Energy Dissipation Hypothesis Applied to Enhance the Affinity of Thrombin Binding Aptamer
Abstract
1. Introduction
2. Materials and Methods
2.1. Analysis of Aptamer–Thrombin Complexes
2.2. Assembly of Aptamer Structure
2.3. Functional Study
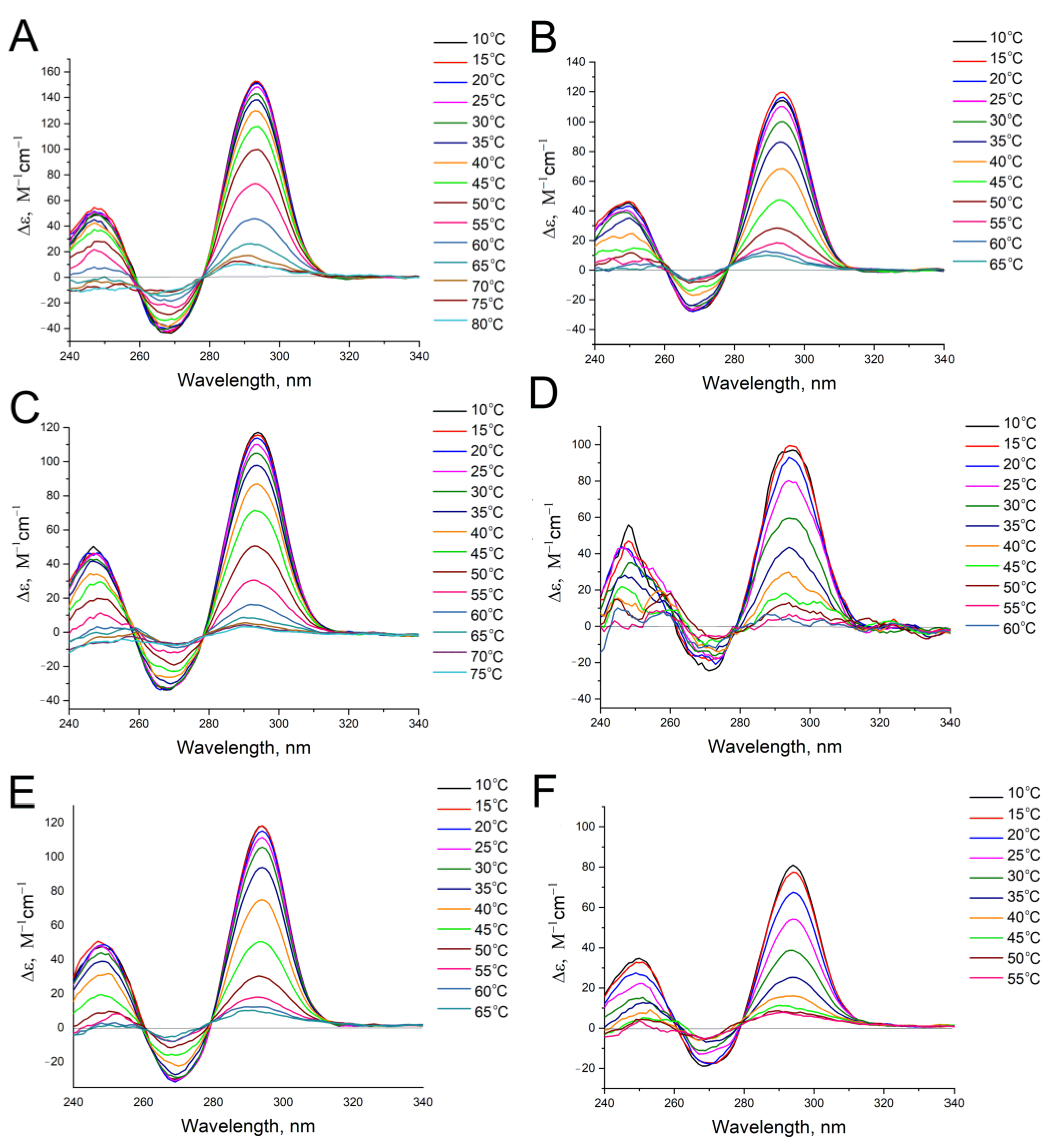
2.4. Circular Dichroism Spectroscopy
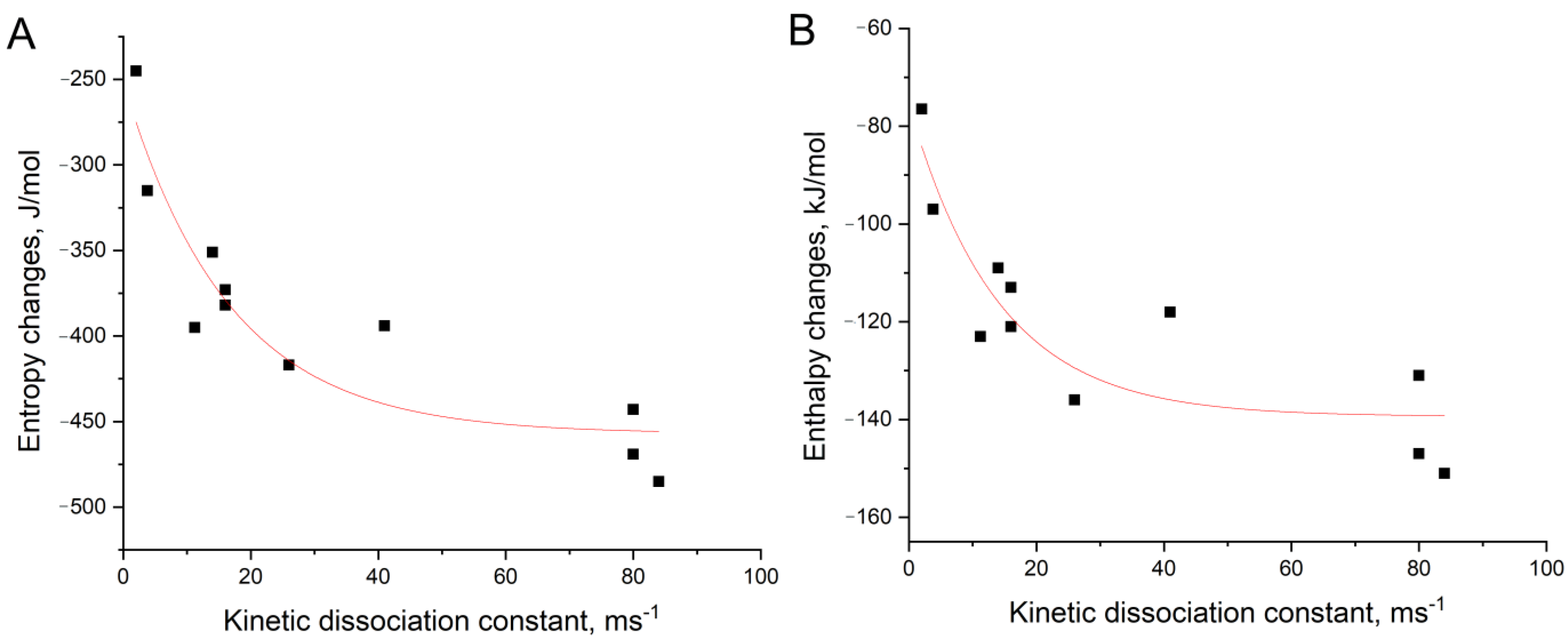
2.5. Determination of Thermodynamic Parameters
3. Results
3.1. Analysis of Nucleic Acid Interface of Aptamers to Thrombin
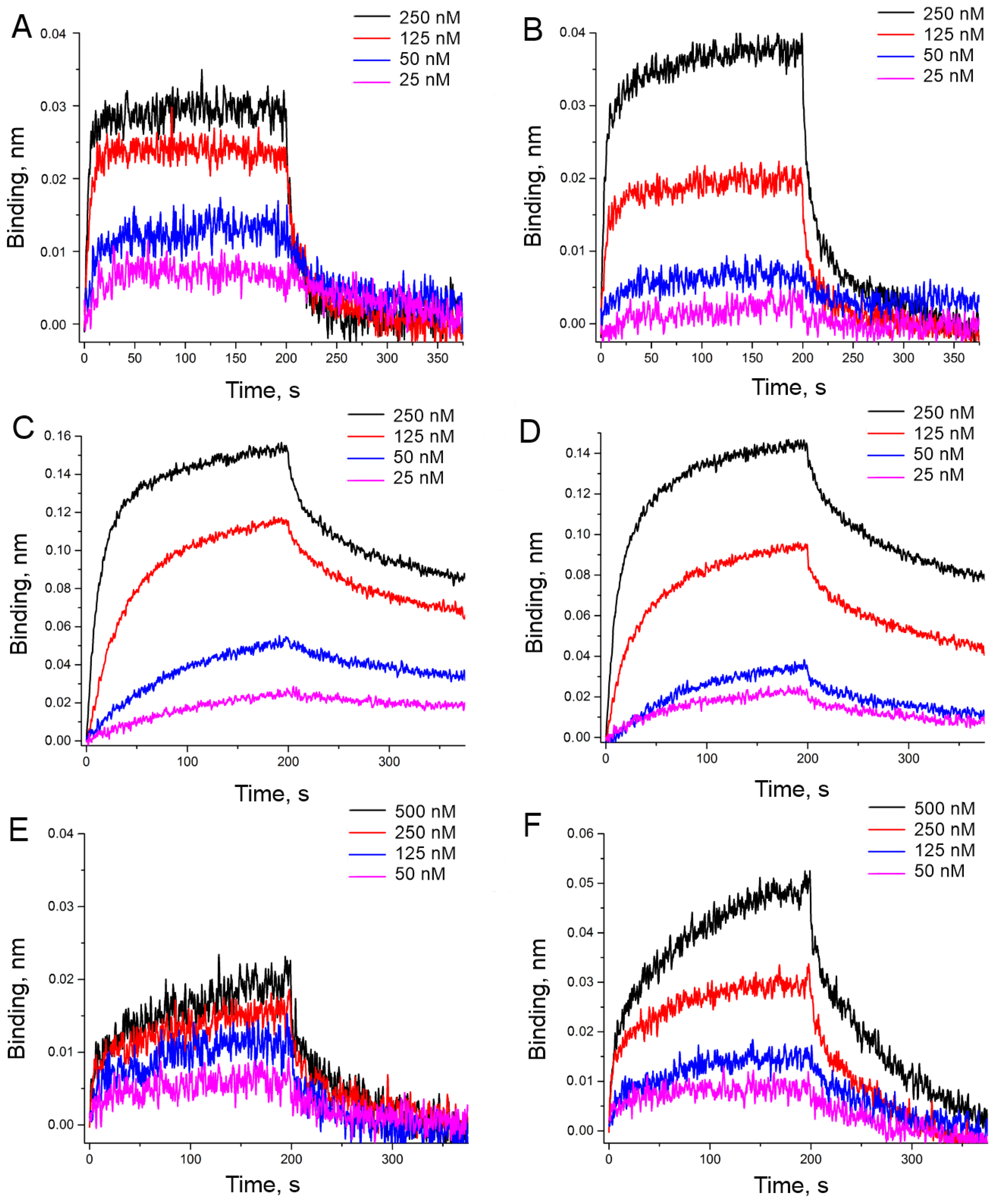
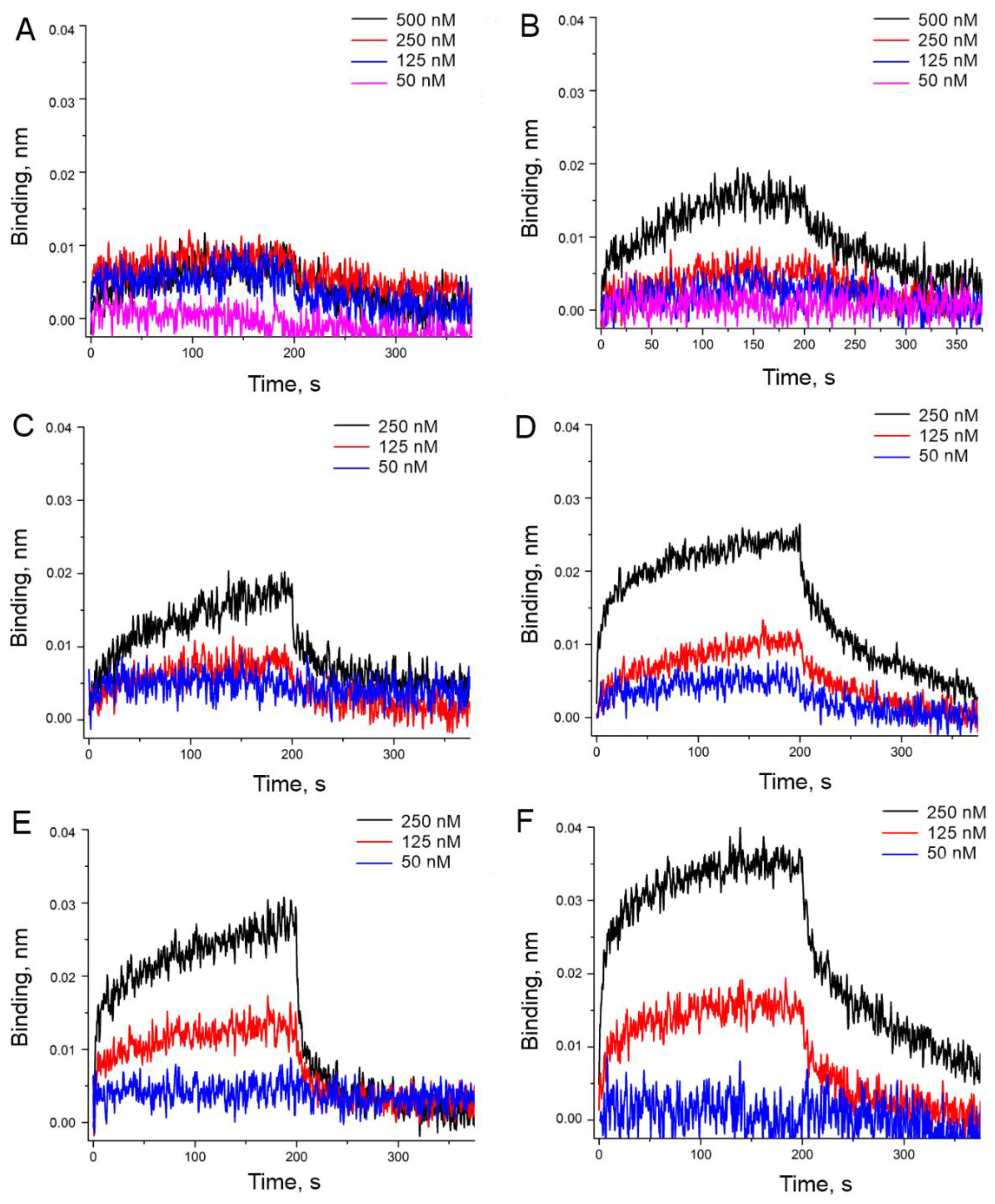
3.2. An Attempt to Experimentally Verify the Energy Dissipation Hypothesis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Blind, M.; Blank, M. Aptamer selection technology and recent advances. Mol. Ther. Nucleic Acids 2015, 4, e223. [Google Scholar] [CrossRef]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent advances in aptamer discovery and applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef]
