Journal Description
Biophysica
Biophysica
is an international, peer-reviewed, open access journal on applying the methods of physics, chemistry, and math to study biological systems, published quarterly online by MDPI.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within ESCI (Web of Science), Scopus, EBSCO, and other databases.
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 22.9 days after submission; acceptance to publication is undertaken in 4.6 days (median values for papers published in this journal in the first half of 2025).
- Recognition of Reviewers: APC discount vouchers, optional signed peer review and reviewer names are published annually in the journal.
- Biophysica is a companion journal of IJMS.
Impact Factor:
1.4 (2024);
5-Year Impact Factor:
1.3 (2024)
Latest Articles
Isolation of an Anti-hG-CSF Nanobody and Its Application in Quantitation and Rapid Detection of hG-CSF in Pharmaceutical Testing
Biophysica 2025, 5(4), 47; https://doi.org/10.3390/biophysica5040047 - 17 Oct 2025
Abstract
►
Show Figures
Human granulocyte colony-stimulating factor (hG-CSF) is primarily used to treat neutropenia induced by cancer chemotherapy and bone marrow transplantation. The current identification test for hG-CSF relies on Western blot (WB), a labor-intensive and technically demanding method. This study aimed to screen and prepare
[...] Read more.
Human granulocyte colony-stimulating factor (hG-CSF) is primarily used to treat neutropenia induced by cancer chemotherapy and bone marrow transplantation. The current identification test for hG-CSF relies on Western blot (WB), a labor-intensive and technically demanding method. This study aimed to screen and prepare an anti-hG-CSF nanobody to identify and quantify hG-CSF, with the ultimate goal of developing colloidal gold-labeled nanobody test strips for rapid identification. An alpaca was immunized with hG-CSF, and the VHH gene sequence encoding the anti-hG-CSF nanobody was obtained through sequencing following phage display library construction and multiple rounds of biopanning. The nanobody C68, obtained from screening, was expressed by E. coli, and its physicochemical properties such as molecular weight, isoelectric point, and affinity were characterized after purification. WB analysis demonstrated excellent performance of the nanobody in identification tests in terms of specificity, limit of detection (LOD), applicability with products from various manufacturers, and thermal stability. Additionally, we established an ELISA method for hG-CSF quantification utilizing the nanobody C68 and conducted methodological validation. Finally, colloidal gold-based test strips were constructed using the nanobody C68, with a LOD of 30 μg/mL, achieving rapid identification for hG-CSF. This study represents a novel application of nanobodies in pharmaceutical testing and offers valuable insights for developing identification tests for other recombinant protein drugs.
Full article
Open AccessArticle
Follow-Up of APSified–BMO-Based Retinal Microcirculation in Patients with Post-COVID-19 Syndrome
by
Cornelius Rosenkranz, Marianna Lucio, Marion Ganslmayer, Thomas Harrer, Jakob Hoffmanns, Charlotte Szewczykowski, Thora Schröder, Franziska Raith, Stephanie Zellinger, Denzel Abelardo, Jule Schumacher, Merle Flecks, Petra Lakatos, Christian Mardin and Bettina Hohberger
Biophysica 2025, 5(4), 46; https://doi.org/10.3390/biophysica5040046 - 16 Oct 2025
Abstract
►▼
Show Figures
Post-COVID-19 syndrome (PCS) is a multifactorial disorder comprising different subgroups. Our study aimed to investigate the longitudinal changes in retinal microcirculation in PCS patients. Eighty PCS patients were recruited at the Department of Ophthalmology at the Friedrich-Alexander University of Erlangen-Nürnberg. Retinal microcirculation was
[...] Read more.
Post-COVID-19 syndrome (PCS) is a multifactorial disorder comprising different subgroups. Our study aimed to investigate the longitudinal changes in retinal microcirculation in PCS patients. Eighty PCS patients were recruited at the Department of Ophthalmology at the Friedrich-Alexander University of Erlangen-Nürnberg. Retinal microcirculation was measured twice using optical coherence tomography angiography (OCT-A) within the superficial vascular plexus (SVP), intermediate capillary plexus (ICP), deep capillary plexus (DCP), and peripapillary region. Vessel density (VD) was calculated using the Erlangen Angio Tool with an APSified and Bruch’s membrane opening-based analyses. The least-squares means (LS-Means) of VD were 30.4 (SE = 0.168) vs. 30.3 (SE = 0.166) (SVP), 22.4 (SE = 0.143) vs. 22.2 (SE = 0.141) (ICP), 23.9 (SE = 0.186) vs. 23.8 (SE = 0.185) (DCP), and 27.4 (SE = 0.226) vs. 27.0 (SE = 0.224) (peripapillary) in patients with PCS at visits 1 and 2, respectively. The study cohort showed physically stable PCS symptoms with PEM/fatigue and concentration disorders as major symptoms and only a slight, clinically irrelevant improvement of the Bell Score. The multivariate longitudinal model confirmed the clinical observations by showing that VD did not change significantly during follow-up (p = 0.46). Strong interdependencies between the macular layers (p < 0.001) were observed. The data of the present study suggests that while overall APSified macular VD and BMO-based APSified peripapillary VD were stable within a PCS cohort of physically stable PCS symptoms, individual patients may experience coordinated microvascular changes, particularly within the macular plexuses. Together, the results support a model of heterogeneous yet biologically consistent microvascular response in PCS pathophysiology.
Full article
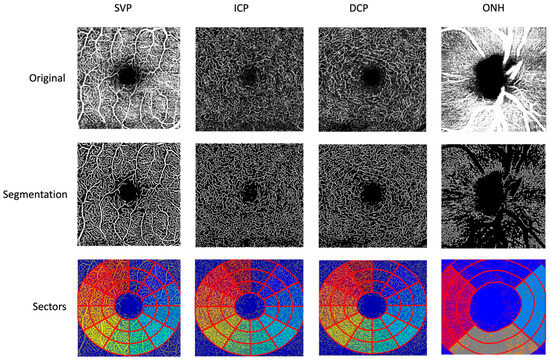
Figure 1
Open AccessReview
Effect of Microgravity and Space Radiation Exposure on Human Oral Health: A Systematic Review
by
Shahnawaz Khijmatgar, Matteo Pellegrini, Martina Ghizzoni and Massimo Del Fabbro
Biophysica 2025, 5(4), 45; https://doi.org/10.3390/biophysica5040045 - 29 Sep 2025
Abstract
►▼
Show Figures
A systematic review was conducted to assess the effects of microgravity and space radiation on astronauts’ oral health. This review aimed to determine if these conditions increase the risk of dental and periodontal diseases, identify pre-mission dental care strategies, and specify relevant dental
[...] Read more.
A systematic review was conducted to assess the effects of microgravity and space radiation on astronauts’ oral health. This review aimed to determine if these conditions increase the risk of dental and periodontal diseases, identify pre-mission dental care strategies, and specify relevant dental emergencies for astronauts to manage during missions. Following PRISMA guidelines, the review was registered on PROSPERO (CRD42023472765). Databases including PubMed, Scopus, Web of Science, Cochrane Library, and OVID Medline were searched. Of the 13 studies identified, 7 were eligible for qualitative synthesis. The included studies revealed that space conditions compromise oral health. Findings indicate changes in saliva composition, with a significant decline in salivary lysozyme levels during missions lasting 28 to 84 days. Salivary IgA levels also increased before and peaked after flights (microgravity alters fluid shear and protein folding). Viral reactivation was a key finding, with latent viruses such as Epstein–Barr virus (EBV), cytomegalovirus (CMV), and varicella zoster virus (VZV) being reactivated during missions (immune suppression and gene expression shifts under spaceflight stress). Data from a study found that 50% of crew members shed viruses in their saliva or urine, and 38% tested positive for herpesviruses. The included studies also documented alterations in the oral microbiome, including increased gastrointestinal and decreased nasal microbial diversity. This suggests alterations in salivary biomarkers, viral shedding, and microbiome changes in astronauts during long-duration missions. These changes appear associated with immune dysregulation and stress, but causality remains uncertain due to observational designs, small heterogeneous samples, and confounding factors. Although current evidence is indicative rather than definitive, these findings highlight the need for preventive dental measures prior to missions and preparedness for managing oral emergencies in-flight. Future studies should address the mechanistic separation of microgravity and radiation effects, with implications for upcoming Moon and Mars missions.
Full article

