Perspective on Quantitative Structure–Toxicity Relationship (QSTR) Models to Predict Hepatic Biotransformation of Xenobiotics
Abstract
1. Introduction
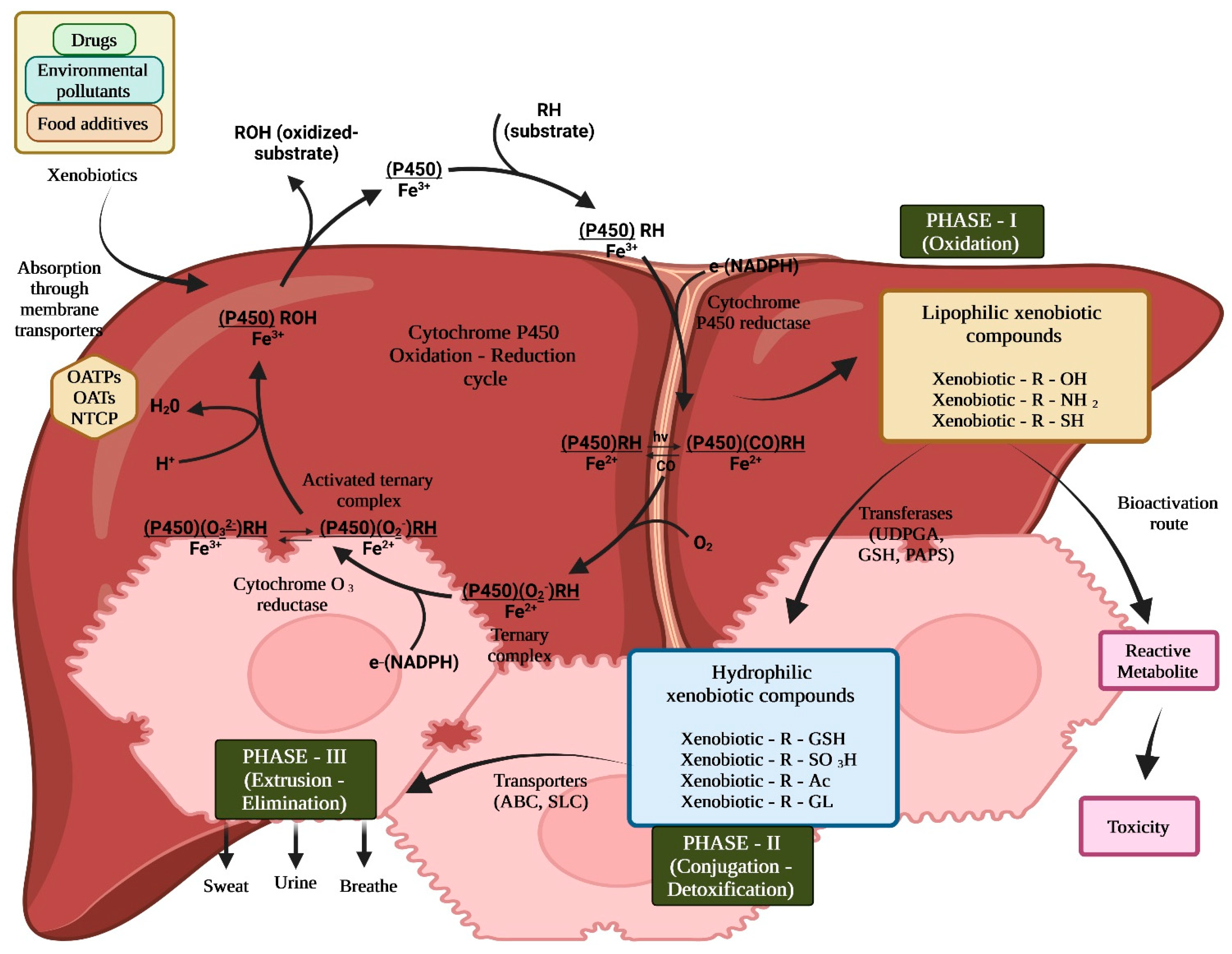
1.1. Exploring the Enigma of Xenobiotic Hepatic Biotransformation
1.2. Progress in QSTR Models for Anticipating Hepatic Biotransformation Pathways
- A method was devised that uses measures of in vitro hepatic biotransformation in animals to predict in vivo hepatic clearance [14]. This method has been applied to chemical risk assessment, evaluating medication candidates, and looking at idiosyncratic drug reactions. Hepatic clearance estimates may be included successfully in compartmental clearance-volume models.