- Kalra, P.; Dhiman, A.; Cho, W.C.; Bruno, J.G.; Sharma, T.K. Simple methods and rational design for enhancing aptamer sensitivity and specificity. Front. Mol. Biosci. 2018, 5, 41. [Google Scholar] [CrossRef]
- Elskens, J.P.; Elskens, J.M.; Madder, A. Chemical modification of aptamers for increased binding affinity in diagnostic applications: Current status and future prospects. Int. J. Mol. Sci. 2020, 21, 4522. [Google Scholar] [CrossRef]
- Hasegawa, H.; Savory, N.; Abe, K.; Ikebukuro, K. Methods for improving aptamer binding affinity. Molecules 2016, 21, 421. [Google Scholar] [CrossRef] [PubMed]
- Gelinas, A.; Davies, D.; Janjic, N. Embracing proteins: Structural themes in aptamer–protein complexes. Curr. Opin. Struct. Biol. 2016, 36, 122–132. [Google Scholar] [CrossRef]
- Yatime, L.; Maasch, C.; Hoehlig, K.; Klussmann, S.; Andersen, G.R.; Vater, A. Structural basis for the targeting of complement anaphylatoxin C5a using a mixed L-RNA/L-DNA aptamer. Nat. Commun. 2015, 6, 6481. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.H.; Fremont, D.H.; Diener, J.L.; Schaub, R.G.; Sadler, J.E. A structural explanation for the antithrombotic activity of ARC1172, a DNA aptamer that binds von Willebrand factor domain A1. Structure 2009, 17, 1476–1484. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Ikeda, H.; Miyakawa, S.; Futakawa, S.; Nonaka, Y.; Fujiwara, M.; Okudaira, S.; Kano, K.; Aoki, J.; Morita, J.; et al. Structural basis for specific inhibition of Autotaxin by a DNA aptamer. Nat. Struct. Mol. Biol. 2016, 23, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Gelinas, A.D.; von Carlowitz, I.; Janjic, N.; Pyle, A.M. Structural basis for IL-1α recognition by a modified DNA aptamer that specifically inhibits IL-1α signaling. Nat. Commun. 2017, 8, 810. [Google Scholar] [CrossRef]
- Jarvis, T.C.; Davies, D.R.; Hisaminato, A.; Resnicow, D.I.; Gupta, S.; Waugh, S.M.; Nagabukuro, A.; Wadatsu, T.; Hishigaki, H.; Gawande, B.; et al. Non-helical DNA triplex forms a unique aptamer scaffold for high affinity recognition of nerve growth factor. Structure 2015, 23, 1293–1304. [Google Scholar] [CrossRef]
- Novoseltseva, A.; Zavyalova, E.; Golovin, A.; Kopylov, A. An insight into aptamer–protein complexes. Aptamers 2018, 2, 1–19. [Google Scholar]
- Zavyalova, E.; Kopylov, A. Energy transfer as a driving force in nucleic acid–protein interactions. Molecules 2019, 24, 1443. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.R.; Gelinas, A.D.; Zhang, C.; Rohloff, J.C.; Carter, J.D.; O’Connell, D.; Waugh, S.M.; Wolk, S.K.; Mayfield, W.S.; Burgin, A.B.; et al. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc. Natl. Acad. Sci. USA 2012, 109, 19971–19976. [Google Scholar] [CrossRef] [PubMed]
- Zavyalova, E.G.; Legatova, V.A.; Alieva, R.S.; Zalevsky, A.O.; Tashlitsky, V.N.; Arutyunyan, A.M.; Kopylov, A.M. Putative mechanisms underlying high inhibitory activities of bimodular DNA aptamers to thrombin. Biomolecules 2019, 9, 41. [Google Scholar] [CrossRef] [PubMed]
- Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB). Available online: https://www.rcsb.org/ (accessed on 15 March 2021).
- SPR-Pages. Available online: https://www.sprpages.nl/ (accessed on 15 March 2021).
- Mergny, J.L.; Lacroix, L. Analysis of thermal melting curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef]
- Russo Krauss, I.; Merlino, A.; Randazzo, A.; Novellino, E.; Mazzarella, L.; Sica, F. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012, 40, 8119–8128. [Google Scholar] [CrossRef]
- Pica, A.; Russo Krauss, I.; Merlino, A.; Nagatoishi, S.; Sugimoto, N.; Sica, F. Dissecting the contribution of thrombin exosite I in the recognition of thrombin binding aptamer. FEBS J. 2013, 280, 6581–6588. [Google Scholar] [CrossRef]
- Nagatoishi, S.; Sugimoto, N. Interaction of water with the G-quadruplex loop contributes to the binding energy of G-quadruplex to protein. Mol. Biosyst. 2012, 8, 2766–2770. [Google Scholar] [CrossRef]
- Zavyalova, E.; Kopylov, A. G-quadruplexes and i-motifs as scaffolds for molecular engineering of DNA aptamers. In G-Quadruplex Structures, Formation and Roles in Biology; Santos, H., Ed.; Nova Publishers: New York, NY, USA, 2016; pp. 53–80. [Google Scholar]
- Troisi, R.; Napolitano, V.; Spiridonova, V.; Russo Krauss, I.; Sica, F. Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer. Nucleic Acids Res. 2018, 46, 12177–12185. [Google Scholar] [CrossRef]
- Zavyalova, E.; Tagiltsev, G.; Reshetnikov, R.; Arutyunyan, A.; Kopylov, A. Cation coordination alters the conformation of a thrombin-binding G-quadruplex DNA aptamer that affects inhibition of thrombin. Nucleic Acid Ther. 2016, 26, 299–308. [Google Scholar] [CrossRef]
- Russo Krauss, I.; Merlino, A.; Giancola, C.; Randazzo, A.; Mazzarella, L.; Sica, F. Thrombin-aptamer recognition: A revealed ambiguity. Nucleic Acids Res. 2011, 39, 7858–7867. [Google Scholar] [CrossRef] [PubMed]
- Pagano, B.; Martino, L.; Randazzo, A.; Giancola, C. Stability and binding properties of a modified thrombin binding aptamer. Biophys. J. 2008, 94, 562–569. [Google Scholar] [CrossRef]
- Dolot, R.; Lam, C.H.; Sierant, M.; Zhao, Q.; Liu, F.W.; Nawrot, B.; Egli, M.; Yang, X. Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity. Nucleic Acids Res. 2018, 46, 4819–4830. [Google Scholar] [CrossRef] [PubMed]
- Russo Krauss, I.; Spiridonova, V.; Pica, A.; Napolitano, V.; Sica, F. Different duplex/quadruplex junctions determine the properties of anti-thrombin aptamers with mixed folding. Nucleic Acids Res. 2016, 44, 983–991. [Google Scholar] [CrossRef]
- Spiridonova, V.A.; Barinova, K.V.; Glinkina, K.A.; Melnichuk, A.V.; Gainutdynov, A.A.; Safenkova, I.V.; Dzantiev, B.B. A family of DNA aptamers with varied duplex region length that forms complexes with thrombin and prothrombin. FEBS Lett. 2015, 589, 2043–2049. [Google Scholar] [CrossRef] [PubMed]
- Zavyalova, E.; Golovin, A.; Pavlova, G.; Kopylov, A. Module-activity relationship of G-quadruplex based DNA aptamers for human thrombin. Curr. Med. Chem. 2013, 20, 4836–4843. [Google Scholar] [CrossRef] [PubMed]
- Russo Krauss, I.; Napolitano, V.; Petraccone, L.; Troisi, R.; Spiridonova, V.; Mattia, C.A.; Sica, F. Duplex/quadruplex oligonucleotides: Role of the duplex domain in the stabilization of a new generation of highly effective anti-thrombin aptamers. Int. J. Biol. Macromol. 2018, 107, 1697–1705. [Google Scholar] [CrossRef]
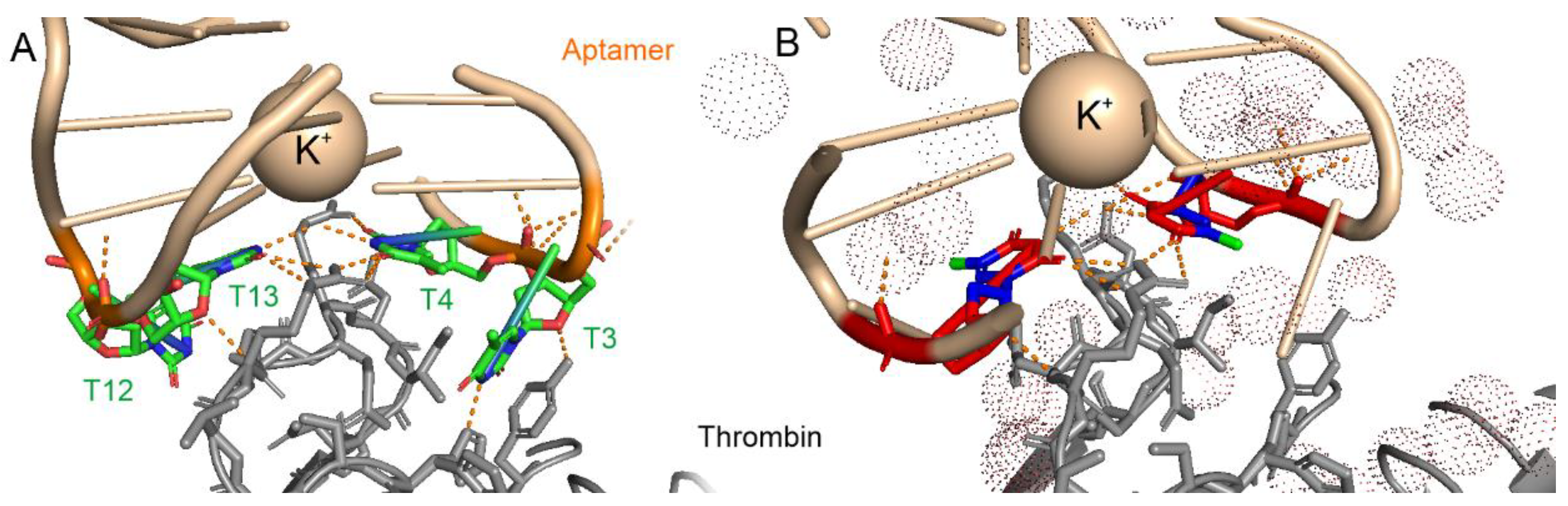

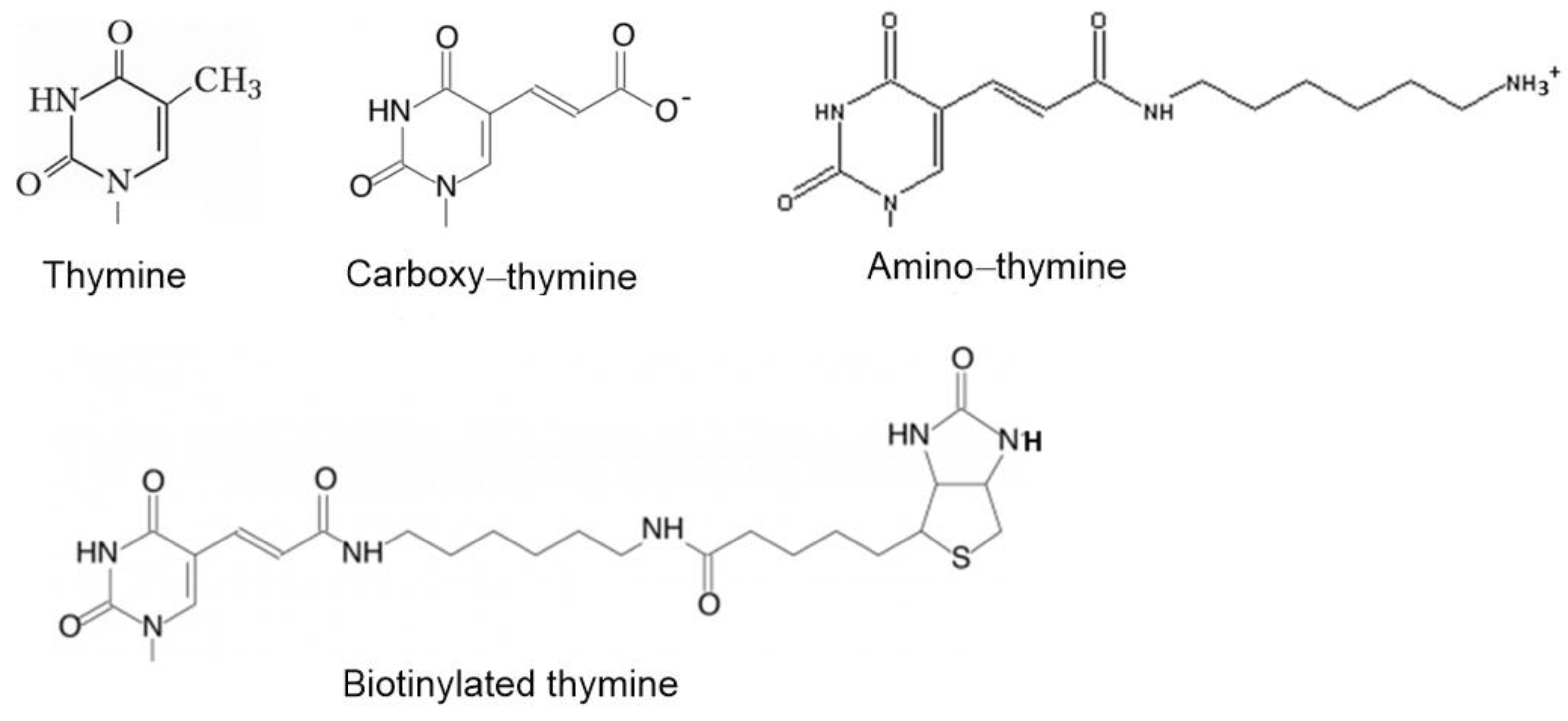

| pdb id | Aptamer-cation | KD, nM | Atoms in Contact with the Solvent | Atoms Shielded from the Solvent | Total Number of Atoms | Fraction of Shielded Atoms |
|---|---|---|---|---|---|---|
| 4lz4 [20] | ΔT3-K+ | 56 [21] | 8 | 12 | 20 | 0.60 |
| 4dih [19] | HD1-Na+ | 50 [22] | 13 | 11 | 24 | 0.46 |
| 4lz1 [20] | ΔT12-K+ | 42 [21] | 9 | 11 | 20 | 0.55 |
| 6gn7 [23] | NU172-Na+ | 35 [24] | 11 | 13 | 24 | 0.54 |
| 3qlp [25] | mTBA-K+ | 25 [26] | 13 | 11 | 24 | 0.46 |
| 4dii [19] | HD1-K+ | 12 [22] | 14 | 10 | 24 | 0.42 |
| 6eo6 [27] | T4W-K+ | 1 [27] | 21 | 14 | 35 | 0.40 |
| 5cmx [28] | RE31-K+ | 0.56 [29] | 11 | 13 | 24 | 0.54 |
| 6eo7 [27] | T4K-K+ | 0.39 [27] | 16 | 10 | 26 | 0.38 |
| 6evv [23] | NU172-K+ | 0.29 [30] | 11 | 13 | 24 | 0.54 |
| Aptamer | Sequence |
|---|---|
| HD1 | 5′-gg-tt-gg-tgt-gg-tt-gg-3′ |
| carboxy-T4,T13 | 5′-gg-t(t-carboxy)-gg-tgt-gg-t(t-carboxy)-gg-3′ |
| biotin-T4,T13 | 5′-gg-t(t-biotin)-gg-tgt-gg-t(t-biotin)-gg-3′ |
| amino-T4,T13 | 5′-gg-t(t-amino)-gg-tgt-gg-t(t-amino)-gg-3′ |
| amino-T13 | 5′-gg-tt-gg-tgt-gg-t(t-amino)-gg-3′ |
| amino-T3,T12 | 5′-gg-(t-amino)t-gg-tgt-gg-(t-amino)t-gg-3′ |
| Aptamer | ‘Potassium Buffer’ | ‘Sodium Buffer’ | ||||
|---|---|---|---|---|---|---|
| KD, nM | kon, µM−1s−1 | koff, ms−1 | KD, nM | kon, µM−1s−1 | koff, ms−1 | |
| HD1 | 32 ± 2 | 2.7 ± 0.4 | 84 ± 6 | 180 ± 16 | 0.43 ± 0.08 | 80 ± 20 |
| carboxy-T4,T13 | 10.1 ± 0.6 | 0.20 ± 0.03 | 2.0 ± 0.3 | 20.0 ± 1.6 | 0.19 ± 0.03 | 3.8 ± 1.2 |
| biotin-T4,T13 | 19 ± 2 | 1.4 ± 0.6 | 26 ± 4 | 25 ± 3 | 0.6 ± 0.2 | 14 ± 2 |
| amino-T4,T13 | >350 | n.d. | n.d. | 210 ± 20 | 0.05 ± 0.02 | 11.2 ± 1.6 |
| amino-T13 | 350 ± 40 | 0.048 ± 0.005 | 16 ± 8 | 64 ± 4 | 0.25 ± 0.04 | 16 ± 3 |
| amino-T3,T12 | 100 ± 20 | 0.8 ± 0.4 | 80 ± 30 | 77 ± 10 | 0.5 ± 0.2 | 41 ± 12 |
| Aptamer | ‘Potassium Buffer’ | ‘Sodium Buffer’ | ||||||
|---|---|---|---|---|---|---|---|---|
| Tm, °C | ΔH°, kJ/mol | ΔS°, J/mol | ΔG°298, kJ/mol | Tm, °C | ΔH°, kJ/mol | ΔS°, J/mol | ΔG°298, kJ/mol | |
| HD1 | 38.7 ± 0.1 | −151 ± 3 | −485 ± 9 | −6 ± 2 | 22.8 ± 0.6 | −131 ± 4 | −443 ± 12 | 1 ± 3 |
| carboxy-T4,T13 | 40.3 ± 0.6 | −77 ± 2 | −245 ± 5 | −3.5 ± 1.1 | 34.0 ± 0.5 | −97 ± 2 | −315 ± 8 | −2.8 ± 1.6 |
| biotin-T4,T13 | 52.6 ± 0.4 | −136 ± 5 | −417 ± 14 | −11 ± 3 | 39.5 ± 0.3 | −109 ± 2 | −351 ± 7 | −4.8 ± 1.5 |
| amino-T4,T13 | 52.6 ± 0.5 | −120 ± 5 | −369 ± 17 | −10 ± 4 | 39.7 ± 0.4 | −123 ± 2 | −395 ± 7 | −5.6 ± 1.7 |
| amino-T13 | 46.0 ± 0.3 | −121 ± 3 | −382 ± 9 | −8 ± 2 | 30.1 ± 0.7 | −113 ± 3 | −373 ± 10 | −2 ± 2 |
| amino-T3,T12 | 40.6 ± 0.2 | −147 ± 3 | −469 ± 9 | −7 ± 2 | 25.8 ± 0.2 | −118 ± 2 | −394 ± 6 | −0.3 ± 1.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhdanov, G.; Arutyunyuan, A.; Kopylov, A.; Zavyalova, E. Energy Dissipation Hypothesis Applied to Enhance the Affinity of Thrombin Binding Aptamer. Biophysica 2021, 1, 179-193. https://doi.org/10.3390/biophysica1020014
Zhdanov G, Arutyunyuan A, Kopylov A, Zavyalova E. Energy Dissipation Hypothesis Applied to Enhance the Affinity of Thrombin Binding Aptamer. Biophysica. 2021; 1(2):179-193. https://doi.org/10.3390/biophysica1020014
Chicago/Turabian StyleZhdanov, Gleb, Alexander Arutyunyuan, Alexey Kopylov, and Elena Zavyalova. 2021. "Energy Dissipation Hypothesis Applied to Enhance the Affinity of Thrombin Binding Aptamer" Biophysica 1, no. 2: 179-193. https://doi.org/10.3390/biophysica1020014
APA StyleZhdanov, G., Arutyunyuan, A., Kopylov, A., & Zavyalova, E. (2021). Energy Dissipation Hypothesis Applied to Enhance the Affinity of Thrombin Binding Aptamer. Biophysica, 1(2), 179-193. https://doi.org/10.3390/biophysica1020014