Figure 1
Open AccessReview
Role of Lipid Composition on the Mechanical and Biochemical Vulnerability of Myelin and Its Implications for Demyelinating Disorders
by
Marcela Ana Morini and Viviana Isabel Pedroni
Biophysica 2025, 5(4), 44; https://doi.org/10.3390/biophysica5040044 - 26 Sep 2025
Abstract
Myelin is a membranous structure critically important for human health. Historically, it was believed that myelin remained largely unchanged in the adult brain. However, recent research has shown that myelin is remarkably dynamic, capable of adjusting axonal conduction velocity and playing a role
[...] Read more.
Myelin is a membranous structure critically important for human health. Historically, it was believed that myelin remained largely unchanged in the adult brain. However, recent research has shown that myelin is remarkably dynamic, capable of adjusting axonal conduction velocity and playing a role in learning, memory, and recovery from injury, in response to both physiological and pathological signals. Axons are more efficiently insulated in myelinated fibers, where segments of the axonal membrane are wrapped by the myelin sheath. Although extensive data are available on the electrical properties of myelin, its structural and mechanical characteristics—as well as the role of its lipid composition—are also relevant, although much less explored. The objective of our review is derived from this point since alterations in lipid components can lead to axonal dysfunction, giving rise to neurological disorders such as multiple sclerosis and other demyelinating conditions. In this review, concerning the lipid composition of myelin, we focus on two distinct classes of lipids: sphingolipids and long-chain fatty acids, emphasizing the differential contributions of saturated versus polyunsaturated species. We analyze studies that correlate the mechanical vulnerability of myelin with its lipid composition, particularly sphingomyelin, thereby underscoring its role in protecting neurons against physical stress and providing a robust microstructural network that reinforces the white matter as a whole. From a biochemical perspective, we examine the susceptibility of myelin to oxidative stress, metabolic disorders, and extreme nutritional deficiencies in relation to the role of long-chain fatty acids. Both perspectives highlight that the aforementioned lipids participate in a complex biomechanical balance that is essential for maintaining the stability of myelin and, consequently, the integrity of the central and peripheral nervous systems.
Full article
(This article belongs to the Collection Feature Papers in Biophysics)
►▼
Show Figures