- Understanding the trajectory of a medication necessitates awareness of the extent of hepatic metabolism and the capability to predict hepatic clearance [15]. Translation of preclinical pharmacokinetic and pharmacodynamic data has improved because of recent advancements in in vitro and in vivo models.
- To parameterize 1-CoTK models, QSARs were created and validated for forecasting in vivo whole-body biotransformation half-lives [16]. These models can be used to forecast chemical toxicity and aid in safer compound development.
- To evaluate possible herb–drug interactions, clearance tests are frequently performed in in vitro hepatic models [17]. These models can aid in the development of safer compounds by offering useful information on substances’ possible toxicities.
- Quantitative structure–pharmacokinetic relationships (QSPRs) that link biological activity to epithelial and hepatic first-pass biotransformation may also be created using QSTR models [18]. These models can be used to forecast drug pharmacokinetics and aid in the creation of more powerful pharmaceuticals.
2. Molecular Descriptors Used in QSTR Models for Hepatic Biotransformation
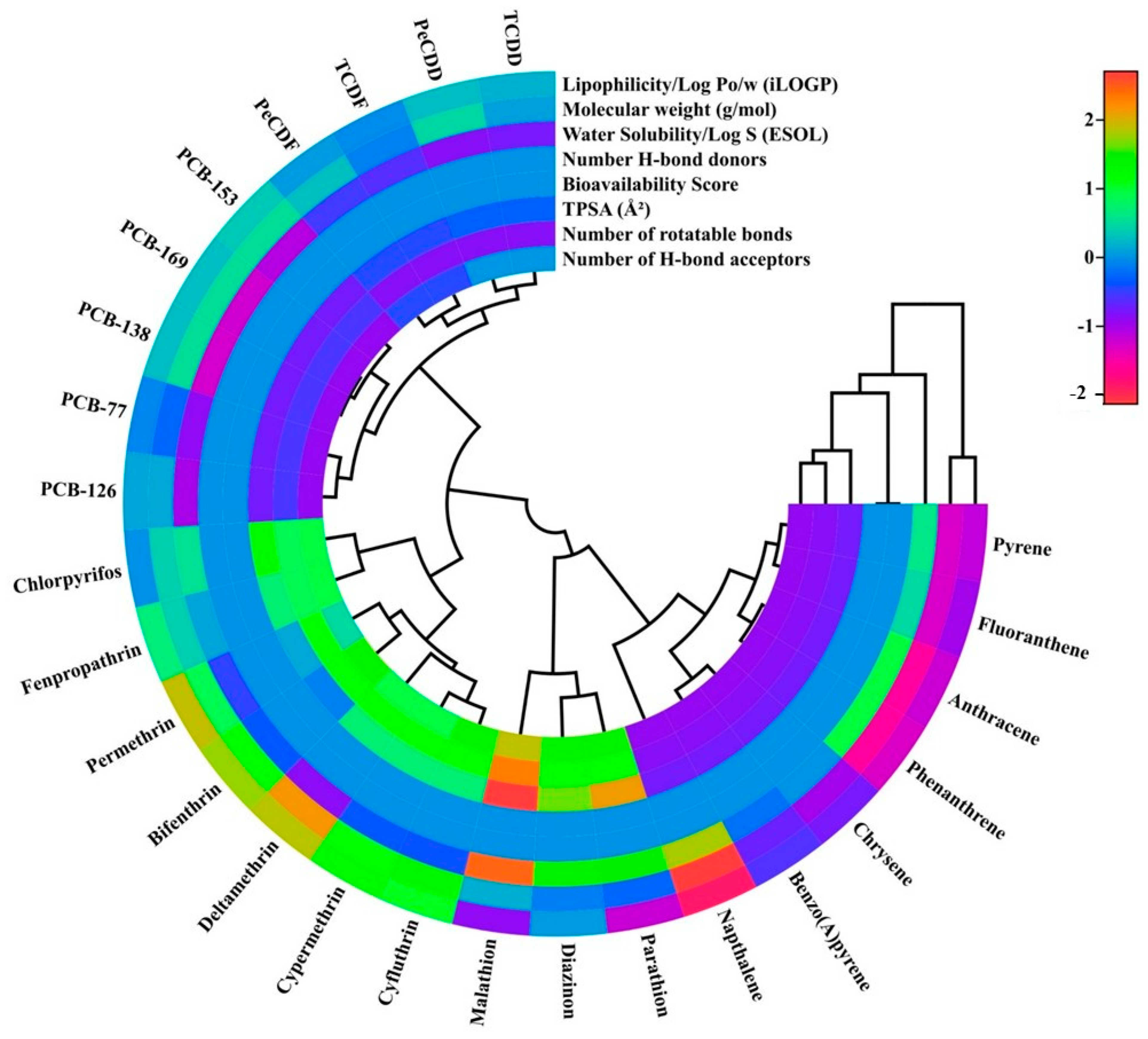
- Lipophilicity: Lipophilicity plays a pivotal role and is commonly considered in quantitative structure-toxicity relationship (QSTR) models. It describes a molecule’s capacity to partition or dissolve into lipid-based environments, such as cell membranes or lipid bilayers. To accurately represent a compound’s hydrophobic properties, lipophilicity is frequently measured experimentally or with a variety of molecular descriptors in QSTR modeling [26]. This term describes a substance’s propensity to dissolve in lipids or fats. Lipophilicity significantly influences the ADME (absorption, distribution, metabolism, and excretion) of xenobiotics. High lipophilicity substances have longer half-lives in the body and accumulate in adipose tissue. In order to anticipate the hepatic biotransformation of xenobiotics, lipophilicity is a crucial molecular descriptor [18].
- Molecular weight: The term “molecular weight” describes how much mass a molecule has. In QSTR modeling, molecular weight is frequently employed as a descriptor to characterize the size and mass of a drug. This might affect its biological activity, pharmacokinetics, and other aspects. Xenobiotics’ physicochemical characteristics, such as solubility, permeability, and bioavailability, are significantly influenced by their molecular weights. High molecular weight substances often have reduced solubility and permeability, which might affect how they are absorbed and distributed by the body. As a result, molecular weight is a crucial molecule descriptor for determining how xenobiotics will be metabolized in the liver [27].
- Polarizability: Polarizability is frequently employed as a descriptor in QSTR modeling to describe compound electrical and structural features. This can influence how it interacts with biological targets and exhibits certain qualities. When exposed to an external electric field, a molecule’s capacity to instantly create dipoles is measured by a property called polarizability. Polarizability is a crucial factor in determining how a molecule interacts with its surroundings, including whether or not it can pass through biological membranes. The capacity to anticipate the hepatic biotransformation of xenobiotics using polarizability is crucial [28].
- Hydrogen bonding: The term “hydrogen bonding” describes how well a molecule creates hydrogen bonds with other molecules. A key factor in determining solubility and reactivity is hydrogen bonding. As a result, hydrogen bonding is a crucial molecular descriptor for determining how xenobiotics will be transformed in the liver [29].
- Topological indices: These are mathematical descriptors that rate the branching, connectedness, and symmetry of molecules as well as other aspects of their topology. The physicochemical characteristics of xenobiotics, such as their solubility, permeability, and bioavailability, are significantly influenced by topological indices. Topological indicators are crucial molecular descriptors for foretelling xenobiotic hepatic biotransformation as a result [30].
- Molecular surface area: This term describes a molecule’s surface area. A key factor in determining how a molecule interacts with its surroundings, such as whether it can pass through biological membranes, is its molecular surface area. In order to anticipate the hepatic biotransformation of xenobiotics, molecular surface area is a crucial molecular descriptor [31].
3. Limitations of QSTR Models for Hepatic Biotransformation
3.1. Limited Ability to Predict Metabolism of Highly Lipophilic Compounds
3.2. Inability to Account for the Structural Complexity of a Molecule
3.3. Limited Ability to Predict Metabolism of Highly Polar Compounds
3.4. Limited Ability to Predict Metabolism of Compounds with Weak Hydrogen Bonding
| Molecular Descriptor | Role in Liver Metabolism | Limitations | How to Overcome Limitations |
|---|---|---|---|
| Lipophilicity | Determines the rate of passive diffusion of a drug across the cell membrane and its distribution in the body. | Limited ability to predict the metabolism of highly lipophilic compounds. | Use other molecular descriptors, such as polarizability and hydrogen bonding [45]. |
| Molecular weight | Affects the rate of metabolism and clearance of a drug. | Inability to account for the structural complexity of a molecule. | Incorporate other molecular descriptors, such as topological indices and molecular surface area [46]. |
| Polarizability | Affects the interaction of a drug with the enzyme and its rate of metabolism. | Limited ability to predict the metabolism of highly polar compounds. | Consider using alternative molecular descriptors, like lipophilicity and hydrogen bonding [47]. |
| Hydrogen bonding | Affects the interaction of a drug with the enzyme and its rate of metabolism. | Limited ability to predict the metabolism of compounds with weak hydrogen bonding. | Explore other molecular descriptors, such as lipophilicity and polarizability [48]. |
| Topological indices | Account for the structural complexity of a molecule and its effect on metabolism. | Limited ability to predict the metabolism of compounds with unusual structures. | Utilize additional molecular descriptors such as molecular weight and molecular surface area [24]. |
| Molecular surface area | Affects the rate of metabolism and clearance of a drug. | Limited ability to predict the metabolism of highly lipophilic compounds. | Consider other molecular descriptors such as polarizability and hydrogen [47]. |
3.5. Limited Ability to Predict Metabolism of Compounds with Unusual Structures
3.6. Limited Ability to Predict Metabolism of Highly Lipophilic Compounds
4. Opportunities to Improve QSTR Models for Hepatic Biotransformation
4.1. Use of Other Molecular Descriptors in Combination with the Ones Mentioned Above
4.2. Development of More Accurate and Reliable In Silico Models of Metabolism
4.3. Consideration of Predicted Small-Molecule Metabolites in Computational Toxicology
4.4. Creation of Complementary Substrate/Non-Substrate Classification Models
4.5. Use of QSAR Approaches to Predict Sites of Metabolism
5. Summary of the Importance of QSTR Models in Predicting Hepatic Biotransformation of Xenobiotics
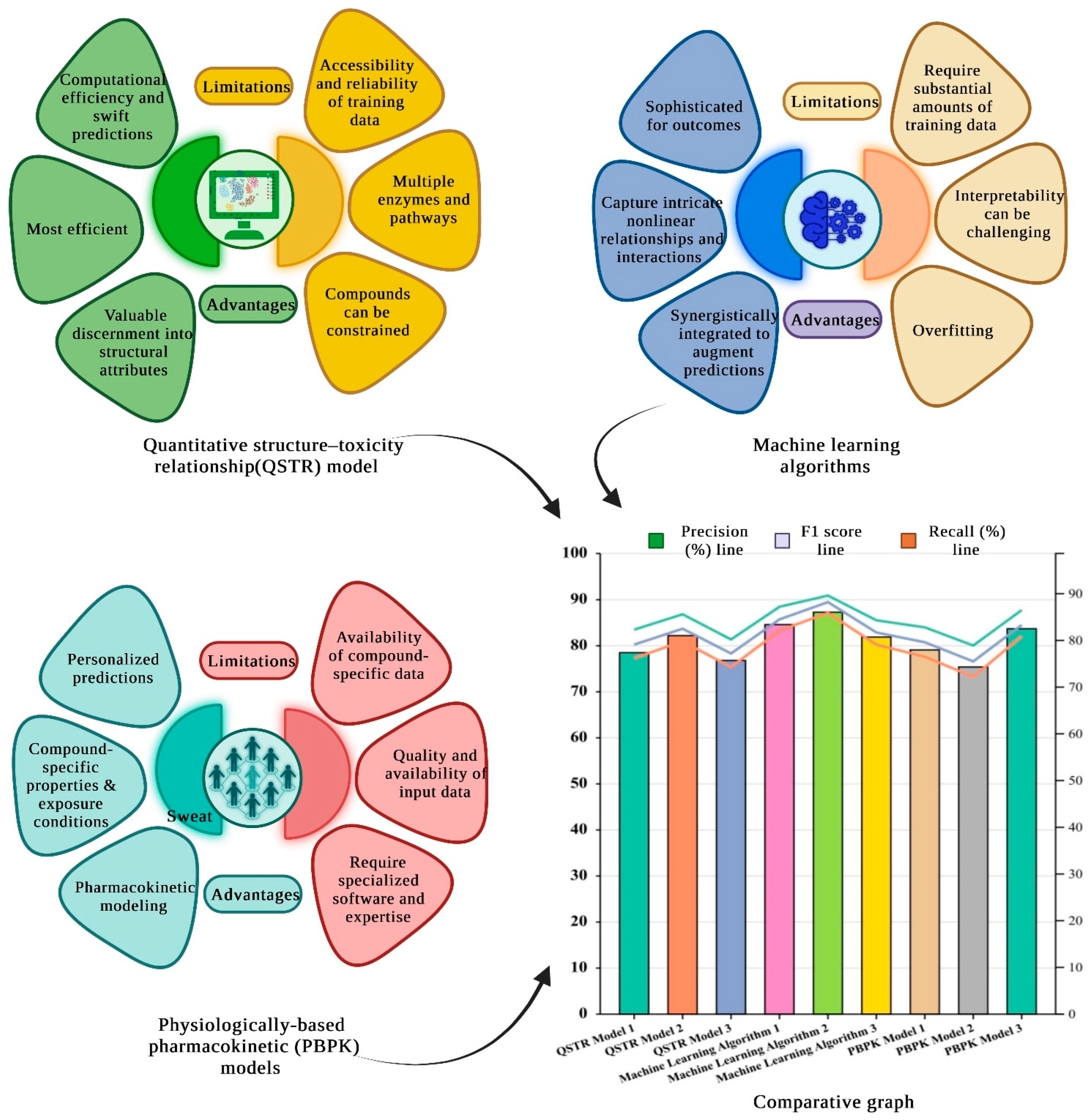
5.1. Predictive Power
5.2. Cost and Time Effectiveness
5.3. Mechanistic Insights
5.4. Structure–Activity Relationships
5.5. Applications for Virtual Screening and Design
6. Discussion of the Potential for Future Improvements in QSTR Models for Hepatic Biotransformation
6.1. Integration of Various Data Sources
6.2. Integration of Enzymes and Metabolic Pathways
6.3. Taking into Account Interindividual Variability
6.4. The Incorporation of Systems Biology Methods
6.5. Expansion of Training Data
6.6. Validation and Openness
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McGinnity, D.F.; Grime, K. Comprehensive Medicinal Chemistry III; Elsevier: Amsterdam, The Netherlands, 2017; Volume 4–8, pp. 34–44. [Google Scholar]
- Singh, A.V.; Kayal, A.; Malik, A.; Maharjan, R.S.; Dietrich, P.; Thissen, A.; Siewert, K.; Curato, C.; Pande, K.; Prahlad, D.; et al. Interfacial water in the SARS spike protein: Investigating the interaction with human ACE2 receptor and in vitro uptake in A549 cells. Langmuir 2022, 38, 7976–7988. [Google Scholar] [CrossRef]
- Johnson, C.H.; Patterson, A.D.; Idle, J.R.; Gonzalez, F.J. Xenobiotic metabolomics: Major impact on the metabolome. Rev. Pharmacol. Toxicol. 2012, 52, 37–56. [Google Scholar] [CrossRef] [PubMed]
- Gregg, C.R. Cytochrome P450; Elsevier: Amsterdam, The Netherlands, 2004. [Google Scholar] [CrossRef]
- Toogood, H.S.; Tait, S.; Jervis, A.; Cheallaigh, A.N.; Humphreys, L.; Takano, E.; Gardiner, J.M.; Scrutton, N.S. Natural Product Biosynthesis in Escherichia coli: Mentha Monoterpenoids. In Methods in Enzymol; Academic Press: Cambridge, MA, USA, 2016; Volume 575, pp. 247–270. [Google Scholar]
- Voutchkova, A.M.; Osimitz, T.G.; Anastas, P.T. Anastas Toward a comprehensive molecular design framework for reduced hazard. Chem. Rev. 2010, 110, 5845–5882. [Google Scholar] [CrossRef] [PubMed]
- Vighi, M.; Altenburger, R.; Arrhenius, Å.; Backhaus, T.; Bödeker, W.; Blanck, H.; Consolaro, F.; Faust, M.; Finizio, A.; Froehner, K.; et al. Water quality objectives for mixtures of toxic chemicals: Problems and perspectives. Ecotoxicol. Environ. Saf. 2003, 54, 139–150. [Google Scholar] [CrossRef]
- Can, A. Quantitative structure–toxicity relationship (QSTR) studies on the organophosphate insecticides. Toxicol. Lett. 2014, 230, 434–443. [Google Scholar] [CrossRef]
- Singh, A.V.; Bansod, G.; Mahajan, M.; Dietrich, P.; Singh, S.P.; Rav, K.; Thissen, A.; Bharde, A.M.; Rothenstein, D.; Kulkarni, S.; et al. Herbal Concoction Unveiled: A Computational Analysis of Phytochemicals’ Pharmacokinetic and Toxicological Profiles using Novel Approach Methodologies (NAMs). ACS Omega 2023, 8, 21377–21390. [Google Scholar] [CrossRef]
- Rott, E.; Kuch, B.; Lange, C.; Richter, P.; Kugele, A.; Minke, R. Removal of Emerging Contaminants and Estrogenic Activity from Wastewater Treatment Plant Effluent with UV/Chlorine and UV/H2O2 Advanced Oxidation Treatment at Pilot Scale. Int. J. Environ. Res. Public Health 2018, 15, 935. [Google Scholar] [CrossRef]
- Khan, Z.G.; Bari, S.B.; Patil, D.D. Lurasidone: A Review of analytical methods for Estimation in Pharmaceutical formulation. Rev. Artic. Int. J. Life Sci. Rev. 2016, 2, 17–22. [Google Scholar]
- Pandith, A.H.; Giri, S.; Chattaraj, P.K. A comparative study of two quantum chemical descriptors in predicting toxicity of aliphatic compounds towards tetrahymena pyriformis. Org. Chem. Int. 2010, 2010, 545087. [Google Scholar] [CrossRef]
- Judson, R.S.; Houck, K.A.; Kavlock, R.J.; Knudsen, T.B.; Martin, M.T.; Mortensen, H.M.; Reif, D.M.; Rotroff, D.M.; Shah, I.; Richard, A.M.; et al. In vitro screening of environmental chemicals for targeted testing prioritization: The ToxCast project. Environ. Health Perspect. 2010, 118, 485–492. [Google Scholar] [CrossRef]
- Ren, Q.; Li, N.; Liu, R.; Ma, X.; Sun, J.; Zeng, J.; Li, Q.; Wang, M.; Chen, X.; Wu, X.; et al. Nitric oxide (NO) involved in Cd tolerance in NHX1 transgenic duckweed during Cd stress. Plant Signal. Behav. 2022, 17, 2065114. [Google Scholar] [CrossRef]
- Yadav, J.; El Hassani, M.; Sodhi, J.; Lauschke, V.M.; Hartman, J.H.; Russell, L.E. Recent developments in in vitro and in vivo models for improved translation of preclinical pharmacokinetics and pharmacodynamics data. Drug Metab. Rev. 2021, 53, 207–233. [Google Scholar] [CrossRef]
- Horst, H.; Ernest, C. Structure-activity relationships in ecotoxicology. Environ. Toxicol. Chem. 1985, 4, 255–257. [Google Scholar]
- Hlengwa, N.; Masilela, C.; Mtambo, T.R.; Sithole, S.; Naidoo, S.; Machaba, K.E.; Shabalala, S.C.; Ntamo, Y.; Dludla, P.V.; Milase, R.N. In Vitro Hepatic Models to Assess Herb–Drug Interactions: Approaches and Challenges. Pharmaceuticals 2023, 16, 409. [Google Scholar]
- Mayer, J.M.; van de Waterbeemd, H. Development of quantitative structure-pharmacokinetic relationships. Environ. Health Perspect. 1985, 61, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; LaPerle, J.L.; Chang, G.; Varma, M.V. Renal clearance in drug discovery and development: Molecular descriptors, drug transporters and disease state. Expert Opin. Drug Metab. Toxicol. 2010, 6, 939–952. [Google Scholar] [CrossRef]
- Pignatello, R.; Musumeci, T.; Basile, L.; Carbone, C.; Puglisi, G. Biomembrane models and drug-biomembrane interaction studies: Involvement in drug design and development. J. Pharm. Bioallied Sci. 2011, 3, 4–14. [Google Scholar] [CrossRef]
- Pajouhesh, H.; Lenz, G.R. Medicinal chemical properties of successful central nervous system drugs. NeuroRX 2005, 2, 541–553. [Google Scholar] [CrossRef]
- Pardridge, W.M. The blood-brain barrier and neurotherapeutics. NeuroRx 2005, 2, 1–2. [Google Scholar] [CrossRef]
- Kidambi, S.; Yarmush, R.S.; Novik, E.; Chao, P.; Yarmush, M.L.; Nahmias, Y. Oxygen-mediated enhancement of primary hepatocyte metabolism, functional polarization, gene expression, and drug clearance. Proc. Natl. Acad. Sci. USA 2009, 106, 15714–15719. [Google Scholar] [CrossRef]
- Crivori, P.; Cruciani, G.; Carrupt, P.A.; Testa, B. Predicting blood—Brain barrier permeation from three-dimensional molecular structure. J. Med. Chem. 2000, 43, 2204–2216. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.; Gironés, X.; Amat, L. 37–41. Available online: https://www.tandfonline.com/doi/abs/10.1080/10629369908033223 (accessed on 15 August 2023).
- Tripathi, D.; Ray, P.; Singh, A.V.; Kishore, V.; Singh, S.L. Durability of Slippery Liquid-Infused Surfaces: Challenges and Advances. Coatings 2023, 13, 1095. [Google Scholar] [CrossRef]
- Acosta-Jiménez, E.H.; Zárate-Hernández, L.A.; Camacho-Mendoza, R.L.; González-Montiel, S.; Alvarado-Rodríguez, J.G.; Gómez-Castro, C.Z.; Pescador-Rojas, M.; Meneses-Viveros, A.; Cruz-Borbolla, J. Modification of the nutritional quality and oxidative stability of lupin (Lupinus mutabilis Sweet) and sacha inchi (Plukenetia volubilis L.) oil blends. Molecules 2022, 27, 7315. [Google Scholar]
- Tandon, H.; Ranjan, P.; Chakraborty, T.; Suhag, V. Polarizability: A promising descriptor to study chemical–biological interactions. Mol. Divers. 2021, 25, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Tinkov, O.V.; Grigorev, V.Y.; Polishchuk, P.G.; Yarkov, A.V.; Raevsky, O.A. QSAR investigation of acute toxicity of organic compounds during oral administration to mice. Biomeditsinskaya Khimiya 2019, 65, 123–132. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gu, X.; Manautou, J.E. Molecular mechanisms underlying chemical liver injury. Expert Rev. Mol. Med. 2012, 14, e4. [Google Scholar] [CrossRef]
- Omiecinski, C.J.; Vanden Heuvel, J.P.; Perdew, G.H.; Peters, J.M. Xenobiotic metabolism, disposition, and regulation by receptors: From biochemical phenomenon to predictors of major toxicities. Toxicol. Sci. 2011, 120, S49–S75. [Google Scholar] [CrossRef]
- Kulkarni, P.G.; Paudel, N.; Magar, S.; Santilli, M.F.; Kashyap, S.; Baranwal, A.K.; Zamboni, P.; Vasavada, P.; Katiyar, A.; Singh, A.V. Overcoming Challenges and Innovations in Orthopedic Prosthesis Design: An Interdisciplinary Perspective. Biomed. Mater. Devices 2023, Volume 1, 1–12. [Google Scholar] [CrossRef]
- Peffers, K.; Tuunanen, T.; Rothenberger, M.A.; Chatterjee, S. A design science research methodology for information systems research. J. Manag. Inf. Syst. 2007, 24, 45–77. [Google Scholar] [CrossRef]
- Singh, A.V.; Rosenkranz, D.; Ansari, M.H.D.; Singh, R.; Kanase, A.; Singh, S.P.; Johnston, B.; Tentschert, J.; Laux, P.; Luch, A. Machine-learning-based approach to decode the influence of nanomaterial properties on their interaction with cells. Adv. Intell. Syst. 2020, 2, 2000084. [Google Scholar] [CrossRef]
- Roy, K.; Ghosh, G. Exploring QSARs with Extended Topochemical Atom (ETA) indices for modeling chemical and drug toxicity. Curr. Pharm. Des. 2010, 16, 2625–2639. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Negi, A.; Gupta, P.K.; Chauhan, M.; Kumar, R. Toxicophore exploration as a screening technology for drug design and discovery: Techniques, scope and limitations. Arch. Toxicol. 2016, 90, 1785–1802. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xing, J.; Xu, Y.; Zhou, N.; Peng, J.; Xiong, Z.; Liu, X.; Luo, X.; Luo, C.; Chen, K.; et al. In silico ADME/T modelling for rational drug design. Q. Rev. Biophys. 2015, 48, 488–515. [Google Scholar] [CrossRef] [PubMed]
- Bohets, H.; Annaert, P.; Mannens, G.; van Beijsterveldt, L.; Anciaux, K.; Verboven, P.; Meuldermans, W.; Lavrijsen, K. Strategies for Absorption Screening in Drug Discovery and Development. Curr. Top. Med. Chem. 2005, 1, 367–383. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.; Roy, K. Ecotoxicological modelling of cosmetics for aquatic organisms: A QSTR approach. SAR QSAR Environ. Res. 2017, 28, 567–594. [Google Scholar]
- Kean, W.F.; Howard-Lock, H.E.; Lock, C.J.L. Chirality in antirheumatic drugs. Lancet 1991, 338, 1565–1568. [Google Scholar] [CrossRef]
- Fiehn, O. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comp. Funct. Genom. 2001, 2, 155–168. [Google Scholar] [CrossRef]
- Phelps, T.J.; Palumbo, A.V.; Beliaev, A.S. Metabolomics and microarrays for improved understanding of phenotypic characteristics controlled by both genomics and environmental constraints. Curr. Opin. Biotechnol. 2002, 13, 20–24. [Google Scholar] [CrossRef]
- Norinder, U.; Bergström, C.A. Prediction of ADMET properties. ChemMedChem 2006, 1, 920–937. [Google Scholar] [CrossRef]
- Garg, D.; Gandhi, T.; Mohan, C.G. Exploring QSTR and toxicophore of hERG K+ channel blockers using GFA and HypoGen techniques. J. Mol. Graph. Model. 2008, 26, 966–976. [Google Scholar] [CrossRef]
- Mannava, M.C.; Garai, A.; Nangia, A.K. Diffusion and Flux Improvement of Drugs through Complexation. Mol. Pharm. 2023, 20, 2293–2316. [Google Scholar] [CrossRef] [PubMed]
- Agatonovic-Kustrin, S.; Ling, L.H.; Tham, S.Y.; Alany, R.G. Molecular descriptors that influence the amount of drugs transfer into human breast milk. J. Pharm. Biomed. Anal. 2002, 29, 103–119. [Google Scholar] [CrossRef]
- Van De Waterbeemd, H.; Gifford, E. ADMET in silico modelling: Towards prediction paradise? Nat. Rev. Drug Discov. 2003, 2, 192–204. [Google Scholar] [CrossRef]
- Varma, M.; Khandavilli, S.; Ashokraj, Y.; Jain, A.; Dhanikula, A.; Sood, A.; Thomas, N.; Pillai, O.; Sharma, P.; Gandhi, R.; et al. Biopharmaceutic Classification System: A Scientific Framework for Pharmacokinetic Optimization in Drug Research. Curr. Drug Metab. 2005, 5, 375–388. [Google Scholar] [CrossRef] [PubMed]
- Pye, C.R.; Bertin, M.J.; Lokey, R.S.; Gerwick, W.H.; Linington, R.G. Retrospective analysis of natural products provides insights for future discovery trends. Proc. Natl. Acad. Sci. USA 2017, 114, 5601–5606. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, I.J. Assessing the absorption of new pharmaceuticals. Curr. Top. Med. Chem. 2005, 1, 385–401. [Google Scholar] [CrossRef]
- Hallifax, D.; Houston, J.B. Uptake and intracellular binding of lipophilic amine drugs by isolated rat hepatocytes and implications for prediction of in vivo metabolic clearance. Drug Metab. Dispos. 2006, 34, 1829–1836. [Google Scholar] [CrossRef] [PubMed]
- Arnott, J.A.; Planey, S.L. The influence of lipophilicity in drug discovery and design. Expert Opin. Drug Discov. 2012, 7, 863–875. [Google Scholar] [CrossRef]
- Klopman, G.; Dimayuga, M.; Talafous, J. META. 1. A program for the evaluation of metabolic transformation of chemicals. J. Chem. Inf. Comput. Sci. 1994, 34, 1320–1325. [Google Scholar] [CrossRef]
- Bruce, E.D.; Autenrieth, R.L.; Burghardt, R.C.; Donnelly, K.C.; McDonald, T.J. Using quantitative structure-activity relationships (QSAR) to predict toxic endpoints for polycyclic aromatic hydrocarbons (PAH). J. Toxicol. Environ. Health Part A Curr. Issues 2008, 71, 1073–1084. [Google Scholar] [CrossRef]
- Pérez Santín, E.; Rodríguez Solana, R.; González García, M.; García Suárez, M.D.M.; Blanco Díaz, G.D.; Cima Cabal, M.D.; Moreno Rojas, J.M.; López Sánchez, J.I. Toxicity prediction based on artificial intelligence: A multidisciplinary overview. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2021, 11, 1–32. [Google Scholar] [CrossRef]
- Maharjan, R.S.; Singh, A.V.; Hanif, J.; Rosenkranz, D.; Haidar, R.; Shelar, A.; Singh, S.P.; Dey, A.; Patil, R.; Zamboni, P.; et al. Investigation of the Associations between a Nanomaterial’s Microrheology and Toxicology. ACS Omega 2022, 7, 13985–13997. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.V.; Chandrasekar, V.; Paudel, N.; Laux, P.; Luch, A.; Gemmati, D.; Tissato, V.; Prabhu, K.S.; Uddin, S.; Dakua, S.P. Integrative toxicogenomics: Advancing precision medicine and toxicology through artificial intelligence and OMICs tech-nology. Biomed. Pharmacother. 2023, 163, 114784. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekar, V.; Singh, A.V.; Maharjan, R.S.; Dakua, S.P.; Balakrishnan, S.; Dash, S.; Laux, P.; Luch, A.; Singh, S.; Pradhan, M.; et al. Perspectives on the Technological Aspects and Biomedical Applications of Virus-Like Particles/Nanoparticles in Reproductive Biology: Insights on the Medicinal and Toxicological Outlook. Adv. NanoBiomed Res. 2022, 2, 2200010. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rai, M.; Paudel, N.; Sakhrie, M.; Gemmati, D.; Khan, I.A.; Tisato, V.; Kanase, A.; Schulz, A.; Singh, A.V. Perspective on Quantitative Structure–Toxicity Relationship (QSTR) Models to Predict Hepatic Biotransformation of Xenobiotics. Livers 2023, 3, 448-462. https://doi.org/10.3390/livers3030032
Rai M, Paudel N, Sakhrie M, Gemmati D, Khan IA, Tisato V, Kanase A, Schulz A, