints in brain tumor clinical trials and enhancing the evaluation of therapeutic impacts in neuro-oncology.
Parallel to these standardization efforts, significant progress has been made in regulating MRI-based DL techniques for brain tumor diagnosis. These efforts also emphasize standardization, improved accuracy, and enhanced diagnostic capabilities. This standardization is vital for improving the reliability of radiographic endpoints in clinical trials and enhancing the assessment of therapeutic impacts. Studies comparing the diagnostic accuracy of neuroradiologists with and without the assistance of these DL systems have shown promising results. One study demonstrated a 12% increase in neuroradiologist accuracy with DL assistance, rising from 63.5% to 75.5% [84]. Results suggest that these systems can match or even surpass the performance of experienced neuroradiologists in identifying and classifying intracranial tumors. In automated tumor classification, one system outperformed neuroradiologists with a 19.9% higher accuracy (73.3% vs. 60.9%), and with system assistance, neuroradiologist classification accuracy increased by 18.9% (from 63.5% to 75.5%) [89].
The regulatory landscape for these DL techniques is also evolving, as evidenced by recent FDA clearances. The FDA clearance process is rigorous. The FDA has granted 510(k) clearance for ClearPoint 2.2, an AI-enabled software (aiMIFY 3.0) designed for the automated segmentation of brain structures in MRI scans [90], or they can undergo the more intensive process if they are novel. For AI-driven tools, this involves validating their safety and efficacy in clinical settings, as well as addressing biases and ensuring robust performance under real-world conditions. For example, ClearPoint 2.2 integrates the Maestro Brain Model for precise brain segmentation, which requires validation against manual expert annotations and open-source tools like FreeSurfer to prove superior accuracy and reproducibility. However, the dynamic nature of AI introduces unique regulatory complexities. Unlike static software, AI models frequently learn and evolve after deployment, posing a challenge to current regulatory frameworks designed for fixed systems.
The FDA is refining its guidance on AI-driven medical imaging tools, with ongoing discussions in the radiology community highlighting the need for standardized evaluation metrics and robust validation datasets to ensure the safety and efficacy of AI models used in brain tumor diagnostics. These combined regulatory advancements and research findings demonstrate a global commitment to improving brain tumor diagnosis through MRI.
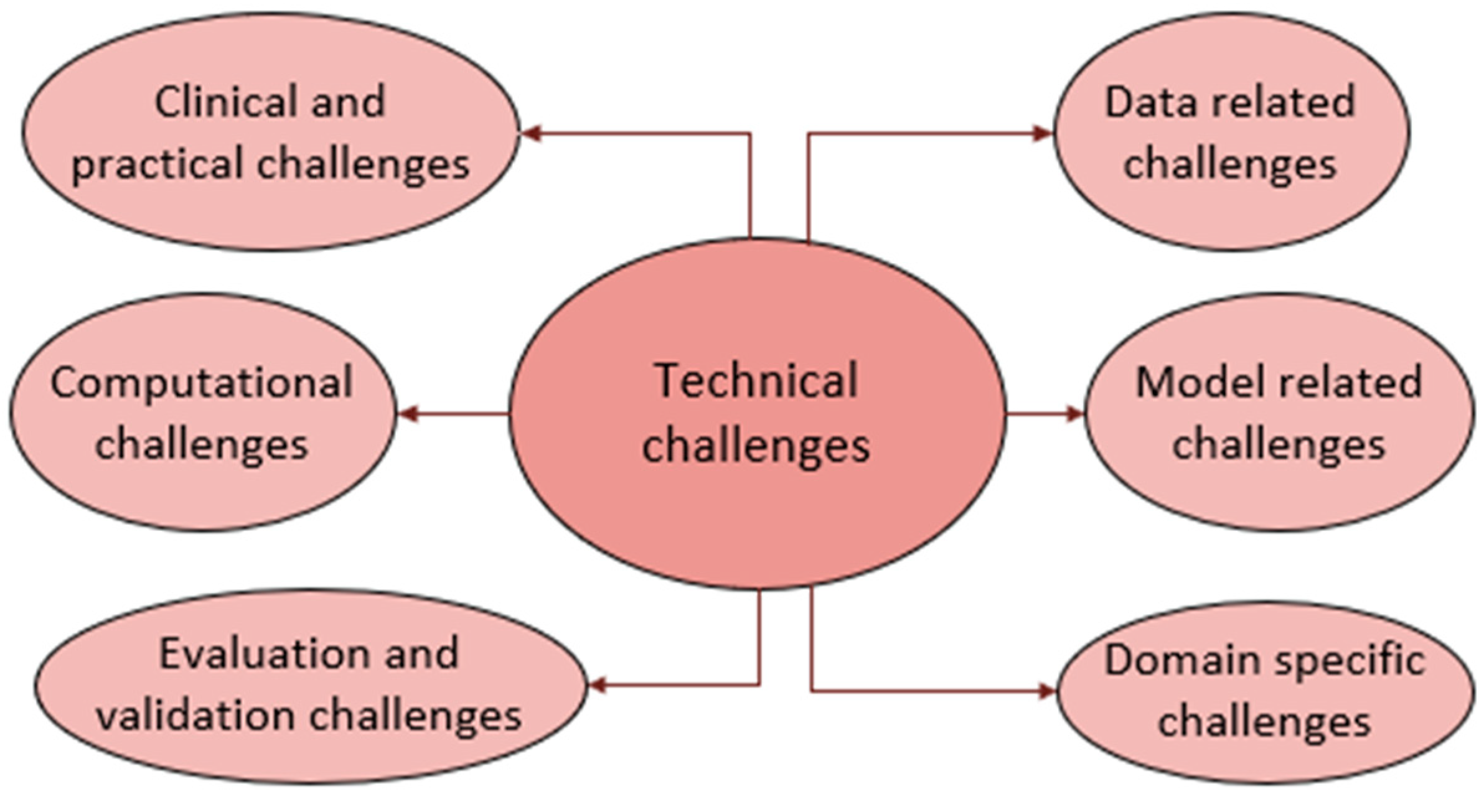
6. Technical Challenges
DL for brain tumor diagnosis using MRI scans is hampered by several obstacles that affect its accuracy, consistency, and real-world use. The obstacles are due to the data, model used, domain, computations involved, inconsistencies in evaluation benchmarks, and clinical challenges, as illustrated in Figure 9. These hurdles in AI-driven brain tumor MRI diagnosis are deeply interconnected. Data variability reduces model robustness, leading to performance drops across datasets due to domain shifts. Computational limitations hinder real-time deployment, while lightweight models sacrifice granularity. Inconsistent benchmarking further complicates cross-study comparisons. These issues collectively impede clinical adoption, creating a cycle where computational inefficiencies and data constraints delay regulatory approval. Below, each of these challenges is discussed separately.
Figure 9.
Technical challenges.
- Data-related challenges
A major problem is the scarcity of large, high-quality, precisely labeled brain tumor datasets. This is due to the infrequency of some tumor types, patient privacy concerns, and the expense of expert radiologist annotation [22]. Furthermore, the data often suffers from class imbalance (some tumor types are much more common than others, creating bias), data heterogeneity (variations in MRI scanners, protocols, resolutions, and image types like T1, T2, FLAIR, and DWI) [91], and the presence of noise and artifacts in the scans [21], all of which can negatively impact model performance.
- b.
- Model-related challenges
Several model-related challenges hinder DL for brain tumor diagnosis. The highly variable shape, size, location, and texture of tumors make it difficult to train models that can robustly identify them. Furthermore, the high dimensionality of MRI data (3D volumes with multiple modalities) necessitates complex architectures for effective processing. Limited training data increases the risk of overfitting, hindering the generalization to unseen data. Integrating information from different MRI modalities (like T1, T2, and FLAIR) for improved performance adds further complexity. The “black box” nature of many DL models limits interpretability and clinician trust. Finally, achieving good generalization across diverse datasets, scanners, and patient populations remains a major hurdle [92].
- c.
- Domain-specific challenges
The heterogeneous nature of brain tumors, often containing regions with distinct histological features like necrosis, edema, and enhancing/non-enhancing areas, makes segmentation and classification difficult. Furthermore, accurately defining tumor boundaries, particularly for infiltrative gliomas, is challenging and can be subjective even for expert radiologists.
- d.
- Evaluation and validation challenges
Further challenges in brain tumor diagnosis using DL include the lack of standardized benchmarks, hindering the comparison of different methods. Models must also demonstrate robustness against variations in tumor appearance, patient demographics, and imaging conditions, a difficult feat requiring extensive validation. Finally, incorporating longitudinal analysis, which involves tracking tumor progression over time, introduces additional complexity as models must effectively handle temporal changes in MRI scans.
- e.
- Computational challenges
MRI data often requires extensive and time-consuming preprocessing steps (like skull stripping, normalization, and registration) to ensure consistency, and these steps can be prone to errors. Training DL models on 3D MRI data demands substantial computational resources, including high-memory GPUs. Finally, effectively fusing information from multiple MRI modalities (such as T1, T2, FLAIR, and DWI) to enhance diagnostic accuracy is a complex undertaking. Researchers are exploring methods to reduce inference times using hardware accelerators like FPGA-based systems, aiming to integrate DL models into clinical workflows for rapid diagnosis.
- f.
- Clinical and practical challenges
Domain shift, caused by variations in datasets (hospital, scanner, demographics), impedes model generalization. The lack of standardized MRI preprocessing pipelines makes comparisons across studies difficult. Clinician acceptance hinges on interpretable and trustworthy AI assistance rather than fully automated solutions.
Significant privacy risks impede the clinical acceptance of brain MRI tumor diagnosis solutions. The sensitive nature of the data, including potentially identifiable facial features, means that re-identification is a serious concern despite anonymization efforts. Data breaches or unauthorized access could result in the misuse, discrimination, or stigmatization of patient information. The reliance on large datasets for AI training also raises questions about patient consent and the adequacy of anonymization [93]. Failure to ensure data anonymity can undermine patient confidence in AI diagnostics. The involvement of third parties in AI development introduces potential risks of data exploitation without patient awareness [94]. Even with metadata removed, advanced facial recognition technology can still potentially identify individuals from MRI images [95]. Sharing MRI data for research exacerbates this issue, as standard de-identification can negatively impact diagnostic analysis [96]. Researchers must prioritize informed consent and implement clear accountability measures when sharing data [97]. Achieving clinical acceptance requires the implementation of strong encryption, strict access controls, and transparent data governance frameworks to mitigate these privacy risks while preserving diagnostic accuracy.
Algorithmic bias in brain MRI tumor diagnosis can undermine clinical trust and adoption by causing inconsistent performance across different patient groups and tumor types. This bias often stems from training data that overemphasizes specific demographics or common tumors, leading to reduced accuracy for underrepresented groups like ethnic minorities or rare tumor variants [98,99]. For example, models trained mainly on large tumors may miss smaller ones (<6 mm), potentially delaying diagnosis [100]. Variations in MRI scanning techniques can also introduce technical bias, limiting how well models generalize [101]. Such biases risk increasing health disparities, with studies showing up to a 10% difference in lesion detection accuracy between patient subgroups [98]. Solutions like federated learning to diversify data and adversarial debiasing during development are being investigated [98]. Clinicians highlight the necessity of robust validation on diverse populations before clinical use to ensure equitable diagnosis [102].
Regulatory initiatives in the U.S. and Europe are increasingly shaping the ethical framework for AI applications in medical imaging. In the U.S., the FDA’s AI/ML-Based Software as a Medical Device Action Plan emphasizes real-world performance monitoring and algorithmic transparency to address bias, particularly in high-risk scenarios such as brain tumor diagnosis [103]—with heightened oversight for adaptive algorithms. Meanwhile, the European AI Act [104] requires high-risk AI systems in healthcare to meet stringent standards, including robust data governance, transparent decision-making processes, human oversight, and the use of geographically diverse validation datasets to mitigate bias. These requirements align with the Medical Device Regulation (MDR) for joint assessments. Both regulatory approaches advocate for diverse training datasets to help reduce demographic disparities. Additionally, collaborative efforts stress the importance of multidisciplinary oversight, incorporating clinical expertise into model development. Despite these proactive steps to balance innovation with accountability, ongoing challenges include standardizing bias detection methods, harmonizing international regulatory standards [105], and clearly defining responsibility for biased AI outcomes—highlighting the need for consistent auditing protocols and well-defined liability frameworks [106].
The following section highlights community initiatives working towards mitigating these challenges for a positive future in this field.
7. Future Outlook
Deep learning (DL) is revolutionizing brain tumor diagnosis, significantly boosting accuracy and efficiency. This progress is driven by innovations in optimized DL models, AI-powered tools, and Explainable AI, all aimed at overcoming data, model, computational, and domain-specific hurdles. Ongoing efforts in benchmarking and standardization are improving evaluation, validation, and generalization. Furthermore, clinical and practical obstacles are being overcome through the integration of DL systems into clinical settings and regulatory approvals. Efforts in key research directions are detailed below.
- Optimized DL models for early detection
DL has become a popular method for automatically detecting and segmenting brain tumors in MRI scans. Models like U-Net, 3D U-Net, DeepMedic, and V-Net have shown great promise in 3D brain tumor segmentation [101]. Adapting models to different image types (like CT to MRI) and combining information from various imaging sources has made these models even more useful. Furthermore, integrating imaging data (MRI, CT), genomic data (DNA/RNA sequencing), and clinical data (patient history, treatment responses) into AI models offers a more complete understanding of tumors. This multimodal approach leads to better tumor classification and allows for personalized treatment plans [107]. Recent studies show that integrating structural MRI sequences (such as T1, T2, and FLAIR) with functional imaging and genomic biomarkers using advanced DL architectures significantly enhances diagnostic performance compared to conventional methods [108]. Multimodal brain MRI tumor models aim to improve diagnostic accuracy by leveraging complementary information across different imaging modalities, offering a means to address data heterogeneity and enrich feature representation. While these models can help mitigate some benchmarking inconsistencies, they do not fully resolve them and may even introduce new challenges. Additionally, although multimodal approaches can contribute to interpretability in specific contexts, they do not inherently overcome the broader issue of model transparency; in fact, the added complexity from multiple data streams can further obscure the decision-making process.
The adoption of federated learning frameworks marks a similarly transformative advancement in neuro-oncology AI. This decentralized approach allows models to be trained collaboratively across multiple institutions without compromising patient privacy. Large-scale efforts like the Global Neuroimaging Consortium have demonstrated the practical viability of federated learning, with participating centers reporting consistent accuracy gains of 15–20% [109] while fully preserving data confidentiality. These distributed systems employ sophisticated methods to manage data heterogeneity, including automated quality control, standardized preprocessing workflows, and adaptive weighting algorithms that adjust for differences in scanner vendors and imaging protocols across sites, thus addressing generalizability.
The increasing use of foundation models trained on very large and diverse datasets is expected to significantly boost the accuracy of DL for brain tumor analysis. These models could allow researchers to study tumor types that were previously difficult to analyze due to limited training data [80].
- b.
- AI-powered diagnostic and prognosis tools
To address computational constraints, lightweight DL models have emerged as effective solutions for brain tumor detection, especially in resource-limited environments. These models enable real-time diagnosis and monitoring on edge devices, supporting timely clinical interventions. AI-assisted tools are also being developed for intraoperative use, aiding in the identification of cancerous tissue during brain tumor surgeries and potentially improving surgical outcomes. One notable example is FastGlioma, which integrates AI with stimulated Raman histology (SRH) to deliver rapid, high-accuracy diagnostic insights from tissue biopsies within seconds [110]. Research shows that streamlined architectures, such as an 8-layer CNN, can achieve up to 99.48% accuracy in binary classification while reducing inference time by 40% compared to conventional models [111]. Similarly, modified U-Net architectures enhanced with spatial attention mechanisms have demonstrated Dice scores of 96% for tumor segmentation, using 60% fewer parameters than standard models [112]. Beyond detection, these models are increasingly being projected to predict clinical outcomes, including patient survival and treatment response. The combination of efficiency and interpretability—particularly when paired with Explainable AI techniques—positions lightweight models as powerful tools for scalable, transparent brain tumor diagnostics.
- c.
- Explainable AI
Alongside efficiency improvements, there is increasing emphasis on explainability and transparency in AI models to build clinician trust. In parallel with efforts to enhance efficiency, there is a growing focus on explainability and transparency in AI models to strengthen clinician trust. Advances in Explainable AI (XAI) have made DL models for brain tumor diagnosis significantly more interpretable, addressing the persistent “black-box” challenge in medical AI. Methods such as Grad-CAM and attention mechanisms highlight the specific image regions the model considers most relevant, thereby improving transparency and fostering confidence in its predictions [113]. For example, one study combined DL with natural language processing to produce descriptive reports of tumor regions, achieving a Dice coefficient of 0.93 for segmentation accuracy [114]—an approach that enhances interpretability and clinician trust. In another case, CNN-TumorNet reached 99% classification accuracy for brain tumors while employing LIME to explain malignant glioma predictions, contributing to diagnostic clarity [115]. Similarly, hybrid models like ViT-GRU combine transformer-based attention maps with post-hoc XAI methods to validate feature relevance, particularly in complex, heterogeneous tumor regions [50]. These explainability frameworks not only improve diagnostic performance but also support regulatory compliance by making AI systems more auditable and transparent in clinical settings.
- d.
- Liquid biopsy and AI-based genetic profiling
Combining DL with liquid biopsies, like circulating tumor DNA (ctDNA) analysis, enables non-invasive monitoring of tumor mutations. AI algorithms analyze genomic data to identify brain tumor-related genetic changes, which helps with early diagnosis and predicting patient outcomes [116].
- e.
- Benchmarking and standardization
Standardization is gaining momentum in the regulatory landscape for AI in healthcare. Public datasets (like BraTS) and organized challenges have fueled innovation and provided benchmarks for comparing various DL methods. Standardizing imaging protocols and annotation guidelines is also improving the consistency and reliability of these models.
- f.
- Integration with clinical workflows
Extensive testing has validated the clinical benefits of DL systems. One study found that neuroradiologists’ diagnostic accuracy improved by 12% (from 63.5% to 75.5%) with AI assistance. Another trial with nearly 280 patients demonstrated that integrating SRH imaging with AI achieved 94.6% accuracy for intraoperative brain tumor diagnosis, matching traditional pathology (93.9%) while reducing diagnosis time to under three minutes [95]. These advancements underscore AI’s potential to enhance clinical decision-making, improve patient outcomes, and support radiologists and oncologists with more comprehensive diagnostic tools.
- g.
- Regulatory success
Regulatory frameworks are anticipated to evolve with stricter guidelines on AI model transparency, bias mitigation, and real-world performance monitoring. Regulatory bodies are prioritizing strong clinical validation across varied populations and imaging conditions before approving AI models for healthcare use. The FDA’s clearance of Vysioneer, Cambridge, MA, Inc.’s VBrain, a DL algorithm for brain tumor contouring [117], represents a significant milestone in AI adoption for brain tumor diagnosis and treatment planning. AI-driven tools are increasingly integrated into hospitals, enhancing precision in radiology and neurosurgery. For instance, AI-powered neuro-navigation improves lesion localization during surgery, reducing risks, while AI-assisted stereotactic radiosurgery ensures accurate detection of brain metastases, aiding in more effective treatment strategies [118].
- h.
- Market growth
The rising incidence of brain cancer in the US, coupled with an aging population, is driving demand for advanced diagnostic and treatment solutions. AI and machine learning are revolutionizing diagnosis through improved imaging analysis and tumor behavior prediction, leading to a rapidly expanding global brain cancer diagnostic market projected to grow from USD 1.55 billion in 2023 to USD 5.21 billion by 2031 at a compound annual growth rate of 18.90% [119], illustrated in Figure 10. This is further fueled by increased pharmaceutical R&D spending.
Figure 10.
Projected brain tumor market growth (2024–2031).
Recent advancements in brain MRI tumor diagnosis have gained momentum through significant funding and collaboration. Case Western Reserve University received a $3.03 million grant from the National Cancer Institute to enhance MRI technology and software for consistent glioblastoma diagnosis [120]. In the UK, brain tumor research is a priority in the upcoming National Cancer Plan, emphasizing clinical trials and interdepartmental cooperation [121]. The American Brain Tumor Association allocated $1.37 million in 2025 grants for research in imaging, biomarkers, and immunotherapy [122]. Additionally, the Brain Tumor Funders’ Collaborative launched a program offering two-year, $500,000 grants to develop liquid biopsy technologies for brain tumors. Collectively, these initiatives aim to improve diagnostic precision and patient outcomes.
While challenges remain, the aforementioned breakthroughs and initiatives are revolutionizing brain tumor diagnosis, making it more accurate, efficient, and accessible. The growing clinical validation of DL models and their regulatory approvals demonstrate increasing trust in AI for this purpose from both the medical community and regulatory bodies. As these technologies continue to demonstrate their safety and effectiveness, their use in clinical practice is expected to expand, ultimately improving patient outcomes and streamlining diagnostic processes.
8. Conclusions
This study highlights the significant progress made in MRI-based brain tumor diagnosis and prognosis through deep learning (DL). Although the reviewed models exhibit impressive capabilities in detecting tumors and predicting their progression, several critical challenges remain. The future of brain MRI tumor diagnosis depends on addressing interconnected issues related to data, model development, and clinical implementation. Data heterogeneity continues to hinder the development of generalizable AI models, especially given the need for large and diverse datasets that encompass underrepresented tumor types. Models must become more adaptable to account for changes in tumor biology and domain shifts. Moreover, the high computational demands of advanced DL methods often conflict with the practical constraints of clinical workflows, and inconsistencies in benchmarking complicate fair performance evaluation. Clinically, the gap between AI tools and real-world adoption persists due to regulatory hurdles and lingering mistrust of “black-box” algorithms. Adaptive AI systems, in particular, face extended FDA approval timelines and skepticism regarding their interpretability and safety.
Although technical hurdles persist, DL is significantly advancing brain tumor diagnostics. To fully realize its potential and ensure equitable clinical impact, existing gaps need to be bridged. This involves establishing harmonized imaging standards, developing lightweight and interpretable models, conducting robust multi-center trials, and creating clinician-centric interfaces within scalable computational frameworks.
As research progresses and standardization efforts evolve, diagnostic protocols are expected to become more refined, paving the way for enhanced patient outcomes and a more efficient diagnostic workflow for brain tumors.
Author Contributions
Conceptualization, Q.A.M. and N.M.; methodology, Q.A.M. and N.M.; investigation, Q.A.M. and N.M.; writing—original draft preparation, Q.A.M. and N.M.; writing—review and editing, Q.A.M., N.M. and M.M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wang, Y.; Pan, Y.; Li, H. What is brain health and why is it important? BMJ 2020, 371, m3683. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Niemelä, S.; Lempinen, L.; Löyttyniemi, E.; Oksi, J.; Jero, J. Bacterial meningitis in adults: A retrospective study among 148 patients in an 8-year period in a university hospital, Finland. BMC Infect. Dis. 2023, 23, 45. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Park, Y.G. Brain tumor rehabilitation: Symptoms, Complications, and Treatment Strategy. Brain Neurorehabil. 2022, 15, e25. [Google Scholar] [CrossRef]
- Ganguly, P.; Soliman, A.; Moustafa, A.A. Holistic management of Schizophrenia symptoms using pharmacological and non-pharmacological treatment. Front. Public Health 2018, 6, 166. [Google Scholar] [CrossRef] [PubMed]
- Torp, S.H.; Solheim, O.; Skjulsvik, A.J. The WHO 2021 classification of central nervous system tumours: A practical update on what neurosurgeons need to know—A minireview. Acta Neurochir. 2022, 164, 2453–2464. [Google Scholar] [CrossRef]
- Go, K.O.; Kim, Y.Z. Brain Invasion and Trends in Molecular Research on Meningioma. Brain Tumor Res. Treat. 2023, 11, 47–58. [Google Scholar] [CrossRef]
- Peddinti, A.S.; Maloji, S.; Manepalli, K. Evolution in diagnosis and detection of brain tumor—Review. J. Phys. Conf. Ser. 2021, 2115, 012039. [Google Scholar] [CrossRef]
- Martucci, M.; Russo, R.; Schimperna, F.; D’apolito, G.; Panfili, M.; Grimaldi, A.; Perna, A.; Ferranti, A.M.; Varcasia, G.; Giordano, C.; et al. Magnetic resonance imaging of primary adult brain tumors: State of the art and future perspectives. Biomedicines 2023, 11, 364. [Google Scholar] [CrossRef]
- Boodman, S.G. Kaiser Health News. Washington Post, 19 May 2014. [Google Scholar]
- Zacharaki, E.I.; Wang, S.; Chawla, S.; Yoo, D.S.; Wolf, R.; Melhem, E.R.; Davatzikos, C. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn. Reson. Med. 2009, 62, 1609–1618. [Google Scholar] [CrossRef] [PubMed]
- Bachir, N.; Memon, Q. Benchmarking YOLOv5 models for improved human detection in search and rescue missions. J. Electron. Sci. Technol. 2024, 22, 1. [Google Scholar] [CrossRef]
- Alameri, M.; Memon, Q. YOLOv5 integrated with recurrent network for object tracking: Experimental results from a hardware platform. IEEE Access 2024, 12, 119733–119742. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, Z.; Wu, L.; Wang, S. A hybrid method for MRI brain image classification. Expert Syst. Appl. 2011, 38, 10049–10053. [Google Scholar] [CrossRef]
- Arakeri, M.P.; Reddy, G.R.M. Computer-aided diagnosis system for tissue characterization of brain tumor on magnetic resonance images. SIViP 2015, 9, 409–425. [Google Scholar] [CrossRef]
- Gumaei, A.; Hassan, M.M.; Hassan, M.R.; Alelaiwi, A.; Fortino, G. A hybrid feature extraction method with regularized extreme learning machine for brain tumor classification. IEEE Access 2019, 7, 36266–36273. [Google Scholar] [CrossRef]
- Islam, R.; Imran, S.; Ashikuzzaman, M.; Khan, M. Detection and classification of brain tumor based on multilevel segmentation with convolutional neural network. J. Biomed. Sci. Eng. 2020, 13, 45–53. [Google Scholar] [CrossRef]
- van Hespen, K.M.; Zwanenburg, J.J.M.; Dankbaar, J.W.; Geerlings, M.I.; Hendrikse, J.; Kuijf, H.J. An anomaly detection approach to identify chronic brain infarcts on MRI. Sci. Rep. 2021, 11, 7714. [Google Scholar] [CrossRef]
- Niyas, S.; Pawan, S.J.; Anand Kumar, M.; Rajan, J. Medical image segmentation with 3D convolutional neural networks: A Survey. Neurocomputing 2022, 493, 397–413. [Google Scholar] [CrossRef]
- Pinaya, W.H.; Tudosiu, P.-D.; Gray, R.; Rees, G.; Nachev, P.; Ourselin, S.; Cardoso, M.J. Unsupervised brain imaging 3D anomaly detection and segmentation with transformers. Med. Image Anal. 2022, 79, 102475. [Google Scholar] [CrossRef]
- Ghorbel, A.; Aldahdooh, A.; Albarqouni, S.; Hamidouche, W. Transformer based models for unsupervised anomaly segmentation in brain MR images. In Proceedings of the 8th International Workshop, BrainLes 2022, MICCAI 2022, Singapore, 18 September 2022; Springer: Cham, Switzerland, 2022. [Google Scholar]
- Yaqub, M.; Jinchao, F.; Ahmed, S.; Mehmood, A.; Chuhan, I.S.; Manan, M.A.; Pathan, M.S. DeepLabV3, IBCO-Based ALCResNet: A fully automated classification, and grading system for brain tumor. Alex. Eng. J. 2023, 76, 609–627. [Google Scholar] [CrossRef]
- Walsh, J.; Othmani, A.; Jain, M.; Dev, S. Using U-Net network for efficient brain tumor segmentation in MRI images. Health Anal. 2022, 2, 100098. [Google Scholar] [CrossRef]
- Zeineldin, R.A.; Karar, M.E.; Coburger, J.; Wirtz, C.R.; Burgert, O. DeepSeg: Deep neural network framework for automatic brain tumor segmentation using magnetic resonance FLAIR images. Int. J. CARS 2020, 15, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Vankdothu, R.; Hameed, M.A.; Fatima, H. A brain tumor identification and classification using deep learning based on CNN-LSTM method. Comput. Electr. Eng. 2022, 101, 107960. [Google Scholar] [CrossRef]
- Aggarwal, M.; Tiwari, A.K.; Sarathi, M.P.; Bijalwan, A. An early detection and segmentation of brain tumor using deep neural network. BMC Med. Inform. Decis. Mak. 2023, 23, 78. [Google Scholar] [CrossRef]
- Aleid, A.M.; Alrasheed, A.S.; Aldanyowi, S.N.; Almalki, S.F. Advanced magnetic resonance imaging for glioblastoma: Oncology-radiology integration. Surg. Neurol. Int. 2024, 15, 309. [Google Scholar] [CrossRef]
- Chaki, J. Brain Tumor MRI Dataset. IEEE DataPort 2023. [Google Scholar] [CrossRef]
- Cheng, J. Brain Tumor Retrieval Dataset. Available online: https://github.com/chengjun583/brainTumorRetrieval (accessed on 1 February 2025).
- Ultralytics. Brain Tumor Dataset. Available online: https://docs.ultralytics.com/datasets/detect/brain-tumor/ (accessed on 1 October 2024).
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv 2019, arXiv:1811.02629. [Google Scholar] [CrossRef]
- The Cancer Imaging Archive (TCIA). Available online: https://www.kaggle.com/datasets/mateuszbuda/lgg-mri-segmentation (accessed on 1 February 2025).
- Baid, U.; Ghodasara, S.; Mohan, S.; Bilello, M.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.; Pati, S.; et al. The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification. arXiv 2021, arXiv:2107.02314. [Google Scholar] [CrossRef]
- Puccio, B.; Pooley, J.P.; Pellman, J.S.; Taverna, E.C.; Craddock, R.C. The preprocessed connectomes project repository of manually corrected skull-stripped T1-weighted anatomical MRI data. GigaScience 2016, 5, 45. [Google Scholar] [CrossRef]
- Hoopes, A.; Mora, J.; Dalca, A.; Fischl, B.; Hoffmann, M. SynthStrip: Skull-Stripping for any brain image. Neuroimage 2022, 260, 119474. [Google Scholar] [CrossRef]
- Commowick, O.; Kain, M.; Casey, R.; Ameli, R.; Ferré, J.-C.; Kerbrat, A.; Tourdias, T.; Cervenansky, F.; Camarasu-Pop, S.; Glatard, T.; et al. Multiple sclerosis lesions segmentation from multiple experts: The MICCAI 2016 Challenge Dataset. Neuroimage 2021, 244, 118589. [Google Scholar] [CrossRef] [PubMed]
- Gusev, Y.; Bhuvaneshwar, K.; Song, L.; Zenklusen, J.-C.; Fine, H.; Madhavan, S. The REMBRANDT study, a large collection of genomic data from brain cancer patients. Sci. Data 2018, 5, 180158. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Duggar, W.N.; Caballero, D.M.; Thomas, T.V.; Adari, N.; Mundra, E.K.; Wang, H. A brain MRI dataset and baseline evaluations for tumor recurrence prediction after Gamma knife radiotherapy. Sci. Data 2023, 10, 785. [Google Scholar] [CrossRef] [PubMed]
- Schmainda, K.M.; Prah, M. Data from Brain-Tumor-Progression. The Cancer Imaging Archive. [CrossRef]
- Cepeda, S.; Garcia-Garcia, S.; Arrese, I.; Herrero, F.; Escudero, T.; Zamora, T.; Sarabia, R. The Río Hortega University Hospital Glioblastoma Dataset: A comprehensive collection of preoperative, early postoperative, and recurrence MRI scans (RHUH-GBM). Data Brief 2023, 50, 109617. [Google Scholar] [CrossRef]
- Gong, Z.; Xu, T.; Peng, N.; Cheng, X.; Niu, C.; Wiestler, B.; Hong, F.; Li, H.B. A Multi-Center, Multi-Parametric MRI Dataset of Primary and Secondary Brain Tumors. Sci. Data 2024, 11, 789. [Google Scholar] [CrossRef]
- ZainEldin, H.; Gamel, S.A.; El-Kenawy, E.-S.M.; Alharbi, A.H.; Khafaga, D.S.; Ibrahim, A.; Talaat, F.M. Brain tumor detection and classification using deep learning and Sine-Cosine fitness grey wolf optimization. Bioengineering 2022, 10, 18. [Google Scholar] [CrossRef]
- Priya, A.; Vasudevan, V. Brain tumor classification and detection via hybrid AlexNet-GRU based on deep learning. Biomed. Signal Process. Control 2024, 89, 105716. [Google Scholar] [CrossRef]
- Sekhar, A.; Biswas, S.; Hazra, R.; Sunaniya, A.K.; Mukherjee, A.; Yang, L. Brain tumor classification using fine-tuned GoogLeNet features and machine learning algorithms: IoMT enabled CAD System. IEEE J. Biomed. Health Inform. 2022, 26, 983–991. [Google Scholar] [CrossRef]
- Malla, N.; Ramesh, K.; Rao, K.S. Efficient brain tumor classification with a hybrid CNN-SVM approach. J. Adv. Inf. Technol. 2024, 15, 340–354. [Google Scholar]
- Khan, M.A.; Ashraf, I.; Alhaisoni, M.; Damaševičius, R.; Scherer, R.; Rehman, A.; Bukhari, S.A.C. Multimodal brain tumor classification using deep learning and robust feature selection: A Machine learning application for radiologists. Diagnostics 2021, 11, 565. [Google Scholar] [CrossRef]
- Rasa, S.M.; Islam, M.; Talukder, A.; Uddin, A.; Khalid, M.; Kazi, M.; Kazi, Z. Brain tumor classification using fine-tuned transfer learning models on Magnetic Resonance Imaging (MRI) Images. Digit. Health 2024, 10, 20552076241286140. [Google Scholar] [CrossRef] [PubMed]
- Vimala, B.B.; Srinivasan, S.; Mathivanan, S.K.; Mahalakshmi; Jayagopal, P.; Dalu, G.T. Detection and classification of brain tumor using hybrid deep learning models. Sci. Rep. 2023, 13, 23029. [Google Scholar]
- Mithun, M.S.; Jawhar, S.J. Detection and classification on MRI images of brain tumor using YOLO NAS deep learning model. J. Radiat. Res. Appl. Sci. 2024, 17, 101113. [Google Scholar] [CrossRef]
- Ahmed, M.; Hossain, M.; Islam, R.; Ali, S.; Nafi, A.A.N.; Ahmed, F.; Ahmed, K.M.; Miah, S.; Rahman, M.; Niu, M.; et al. Brain tumor detection and classification in MRI using hybrid ViT and GRU model with Explainable AI in southern Bangladesh. Sci. Rep. 2024, 14, 22797. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.K.; Sonaimuthu, S.; Murugesan, S.; Rajadurai, H.; Shivahare, B.D.; Shah, M.A. Employing deep learning and transfer learning for accurate brain tumor detection. Sci. Rep. 2024, 14, 7232. [Google Scholar] [CrossRef]
- Yousef, R.; Khan, S.; Gupta, G.; Siddiqui, T.; Albahlal, B.M.; Alajlan, S.A.; Haq, M.A. U-Net-based models towards optimal MR brain image segmentation. Diagnostics 2023, 13, 1624. [Google Scholar] [CrossRef]
- Musthafa, N.; Masud, M.M.; Memon, Q. Advancing early-stage brain tumor detection with segmentation by modified U-Net. In Proceedings of the 8th International Conference on Medical and Health Informatics, (ICMHI’24), Yokohama, Japan, 17–19 May 2024; Association for Computing Machinery: New York, NY, USA, 2024; pp. 52–57. [Google Scholar]
- Li, Z.; Wu, X.; Yang, X. A multi brain tumor region segmentation model based on 3D U-Net. Appl. Sci. 2023, 13, 9282. [Google Scholar] [CrossRef]
- Huang, A.; Jiang, L.; Zhang, J.; Wang, Q. Attention-VGG16-UNet: A novel deep learning approach for automatic segmentation of the median nerve in ultrasound images. Quant. Imaging Med. Surg. 2022, 12, 3138–3150. [Google Scholar] [CrossRef]
- Iriawan, N.; Pravitasari, A.; Nuraini, S.; Nirmalasari, I.; Azmi, T.; Nasrudin, M.; Fandisyah, A.; Fithriasari, K.; Purnami, W.; Irhamah, W. YOLO-UNet architecture for detecting and segmenting the localized MRI brain tumor image. Appl. Comput. Intell. Soft Comput. 2024, 2024, 3819801. [Google Scholar] [CrossRef]
- Sami, U.; Attique, K.; Anum, M.; Olfa, M.; Oumaima, S.; Nazik, A. Brain tumor classification from MRI Scans: A framework of hybrid deep learning model with Bayesian optimization and quantum theory-based marine predator algorithm. Front. Oncol. 2024, 14, 1335740. [Google Scholar]
- Shafi, A.S.M.; Rahman, M.B.; Anwar, T.; Halder, R.S.; Kays, H.E. Classification of brain tumors and autoimmune disease using ensemble learning. Inform. Med. Unlocked 2021, 24, 100608. [Google Scholar] [CrossRef]
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans. Med. Imaging 2016, 35, 1240–1251. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, S.; Panigrahi, B.K.; Gandhi, T.K. Enhanced performance of Dark-Nets for brain tumor classification and segmentation using colormap-based superpixel techniques. Mach. Learn. Appl. 2022, 7, 100212. [Google Scholar] [CrossRef]
- Pereira, S.; Meier, R.; Alves, V.; Reyes, M.; Silva, C.A. Automatic brain tumor grading from MRI data using convolutional neural networks and quality assessment. In Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 11038, pp. 106–114. [Google Scholar]
- Tandel, G.S.; Tiwari, A.; Kakde, O.G. Performance optimisation of deep learning models using majority voting algorithm for brain tumor classification. Comput. Biol. Med. 2021, 135, 104564. [Google Scholar] [CrossRef]
- Komaki, K.; Sano, N.; Tangoku, A. Problems in histological grading of malignancy and its clinical significance in patients with operable breast cancer. Breast Cancer 2006, 13, 249–253. [Google Scholar] [CrossRef]
- Abdusalomov, A.B.; Mukhiddinov, M.; Whangbo, T.K. Brain tumor detection based on deep learning approaches and magnetic resonance imaging. Cancers 2023, 15, 4172. [Google Scholar] [CrossRef]
- Azhar, A. Brain MRI sequence and view plane identification using deep learning. Front. Neuroinform. 2024, 18, 1373502. [Google Scholar]
- Mishra, S.; Singh, K.; Kumar, R.; Kumar, P. Use of 3D coronal and sagittal images to improve the diagnosis of brain tumor. ECS Trans. 2022, 107, 171. [Google Scholar] [CrossRef]
- Saboor, A.; Li, J.P.; Haq, A.U.; Shehzad, U.; Khan, S.; Aotaibi, R.M.; Alajlan, S.A. DDFC: Deep learning approach for deep feature extraction and classification of brain tumors using magnetic resonance imaging in E-Healthcare system. Sci. Rep. 2024, 14, 6425. [Google Scholar] [CrossRef]
- Tabatabaei, S.; Rezaee, K.; Zhu, M. Attention transformer mechanism and fusion-based deep learning architecture for MRI brain tumor classification system. Biomed. Signal Process. Control 2023, 86, 105119. [Google Scholar] [CrossRef]
- Li, Y.; Ma, Y.; Wu, Z.; Xie, R.; Zeng, F.; Cai, H.; Lui, S.; Song, B.; Chen, L.; Wu, M. Advanced imaging techniques for differentiating pseudoprogression and tumor recurrence after immunotherapy for Glioblastoma. Front. Immunol. 2021, 12, 790674. [Google Scholar] [CrossRef]
- Meaney, C.; Das, S.; Colak, E.; Kohandel, M. Deep learning characterization of brain tumors with diffusion-weighted imaging. J. Theor. Biol. 2023, 557, 111342. [Google Scholar] [CrossRef]
- Sasank, V.; Venkateswarlu, S. An automatic tumor growth prediction-based segmentation using full resolution convolutional network for brain tumor. Biomed. Signal Process. Control 2022, 71, 103090. [Google Scholar] [CrossRef]
- Vinurajkumar, S.; Anandhavelu, S. An enhanced fuzzy segmentation framework for extracting white matter from T1-weighted MR images. Biomed. Signal Process. Control 2022, 71, 103093. [Google Scholar] [CrossRef]
- Noviantri, V.; Tomy, T.; Chowanda, A. Linear and nonlinear model of brain tumor growth simulation using finite difference method. Procedia Comput. Sci. 2021, 179, 297–304. [Google Scholar] [CrossRef]
- Sigal, D.; Przedborski, M.; Sivaloganathan, D.; Kohandel, M. Mathematical modelling of cancer stem cell-targeted immunotherapy. Math. Biosci. 2019, 318, 108269. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.I.; Kanaly, C.W.; Friedman, A.H.; Bigner, D.D.; Sampson, J.H. Monitoring radiographic brain tumor progression. Toxins 2011, 3, 191–200. [Google Scholar] [CrossRef]
- Yan, J.L.; Toh, C.H.; Ko, L.; Wei, K.C.; Chen, P.Y. A neural network approach to identify Glioblastoma progression Phenotype from Multimodal MRI. Cancers 2021, 13, 2006. [Google Scholar] [CrossRef]
- Mehdi, A.; Guang, Y.; Yousuf, Z.; Iuliana, T.; Örjan, S.; Chunliang, W. A comparative study of radiomics and deep-learning based methods for pulmonary nodule malignancy prediction in low dose CT images. Front. Oncol. 2021, 11, 737368. [Google Scholar]
- Farzana, W.; Basree, M.M.; Diawara, N.; Shboul, Z.A.; Dubey, S.; Lockhart, M.M.; Hamza, M.; Palmer, J.D.; Iftekharuddin, K.M. Prediction of rapid early progression and survival risk with pre-radiation MRI in WHO Grade 4 Glioma patients. Cancers 2023, 15, 4636. [Google Scholar] [CrossRef] [PubMed]
- Dorfner, F.J.; Patel, J.B.; Kalpathy-Cramer, J.; Gerstner, E.R.; Bridge, C.P. A review of deep learning for brain tumor analysis in MRI. Npj Precis. Oncol. 2025, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Kiran, L.; Zeb, A.; Rehman, Q.N.U.; Rahman, T.; Shehzad, M.; Ahmad, S.; Irfan, M.; Naeem, M.; Huda, S.; Mahmoud, H. An enhanced pattern detection and segmentation of brain tumors in MRI images using deep learning technique. Frontiers. Comput. Neurosci. 2024, 18, 1418280. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Jha, A.K.; Mithun, S.; Sherkhane, U.B.; Dwivedi, P.; Puts, S.; Osong, B.; Traverso, A.; Purandare, N.; Wee, L.; Rangarajan, V.; et al. Emerging role of quantitative imaging (Radiomics) and artificial intelligence in precision oncology. Explor. Target Antitumor Ther. 2023, 4, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Irjudson. BRaTS 2012—Multimodal Brain Tumor Segmentation Challenge. Available online: https://competitions.codalab.org/competitions/191 (accessed on 1 February 2025).
- Moawad, A.W.; Janas, A.; Baid, U.; Ramakrishnan, D.; Saluja, R.; Ashraf, N.; Maleki, N.; Jekel, L.; Yordanov, N.; Fehringer, P.; et al. The brain tumor segmentation (BraTS-METS) challenge 2023: Brain metastasis segmentation on pre-treatment MRI. arXiv 2024, arXiv:2306.00838v3. [Google Scholar]
- Pati, S.; Baid, U.; Zenk, M.; Edwards, B.; Sheller, M.; Reina, G.A.; Foley, P.; Gruzdev, A.; Martin, J.; Albarqouni, S.; et al. The federated tumor segmentation (FeTS) challenge. arXiv 2021, arXiv:2105.05874. [Google Scholar]
- Flanders, A.; Carr, C.; Calabrese, E.; Kitamura, F.; Rudie, J.; Mongan, J.; Elliott, J.; Prevedello, L.; Riopel, M.; Bakas, S.; et al. RSNA-MICCAI Brain Tumor Radiogenomic Classification. Kaggle. 2021. Available online: https://kaggle.com/competitions/rsna-miccai-brain-tumor-radiogenomic-classification (accessed on 3 February 2025).
- Gatidis, S.; Kuestner, T. A whole-body FDG-PET/CT dataset with manually annotated tumor lesions (FDG-PET-CT-Lesions). Cancer Imaging Arch. 2022, 9, 601. [Google Scholar] [CrossRef]
- Gao, P.; Shan, W.; Guo, Y.; Wang, Y.; Sun, R.; Cai, J.; Li, H.; Chan, W.S.; Liu, P.; Yi, L.; et al. Development and validation of a deep learning model for brain tumor diagnosis and classification using magnetic resonance imaging. JAMA Netw. Open 2022, 5, e2225608. [Google Scholar] [CrossRef]
- Hall, J. FDA Clears Emerging AI-Powered Software for Brain MRI. Available online: https://www.diagnosticimaging.com/view/fda-clears-emerging-ai-powered-software-for-brain-mri (accessed on 28 January 2025).
- Swathi, V.; Sinduja, K.; Kumar, V.; Mahendar, A.; Prasad, G.; Samya, B. Deep learning-based brain tumor detection: An MRI segmentation approach. MATEC Web Conf. 2024, 392, 01157. [Google Scholar] [CrossRef]
- Guo, X.; Liu, T.; Chi, Q. Brain tumor diagnosis in MRI scans images using residual/shuffle network optimized by augmented falcon finch optimization. Sci. Rep. 2024, 14, 27911. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Esteva, A.; Chou, K.; Yeung, S.; Naik, N.; Madani, A.; Mottaghi, A.; Liu, Y.; Topol, E.; Dean, J.; Socher, R. Deep learning-enabled medical computer vision. NPJ Digit. Med. 2021, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Kapkiç, M.; Sağıroğlu, Ş. Privacy Issues in Magnetic Resonance Images. Int. J. Inf. Secur. Sci. 2022, 12, 21–31. [Google Scholar] [CrossRef]
- Garbuzova, E. Ethical Concerns About Reidentification of Individuals from MRI Neuroimages. Voices Bioeth. 2021, 7. [Google Scholar] [CrossRef]
- Shen, F.X.; Wolf, S.M.; Gonzalez, R.G.; Garwood, M. Ethical Issues Posed by Field Research Using Highly Portable and Cloud-Enabled Neuroimaging. Neuron 2020, 105, 771–775. [Google Scholar] [CrossRef]
- Koçak, B.; Ponsiglione, A.; Stanzione, A.; Bluethgen, C.; Santinha, J.; Ugga, L.; Huisman, M.; Klontzas, M.E.; Cannella, R.; Cuocolo, R. Bias in artificial intelligence for medical imaging: Fundamentals, detection, avoidance, mitigation, challenges, ethics, and prospects. Diagn. Interv. Radiol. 2025, 31, 75–88. [Google Scholar] [CrossRef]
- Larrazabal, A.J.; Nieto, N.; Peterson, V.; Milone, D.H.; Ferrante, E. Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis. Proc. Natl. Acad. Sci. USA 2020, 117, 12592–12594. [Google Scholar] [CrossRef]
- Choi, K.S.; Sunwoo, L. Artificial Intelligence in Neuroimaging: Clinical Applications. Investig. Magn. Reson. Imaging 2022, 26, 1–9. [Google Scholar] [CrossRef]
- Khalighi, S.; Reddy, K.; Midya, A.; Pandav, K.B.; Madabhushi, A.; Abedalthagafi, M. Artificial intelligence in neuro-oncology: Advances and challenges in brain tumor diagnosis, prognosis, and precision treatment. Npj Precis. Oncol. 2024, 8, 80. [Google Scholar] [CrossRef]
- Ali, S.; Tejani, Y.; Seng, N.; Yin, X.; Rayan, J.C. Understanding and Mitigating Bias in Imaging Artificial Intelligence. Radiographics 2024, 44, 5. [Google Scholar]
- Brady, A.P.; Allen, B.; Chong, J.; Kotter, E.; Kottler, N.; Mongan, J.; Oakden-Rayner, L.; dos Santos, D.P.; Tang, A.; Wald, C.; et al. Developing, Purchasing, Implementing and Monitoring AI Tools in Radiology: Practical Considerations. A Multi-Society Statement from the ACR, CAR, ESR, RANZCR and RSNA. Can. Assoc. Radiol. J. 2024, 75, 226–244. [Google Scholar] [CrossRef]
- Heikkiläarchive, M. Five Things you Need to Know About the EU’s New AI Act. MIT Technology Review. 2023. Available online: https://www.technologyreview.com/2023/12/11/1084942/five-things-you-need-to-know-about-the-eus-new-ai-act/ (accessed on 3 February 2025).
- Herington, J.; McCradden, M.D.; Creel, K.; Boellaard, R.; Jones, E.C.; Jha, A.K.; Rahmim, A.; Scott, P.J.; Sunderland, J.J.; Wahl, R.L.; et al. Ethical Considerations for Artificial Intelligence in Medical Imaging: Deployment and Governance. J. Nucl. Med. 2023, 64, 1509–1515. [Google Scholar] [CrossRef]
- Price, W.N.; Sendak, M.; Balu, S.; Singh, K. Enabling collaborative governance of medical AI. Nat. Mach. Intell. 2023, 5, 821–823. [Google Scholar] [CrossRef]
- Usha, M.P.; Kannan, G.; Ramamoorthy, M. Multimodal brain tumor classification using convolutional Tumnet architecture. Behav. Neurol. 2024, 2024, 4678554. [Google Scholar] [CrossRef]
- Liu, Y.; Mu, F.; Shi, Y.; Cheng, J.; Li, C.; Chen, X. Brain tumor segmentation in multimodal MRI via pixel-level and feature-level image fusion. Front. Neurosci. 2022, 16, 1000587. [Google Scholar] [CrossRef]
- Pati, S.; Baid, U.; Edwards, B.; Sheller, M.; Wang, S.-H.; Reina, G.A.; Foley, P.; Gruzdev, A.; Karkada, D.; Davatzikos, C.; et al. Federated learning enables big data for rare cancer boundary detection. Nat. Commun. 2022, 13, 7346. [Google Scholar] [CrossRef]
- Kondepudi, A.; Pekmezci, M.; Hou, X.; Scotford, K.; Jiang, C.; Rao, A.; Harake, E.S.; Chowdury, A.; Al-Holou, W.; Wang, L.; et al. Foundation models for fast, label-free detection of Glioma infiltration. Nature 2025, 637, 439–445. [Google Scholar] [CrossRef]
- Hammad, M.; ElAffendi, M.; Ateya, A.A.; Abd El-Latif, A.A. Efficient brain tumor detection with lightweight end-to-end deep learning model. Cancers 2023, 15, 2837. [Google Scholar] [CrossRef]
- Avazov, K.; Mirzakhalilov, S.; Umirzakova, S.; Abdusalomov, A.; Cho, Y.I. Dynamic focus on tumor boundaries: A Lightweight U-Net for MRI brain tumor segmentation. Bioengineering 2024, 11, 1302. [Google Scholar] [CrossRef]
- Li, Z.; Dib, O. Empowering brain tumor diagnosis through explainable deep learning. Mach. Learn. Knowl. Extr. 2025, 6, 2248–2281. [Google Scholar] [CrossRef]
- Rawas, S.; Tafran, C.; AlSaeed, D. ChatGPT-powered deep learning: Elevating brain tumor detection in MRI scans. Appl. Comput. Inform. 2024, 2024, 4678554. [Google Scholar] [CrossRef]
- Rasool, N.; Wani, N.A.; Bhat, J.I.; Saharan, S.; Sharma, V.K.; Alsulami, B.S.; Alsharif, H.; Lytras, M.D. CNN-TumorNet: Leveraging explainability in deep learning for precise brain tumor diagnosis on MRI images. Front. Oncol. 2025, 15, 1554559. [Google Scholar] [CrossRef]
- Vavoulis, D.V.; Cutts, A.; Thota, N.; Brown, J.; Sugar, R.; Rueda, A.; Ardalan, A.; Howard, K.; Santo, F.M.; Sannasiddappa, T.; et al. Multimodal cell-free DNA whole-genome TAPS is sensitive and reveals specific cancer signals. Nat. Commun. 2025, 16, 430. [Google Scholar] [CrossRef]
- Wang, J.-Y.; Qu, V.; Hui, C.; Sandhu, N.; Mendoza, M.G.; Panjwani, N.; Chang, Y.-C.; Liang, C.-H.; Lu, J.-T.; Wang, L.; et al. Stratified assessment of an FDA-cleared deep learning algorithm for automated detection and contouring of metastatic brain tumors in stereotactic radiosurgery. Radiat. Oncol. 2023, 18, 61. [Google Scholar] [CrossRef]
- Lu, S.-L.; Xiao, F.-R.; Cheng, J.C.-H.; Yang, W.-C.; Cheng, Y.-H.; Chang, Y.-C.; Lin, J.-Y.; Liang, C.-H.; Lu, J.-T.; Chen, Y.-F.; et al. Randomized multi-reader evaluation of automated detection and segmentation of brain tumors in stereotactic radiosurgery with deep neural networks. Neuro-Oncology 2021, 23, 1560–1568. [Google Scholar] [CrossRef]
- Global Brain Cancer Diagnostic Market Industry Overview and Forecasts to 2031—Market Analysis and Market Share. Available online: https://www.databridgemarketresearch.com/reports/global-brain-cancer-diagnostic-market (accessed on 12 February 2025).
- CWRU Center for Imaging Research: $3M Grant to Advance MRI Scan. Available online: https://case.edu/medicine/ccir/news-articles/case-western-reserve-university-awarded-3m-grant-advance-mri-scan-and-software (accessed on 3 February 2025).
- Hugh Adams, National Cancer Plan in 2025. Available online: https://braintumourresearch.org/blogs/research-campaigning-news/national-cancer-plan-in-2025 (accessed on 1 March 2025).
- American Brain Tumor Association. Available online: https://www.abta.org/research/research-initiatives/ (accessed on 1 March 2025).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).