Figure 1
Open AccessReview
AI-Enhanced Morphological Phenotyping in Humanized Mouse Models: A Transformative Approach to Infectious Disease Research
by
Asim Muhammad, Xin-Yu Zheng, Hui-Lin Gan, Yu-Xin Guo, Jia-Hong Xie, Yan-Jun Chen and Jin-Jun Chen
Biophysica 2025, 5(4), 43; https://doi.org/10.3390/biophysica5040043 - 24 Sep 2025
Abstract
Humanized mouse models offer human-specific platforms for investigating complex host–pathogen interactions, addressing shortcomings of conventional preclinical models that often fail to replicate human immune responses accurately. This integrative review examines the intersection of advanced morphological phenotyping and artificial intelligence (AI) to enhance predictive
[...] Read more.
Humanized mouse models offer human-specific platforms for investigating complex host–pathogen interactions, addressing shortcomings of conventional preclinical models that often fail to replicate human immune responses accurately. This integrative review examines the intersection of advanced morphological phenotyping and artificial intelligence (AI) to enhance predictive capacity and translational relevance in infectious disease research. A structured literature search was conducted across PubMed, Scopus, and Web of Science (2010–2025), applying defined inclusion and exclusion criteria. Evidence synthesis highlights imaging modalities, AI-driven phenotyping, and standardization strategies, supported by comparative analyses and quality considerations. Persistent challenges include variability in engraftment, lack of harmonized scoring systems, and ethical governance. We propose recommendations for standardized protocols, risk-of-bias mitigation, and collaborative training frameworks to accelerate adoption of these technologies in translational medicine.
Full article
(This article belongs to the Special Issue Advances in Computational Biophysics)
Open AccessPerspective
Rethinking Metabolic Imaging: From Static Snapshots to Metabolic Intelligence
by
Giuseppe Maulucci
Biophysica 2025, 5(3), 42; https://doi.org/10.3390/biophysica5030042 - 19 Sep 2025
Abstract
Metabolic imaging is undergoing a fundamental transformation. Traditionally confined to static representations of metabolite distribution through modalities such as PET, MRS, and MSOT, imaging has offered only partial glimpses into the dynamic and systemic nature of metabolism. This Perspective envisions a shift toward
[...] Read more.
Metabolic imaging is undergoing a fundamental transformation. Traditionally confined to static representations of metabolite distribution through modalities such as PET, MRS, and MSOT, imaging has offered only partial glimpses into the dynamic and systemic nature of metabolism. This Perspective envisions a shift toward dynamic metabolic intelligence—an integrated framework where real-time imaging is fused with physics-informed models, artificial intelligence, and wearable data to create adaptive, predictive representations of metabolic function. We explore how novel technologies like hyperpolarized MRI and time-resolved optoacoustics can serve as dynamic inputs into digital twin systems, enabling closed-loop feedback that not only visualizes but actively guides clinical decisions. From early detection of metabolic drift to in silico therapy simulation, we highlight translational pathways across oncology, cardiology, neurology, and space medicine. Finally, we call for a cross-disciplinary effort to standardize, validate, and ethically implement these systems, marking the emergence of a new paradigm: metabolism as a navigable, model-informed space of precision medicine.
Full article
(This article belongs to the Collection Feature Papers in Biophysics)
►▼
Show Figures

Figure 1
Open AccessReview
Resistance of Nitric Oxide Dioxygenase and Cytochrome c Oxidase to Inhibition by Nitric Oxide and Other Indications of the Spintronic Control of Electron Transfer
by
Paul R. Gardner
Biophysica 2025, 5(3), 41; https://doi.org/10.3390/biophysica5030041 - 9 Sep 2025
Abstract
Heme enzymes that bind and reduce O2 are susceptible to poisoning by NO. The high reactivity and affinity of NO for ferrous heme produces stable ferrous-NO complexes, which in theory should preclude O2 binding and turnover. However, NO inhibition is often
[...] Read more.
Heme enzymes that bind and reduce O2 are susceptible to poisoning by NO. The high reactivity and affinity of NO for ferrous heme produces stable ferrous-NO complexes, which in theory should preclude O2 binding and turnover. However, NO inhibition is often competitive with respect to O2 and rapidly reversible, thus providing cellular and organismal survival advantages. This kinetic paradox has prompted a search for mechanisms for reversal and hence resistance. Here, I critically review proposed resistance mechanisms for NO dioxygenase (NOD) and cytochrome c oxidase (CcO), which substantiate reduction or oxidation of the tightly bound NO but nevertheless fail to provide kinetically viable solutions. A ferrous heme intermediate is clearly not available during rapid steady-state turnover. Reversible inhibition can be attributed to NO competing with O2 in transient low-affinity interactions with either the ferric heme in NOD or the ferric heme-cupric center in CcO. Toward resolution, I review the underlying principles and evidence for kinetic control of ferric heme reduction via an O2-triggered ferric heme spin crossover and an electronically-forced motion of the heme and structurally-linked protein side chains that elicit electron transfer and activate O2 in the flavohemoglobin-type NOD. For CcO, kinetics, structures, and density functional theory point to the existence of an analogous O2 and reduced oxygen intermediate-controlled electron-transfer gate with a linked proton pump function. A catalytic cycle and mechanism for CcO is finally at hand that links each of the four O2-reducing electrons to each of the four pumped protons in time and space. A novel proton-conducting tunnel and channel, electron path, and pump mechanism, most notably first hypothesized by Mårten Wikström in 1977 and pursued since, are laid out for further scrutiny. In both models, low-energy spin-orbit couplings or ‘spintronic’ interactions with O2 and NO or copper trigger the electronic motions within heme that activate electron transfer to O2, and the exergonic reactions of transient reactive oxygen intermediates ultimately drive all enzyme, electron, and proton motions.
Full article
(This article belongs to the Special Issue Investigations into Protein Structure)
►▼
Show Figures

Figure 1
Open AccessReview
Advancing Precision Neurology and Wearable Electrophysiology: A Review on the Pivotal Role of Medical Physicists in Signal Processing, AI, and Prognostic Modeling
by
Constantinos Koutsojannis, Athanasios Fouras and Dionysia Chrysanthakopoulou
Biophysica 2025, 5(3), 40; https://doi.org/10.3390/biophysica5030040 - 5 Sep 2025
Abstract
►▼
Show Figures
Medical physicists are transforming physiological measurements and electrophysiological applications by addressing challenges like motion artifacts and regulatory compliance through advanced signal processing, artificial intelligence (AI), and statistical rigor. Their innovations in wearable electrophysiology achieve 8–12 dB signal-to-noise ratio (SNR) improvements in EEG, 60%
[...] Read more.
Medical physicists are transforming physiological measurements and electrophysiological applications by addressing challenges like motion artifacts and regulatory compliance through advanced signal processing, artificial intelligence (AI), and statistical rigor. Their innovations in wearable electrophysiology achieve 8–12 dB signal-to-noise ratio (SNR) improvements in EEG, 60% motion artifact reduction, and 94.2% accurate AI-driven arrhythmia detection at 12 μW power. In precision neurology, machine learning (ML) with evoked potentials (EPs) predicts spinal cord injury (SCI) recovery and multiple sclerosis (MS) progression with 79.2% accuracy based on retrospective data from 560 SCI/MS patients. By integrating multimodal data (EPs, MRI), developing quantum sensors, and employing federated learning, these can enhance diagnostic precision and prognostic accuracy. Clinical applications span epilepsy, stroke, cardiac monitoring, and chronic pain management, reducing diagnostic errors by 28% and optimizing treatments like deep brain stimulation (DBS). In this paper, we review the current state of wearable devices and provide some insight into possible future directions. Embedding medical physicists into standardization efforts is critical to overcoming barriers like quantum sensor power consumption, advancing personalized, evidence-based healthcare.
Full article

Figure 1
Open AccessArticle
Exploring the Bottleneck in Cryo-EM Dynamic Disorder Feature and Advanced Hybrid Prediction Model
by
Sen Zheng
Biophysica 2025, 5(3), 39; https://doi.org/10.3390/biophysica5030039 - 29 Aug 2025
Abstract
►▼
Show Figures
Cryo-electron microscopy single-particle analysis (cryo-EM SPA) has advanced three-dimensional protein structure determination, yet resolving intrinsically disordered proteins and regions (IDPs/IDRs) remains challenging due to conformational heterogeneity. This research evaluates cryo-EM’s capacity to map dynamic regions, assesses the adaptability of disorder prediction tools, and
[...] Read more.
Cryo-electron microscopy single-particle analysis (cryo-EM SPA) has advanced three-dimensional protein structure determination, yet resolving intrinsically disordered proteins and regions (IDPs/IDRs) remains challenging due to conformational heterogeneity. This research evaluates cryo-EM’s capacity to map dynamic regions, assesses the adaptability of disorder prediction tools, and explores optimization strategies for dynamic structure prediction. Cryo-EM SPA datasets from 2000 to 2024 were categorized into different periods, forming a database integrating sequence data and disorder indices. Established prediction tools—AlphaFold2 (pLDDT), flDPnn, and IUPred—were evaluated for transferability, while a multi-level CLTC hybrid model (combining CNN, LSTM, Transformer, and CRF architectures) was developed to link local conformational fluctuations with global sequence contexts. Analyses revealed consistent advancements in average resolution and model counts over the past decade, although mapping disordered regions remained technically demanding. Both the adapted AlphaFold pLDDT and the CLTC model demonstrated efficacy in predicting structurally variable and poorly resolved regions. A subset of the cryo-EM missing residues exhibited intermediate conformational features, suggesting classification ambiguities potentially influenced by experimental conditions. These findings systematically outline the evolving capabilities of cryo-EM in resolving dynamic regions, benchmark the adaptability of computational tools, and introduce a hybrid model to enhance prediction accuracy. This study provides a framework for addressing conformational heterogeneity, contributing to methodological advancements in structural biology.
Full article

Figure 1
Open AccessReview
Organs-on-Chips: Revolutionizing Biomedical Research
by
Ankit Monga, Khush Jain, Harvinder Popli, Prashik Telgote, Ginpreet Kaur, Fariah Rizwani, Ritu Chauhan, Damandeep Kaur, Abhishek Chauhan and Hardeep Singh Tuli
Biophysica 2025, 5(3), 38; https://doi.org/10.3390/biophysica5030038 - 26 Aug 2025
Abstract
►▼
Show Figures
Organs-on-Chips (OoC) technology has begun to be considered a pragmatic tool for drug evaluation, offering researchers an opportunity to move beyond the less physiologically relevant animal models. OoCs are microfluidic structures that imitate the functionalities of individual human organs, serving as mimicry tools
[...] Read more.
Organs-on-Chips (OoC) technology has begun to be considered a pragmatic tool for drug evaluation, offering researchers an opportunity to move beyond the less physiologically relevant animal models. OoCs are microfluidic structures that imitate the functionalities of individual human organs, serving as mimicry tools for drug response and reproducibility studies. On the one hand, companies producing OoCs find managing and analyzing the large amounts of data generated challenging. This is where artificial intelligence (AI) can be deployed to address such problems. This paper will present the state-of-the-art of current OoC technology and AI, discussing the benefits and threats of combining these approaches. AI can be applied to optimize the process of OoC fabrication and operation, as well as for the big data analysis of OoC devices. By combining these technologies, scientists gain a powerful tool for drug development that is more efficient and accurate. However, processing the vast datasets generated by OoC systems often requires specialized AI expertise and computational resources. Despite the numerous possible benefits of amalgamating OoC technology with AI, several challenges and limitations need to be addressed. The large datasets generated by OoC systems can be difficult to process and analyze, which is a task that may require specialized AI expertise. Additionally, limitations of OoC systems include issues with reproducibility, as the devices are sensitive to perturbations in experimental conditions. Furthermore, the development and implementation of AI algorithms require significant computational resources and expertise, which may not be readily available to all research institutions. To overcome these challenges, interdisciplinary collaboration between biologists, engineers, data scientists, and AI experts is essential. Continued advancements in both OoC technology and AI will likely lead to more robust and versatile platforms for biomedical research and drug development, ultimately contributing to the advancement of personalized medicine and the reduction of reliance on animal testing.
Full article

Figure 1
Open AccessArticle
Exploring Therapeutic Dynamics: Mathematical Modeling and Analysis of Type 2 Diabetes Incorporating Metformin Dynamics
by
Alireza Mirzaee and Shantia Yarahmadian
Biophysica 2025, 5(3), 37; https://doi.org/10.3390/biophysica5030037 - 14 Aug 2025
Abstract
►▼
Show Figures
Type 2 diabetes (T2D) is a chronic metabolic disorder requiring effective management to avoid complications. Metformin is a first-line drug agent and is routinely prescribed for the control of glycemia, but its underlying dynamics are complicated and not fully quantified. This paper formulates
[...] Read more.
Type 2 diabetes (T2D) is a chronic metabolic disorder requiring effective management to avoid complications. Metformin is a first-line drug agent and is routinely prescribed for the control of glycemia, but its underlying dynamics are complicated and not fully quantified. This paper formulates a control-oriented and interpretable mathematical model that integrates metformin dynamics into a classic beta-cell–insulin–glucose (BIG) regulation system. The paper’s applicability to theoretical and clinical settings is enhanced by rigorous mathematical analysis, which guarantees the model is globally bounded, well-posed, and biologically meaningful. One of the key features of the study is its global stability analysis using Lyapunov functions, which demonstrates the asymptotic stability of critical equilibrium points under realistic physiological constraints. These findings support the predictive reliability of the model in explaining long-term glycemic regulation. Bifurcation analysis also clarifies the dynamic interplay between glucose production and utilization by identifying parameter thresholds that signify transitions between homeostasis and pathological states. Residual analysis, which detects Gaussian-distributed errors, underlines the robustness of the fitting process and suggests possible refinements by including temporal effects. Sensitivity analysis highlights the predominant effect of the initial dose of metformin on long-term glucose regulation and provides practical guidance for optimizing individual treatment. Furthermore, changing the two considered metformin parameters from their optimal values—altering the dose by ±50% and the decay rate by ±20%—demonstrates the flexibility of the model in simulating glycemic responses, confirming its adaptability and its potential for optimizing personalized treatment strategies.
Full article

Figure 1
Open AccessArticle
Biophysical Insights into the Binding Interactions of Inhibitors (ICA-1S/1T) Targeting Protein Kinase C-ι
by
Radwan Ebna Noor, Shahedul Islam, Tracess Smalley, Katarzyna Mizgalska, Mark Eschenfelder, Dimitra Keramisanou, Aaron Joshua Astalos, James William Leahy, Wayne Charles Guida, Aleksandra Karolak, Ioannis Gelis and Mildred Acevedo-Duncan
Biophysica 2025, 5(3), 36; https://doi.org/10.3390/biophysica5030036 - 11 Aug 2025
Abstract
The overexpression of atypical protein kinase C-iota (PKC-ι) is a biomarker for carcinogenesis in various cell types, such as glioma, ovarian, renal, etc., manifesting as a potential drug target. In previous in vitro studies, ICA-1S and ICA-1T, experimental candidates for inhibiting PKC-ι, have
[...] Read more.
The overexpression of atypical protein kinase C-iota (PKC-ι) is a biomarker for carcinogenesis in various cell types, such as glioma, ovarian, renal, etc., manifesting as a potential drug target. In previous in vitro studies, ICA-1S and ICA-1T, experimental candidates for inhibiting PKC-ι, have demonstrated their specificity and promising efficacy against various cancers. Moreover, the in vivo studies have demonstrated low toxicity levels in acute and chronic murine models. Despite these prior developments, the binding affinities of the inhibitors were never thoroughly explored from a biophysical perspective. Here, we present the biophysical characterizations of PKC-ι in combination with ICA-1S/1T. Various methods based on molecular docking, light scattering, intrinsic fluorescence, thermal denaturation, and heat exchange were applied. The biophysical characteristics including particle sizing, thermal unfolding, aggregation profiles, enthalpy, entropy, free energy changes, and binding affinity (Kd) of the PKC-ι in the presence of ICA-1S were observed. The studies indicate the presence of domain-specific stabilities in the protein–ligand complex. Moreover, the results indicate a spontaneous reaction with an entropic gain, resulting in a possible entropy-driven hydrophobic interaction and hydrogen bonds in the binding pocket. Altogether, these biophysical studies reveal important insights into the binding interactions of PKC-ι and its inhibitors ICA-1S/1T.
Full article
(This article belongs to the Collection Feature Papers in Biophysics)
►▼
Show Figures

Figure 1
Open AccessArticle
A Novel Purification Process of Sardine Lipases Using Protein Ultrafiltration and Dye Ligand Affinity Chromatography
by
Juan Antonio Noriega-Rodríguez, Armando Tejeda-Mansir and Hugo Sergio García
Biophysica 2025, 5(3), 35; https://doi.org/10.3390/biophysica5030035 - 10 Aug 2025
Abstract
Protein purification is often performed for various applications. However, enzyme purification processes typically involve multiple steps that reduce yield and increase production costs. To overcome these challenges, we developed a novel three-step process to purify a lipase from whole sardine viscera (WSV), leveraging
[...] Read more.
Protein purification is often performed for various applications. However, enzyme purification processes typically involve multiple steps that reduce yield and increase production costs. To overcome these challenges, we developed a novel three-step process to purify a lipase from whole sardine viscera (WSV), leveraging protein properties and the structural affinity of lipases for dye ligands. A crude extract of the viscera (CEV) was obtained by grinding the whole viscera in 50 mM phosphate buffer (pH 7.0, Solution B) followed by centrifugation (6000× g; 30 min, 0 °C). Lipolytic activity (3.3 U/mg) was recorded only in the supernatant. The purification process began with ammonium sulfate fractionation (30–50% saturation), increasing lipolytic activity in the precipitate (PF30-50) to 32.9 U/mg. PF30-50 was then ultrafiltered using a 30 KDa MWCO membrane, where 5% of semi-purified lipases (SPLSV) was retained with an activity of 156.5 U/mg (UF30). Finally, the SPLSV was injected into a column packed with dye ligand affinity adsorbent, pre-equilibrated with 1.0 M ammonium sulfate in buffer A. The WSV lipase was eluted using a step gradient to progressively reduce salt concentration. SDS-PAGE analysis revealed a single band of purified lipase from sardine viscera (PLSV) corresponding to a molecular weight of 123.4 kDa, with a specific activity of 266.4 U/mg. The combination of ammonium sulfate precipitation, ultrafiltration, and dye-ligand affinity chromatography provides a scalable and reproducible approach with potential industrial relevance, particularly in biocatalysis and waste valorization contexts.
Full article
(This article belongs to the Special Issue Advances in Enzyme Inhibition: Biophysical and Experimental Approaches)
►▼
Show Figures

Figure 1
Open AccessArticle
Protein Polarimetry, Perfected: Specific Rotation Measurement for the Refracto-Polarimetric Detection of Cryptic Protein Denaturation
by
Lisa Riedlsperger, Heinz Anderle, Andreas Schwaighofer and Martin Lemmerer
Biophysica 2025, 5(3), 34; https://doi.org/10.3390/biophysica5030034 - 7 Aug 2025
Abstract
Protein polarimetry has been evaluated as a simple and straightforward technique to detect the cryptic denaturation of exemplary proteins. The general rules of rotation vs. amino acid and structural composition and the respective knowledge gaps were reviewed, and the specific rotation of cystine
[...] Read more.
Protein polarimetry has been evaluated as a simple and straightforward technique to detect the cryptic denaturation of exemplary proteins. The general rules of rotation vs. amino acid and structural composition and the respective knowledge gaps were reviewed, and the specific rotation of cystine was determined in 4 M NaCl solution as [α]D20 = –302.5°. The specific rotations at 589 nm and 436 nm and the ratio were measured for several model proteins, some purified plasma-derived proteins and for three monoclonal antibodies. The immunoglobulin G concentrates all showed a narrow ratio range likely characteristic for this protein class. Heat denaturation experiments were conducted at temperatures between 50 and 85 °C both for short-time (10 min) and for prolonged periods of heat exposure (up to 210 min). Denaturation by heat resulted not only in the known levorotatory shift, but also in a shift in the specific rotation ratio. The stabilizing effect of fatty acids in bovine serum could be demonstrated by this parameter. Polarimetry thus appears to be a particularly sensitive and simple method for the characterization of the identity and the thermal stability of proteins and should therefore be added again as a complimentary method to the toolbox of protein chemistry.
Full article
(This article belongs to the Special Issue Investigations into Protein Structure)
►▼
Show Figures

Figure 1
Open AccessArticle
Modulating Enzyme–Ligand Binding with External Fields
by
Pedro Ojeda-May
Biophysica 2025, 5(3), 33; https://doi.org/10.3390/biophysica5030033 - 6 Aug 2025
Abstract
Protein enzymes are highly efficient catalysts that exhibit adaptability and selectivity under diverse biological conditions. In some organisms, such as bacteria, structurally similar enzymes, for instance, shikimate kinase (SK) and adenylate kinase (AK), coexist and act on chemically related ligands. This raises the
[...] Read more.
Protein enzymes are highly efficient catalysts that exhibit adaptability and selectivity under diverse biological conditions. In some organisms, such as bacteria, structurally similar enzymes, for instance, shikimate kinase (SK) and adenylate kinase (AK), coexist and act on chemically related ligands. This raises the question of whether these enzymes can accommodate and potentially react with each other’s ligands. In this study, we investigate the stability of non-cognate ligand binding in SK and explore whether external electric fields (EFs) can modulate this interaction, leading to cross-reactivity in SK. Using molecular dynamics simulations, we assess the structural integrity of SK and the binding behavior of ATP and AMP under EF-off and EF-on cases. Our results show that EFs enhance protein structure stability, stabilize non-cognate ligands in the binding pocket, and reduce local energetic frustration near the R116 residue located in the binding site. In addition to this, dimensionality reduction analyses reveal that EFs induce more coherent protein motions and reduce the number of metastable states. Together, these findings suggest that external EFs can reshape enzyme–ligand interactions and may serve as a tool to modulate enzymatic specificity and functional promiscuity. Thus, we provide computational evidence that supports the concept of using an EF as a tunable parameter in enzyme engineering and synthetic biology. However, further experimental investigation would be valuable to assess the reliability of our computational predictions.
Full article
(This article belongs to the Collection Feature Papers in Biophysics)
►▼
Show Figures

Figure 1
Open AccessArticle
Probing the Interaction Between Icariin and Proteinase K: A Combined Spectroscopic and Molecular Modeling Study
by
Zhongbao Han, Huizi Zheng, Yimeng Qi, Dilshadbek T. Usmanov, Liyan Liu and Zhan Yu
Biophysica 2025, 5(3), 32; https://doi.org/10.3390/biophysica5030032 - 28 Jul 2025
Abstract
Icariin (ICA) is widely recognized for its health benefits. In this work, we examined the intermolecular interactions between ICA and proteinase K (PK) via multi-spectroscopic techniques and molecular simulations. The experimental findings revealed that ICA quenched the fluorescence emission of PK by forming
[...] Read more.
Icariin (ICA) is widely recognized for its health benefits. In this work, we examined the intermolecular interactions between ICA and proteinase K (PK) via multi-spectroscopic techniques and molecular simulations. The experimental findings revealed that ICA quenched the fluorescence emission of PK by forming a noncovalent complex. Both hydrogen bonding and van der Waals interactions are essential for the complex’s formation. Then Förster resonance energy transfer (FRET), competitive experiments, and synchronous fluorescence spectroscopy were adopted to verify the formation of the complex. Molecular docking studies demonstrated that ICA could spontaneously bind to PK by hydrogen bonding and hydrophobic interactions, which is consistent with the spectroscopic results. The PK-ICA complex’s dynamic stability was evaluated using a 50 ns molecular dynamics (MD) simulation. The simulation results revealed no significant structural deformation or positional changes throughout the entire simulation period. The complex appears to be rather stable, as seen by the average root-mean-square deviation (RMSD) fluctuations for the host protein in the PK-ICA complex of 1.08 Å and 3.09 Å. These outcomes of molecular simulations suggest that ICA interacts spontaneously and tightly with PK, consistent with the spectroscopic findings. The approach employed in this research presents a pragmatic and advantageous method for examining protein–ligand interactions, as evidenced by the concordance between empirical and theoretical findings.
Full article
(This article belongs to the Special Issue Biomedical Optics: 3rd Edition)
►▼
Show Figures

Figure 1
Open AccessArticle
Imaging of Laser-Induced Thermal Convection and Conduction in Artificial Vitreous Humor
by
Jack Pelzel, Reese Anderson, Darin J. Ulness and Krys Strand
Biophysica 2025, 5(3), 31; https://doi.org/10.3390/biophysica5030031 - 27 Jul 2025
Abstract
This study extends the application of photothermal spectroscopy to explore heat transfer dynamics in biological fluids, focusing on the examination of artificial vitreous humor (VH) models of human VH and an endogenous sample of cervine (deer) VH. The research integrates previously established methods
[...] Read more.
This study extends the application of photothermal spectroscopy to explore heat transfer dynamics in biological fluids, focusing on the examination of artificial vitreous humor (VH) models of human VH and an endogenous sample of cervine (deer) VH. The research integrates previously established methods for analyzing thermal lensing through photothermal deflection. By visualizing convective and conductive heat transfer processes in the artificial components of human VH, one gains insights into the dynamic behavior of heat transfer in the VH. Relevance extends to clinical cases where pathology requires replacement of endogenous VH with an artificial VH substitute. Several VH substitutes identified in the literature were chosen for this study based on their physical properties and relative abundance in the VH. Individual component fluids, and mixtures of these components, were analyzed at various concentrations based on their physiological concentration ranges in the human VH as they varied with age, sex, and certain disease states. By way of comparison to endogenous biological VH, a sample of VH obtained from a female white-tailed deer eye was analyzed, enhancing the understanding of heat transfer in artificial components of the VH compared to endogenous VH. There is a vast array of ophthalmological procedures that utilize an external heat source interacting with endogenous or artificial VH. The data found in this study will progress the understanding of heat transfer within artificial VH components in comparison to endogenous VH and contribute to the advancement of certain ophthalmological procedures.
Full article
(This article belongs to the Special Issue Biomedical Optics: 3rd Edition)
►▼
Show Figures

Figure 1
Open AccessArticle
Optimizing Single-Particle Analysis Workflow: Comparative Analysis of the Symmetry Parameter and Particle Quantity upon Reconstruction of the Molecular Complex
by
Myeong Seon Jeong, Han-ul Kim, Mi Young An, Yoon Ho Park, Sun Hee Park, Sang J. Chung, Yoon-Sun Yi, Sangmi Jun, Young Kwan Kim and Hyun Suk Jung
Biophysica 2025, 5(3), 30; https://doi.org/10.3390/biophysica5030030 - 22 Jul 2025
Abstract
Recent major advancements in cryo-electron microscopy (cryo-EM) have enabled high-resolution structural analysis, accompanied by developments in image processing software packages for single-particle analysis (SPA). SPA facilitates the 3D reconstruction of proteins and macromolecular complexes from numerous individual particles. In this study, we systematically
[...] Read more.
Recent major advancements in cryo-electron microscopy (cryo-EM) have enabled high-resolution structural analysis, accompanied by developments in image processing software packages for single-particle analysis (SPA). SPA facilitates the 3D reconstruction of proteins and macromolecular complexes from numerous individual particles. In this study, we systematically evaluated the impact of symmetry parameters and particle quantity on the 3D reconstruction efficiency using the dihydrolipoyl acetyltransferase (E2) inner core of the pyruvate dehydrogenase complex (PDC). We specifically examined how inappropriate symmetry constraints can introduce structural artifacts and distortions, underscoring the necessity for accurate symmetry determination through rigorous validation methods such as directional Fourier shell correlation (FSC) and local-resolution mapping. Additionally, our analysis demonstrates that efficient reconstructions can be achieved with a moderate particle number, significantly reducing computational costs without compromising structural accuracy. We further contextualize these results by discussing recent developments in SPA workflows and hardware optimization, highlighting their roles in enhancing reconstruction accuracy and computational efficiency. Overall, our comprehensive benchmarking provides strategic insights that will facilitate the optimization of SPA experiments, particularly in resource-limited settings, and offers practical guidelines for accurately determining symmetry and particle quantity during cryo-EM data processing.
Full article
(This article belongs to the Special Issue Investigations into Protein Structure)
►▼
Show Figures

Figure 1
Open AccessArticle
How to Improve the Repeatability, Reproducibility and Accuracy in the Dynamic Structuration of Water by Electromagnetic Waves?
by
Marie-Valérie Moreno, Sid Ahmed Ben Mansour and Frédéric Roscop
Biophysica 2025, 5(3), 29; https://doi.org/10.3390/biophysica5030029 - 21 Jul 2025
Abstract
►▼
Show Figures
This study represents a first step toward improving the repeatability, reproducibility, and accuracy of a process designed to enhance dynamic water structuring. We aim is to investigate the optical reflectivity of a watery magnesium chloride solution treated with electromagnetic waves, we employ a
[...] Read more.
This study represents a first step toward improving the repeatability, reproducibility, and accuracy of a process designed to enhance dynamic water structuring. We aim is to investigate the optical reflectivity of a watery magnesium chloride solution treated with electromagnetic waves, we employ a novel methodology derived from human plethysmography (PPG) with three wavelengths spanning the visible and infrared spectra. We measured the reflectance of 17 flasks at 536 nm, 660 nm, and 940 nm before and after treatment, first using the succussion method (control) and second using a 50 Hz signal. The observed variability was acceptable, with repeatability errors below 0.15% and reproducibility errors below 3.5% across all wavelengths before and after treatment. Out of 51 samples dynamically structured using the succussion method, we obtained two false negatives, while one false negative was recorded out of 51 samples dynamically structured using the electromagnetic (EM) method. PPG appears to be a relevant sensor, as it correctly detected dynamically structured water in 99 out of 102 cases, using either the succussion or electromagnetic method. Our results show significant differences in reflectance (supposedly correlated with water’s structured status) at 536 nm between dynamically structured and dynamic non-structured samples (p < 0.001). Future improvements will include a validation protocol against gold-standard spectrophotometry with a larger sample size.
Full article

Figure 1
Open AccessArticle
Calibration and Detection of Phosphine Using a Corrosion-Resistant Ion Trap Mass Spectrometer
by
Dragan Nikolić and Xu Zhang
Biophysica 2025, 5(3), 28; https://doi.org/10.3390/biophysica5030028 - 17 Jul 2025
Abstract
We present a corrosion-resistant quadrupole ion trap mass spectrometer (QIT-MS) designed for trace detection of volatiles in sulfuric acid aerosols, with a specific focus on phosphine (PH3). Here, we detail the gas calibration methodology using permeation tube technology for generating certified
[...] Read more.
We present a corrosion-resistant quadrupole ion trap mass spectrometer (QIT-MS) designed for trace detection of volatiles in sulfuric acid aerosols, with a specific focus on phosphine (PH3). Here, we detail the gas calibration methodology using permeation tube technology for generating certified ppb-level PH3/H2S/CO2 mixtures, and report results from mass spectra with sufficient resolution to distinguish isotopic envelopes that validate the detection of PH3 at a concentration of 62 ppb. Fragmentation patterns for PH3 and H2S agree with NIST data, and signal-to-noise performance confirms ppb sensitivity over 2.6 h acquisition periods. We further assess spectral interferences from oxygen isotopes and propose a detection scheme based on isolated phosphorus ions (P+) to enable specific and interference-resistant identification of PH3 and other reduced phosphorus species of astrobiological interest in Venus-like environments. This work extends the capabilities of QIT-MS for trace gas analysis in chemically aggressive atmospheric conditions.
Full article
(This article belongs to the Special Issue Mass Spectrometry Applications in Biology Research)
►▼
Show Figures

Figure 1
Highly Accessed Articles
Latest Books
E-Mail Alert
News
Topics
Topic in
Biophysica, CIMB, Diagnostics, IJMS, IJTM
Molecular Radiobiology of Protons Compared to Other Low Linear Energy Transfer (LET) Radiation
Topic Editors: Francis Cucinotta, Jacob RaberDeadline: 20 December 2025
Topic in
Biomedicines, JCM, Pharmaceuticals, Pharmaceutics, Pharmacy, IJMS, Molecules, Biophysica
Peptoids and Peptide Based Drugs
Topic Editors: Laura Zaccaro, Annarita Del Gatto, Galia MaayanDeadline: 31 December 2025
Topic in
Biophysica, CIMB, Biology, Membranes, IJMS
New Insights into Cytoskeleton
Topic Editors: Malgorzata Kloc, Jacek KubiakDeadline: 31 March 2026
Topic in
Biophysica, CIMB, IJMS, IJTM, Lymphatics, Sci. Pharm.
Smart Delivery Systems for Biomolecular Therapeutics
Topic Editors: Biana Godin, Vivek GuptaDeadline: 31 October 2026

Special Issues
Special Issue in
Biophysica
Advances in Computational Biophysics
Guest Editor: Aman UllahDeadline: 31 October 2025
Special Issue in
Biophysica
Biomedical Optics: 3rd Edition
Guest Editor: Herbert SchneckenburgerDeadline: 31 October 2025
Special Issue in
Biophysica
Mass Spectrometry Applications in Biology Research
Guest Editor: Dragan NikolicDeadline: 25 November 2025
Special Issue in
Biophysica
Mechanobiology of Regeneration: From Physical Aspects
Guest Editors: Christopher Robinson, Alan David KayeDeadline: 30 November 2025
Topical Collections
Topical Collection in
Biophysica
Feature Papers in Biophysics
Collection Editors: Paul C. Whitford, Chandra Kothapalli









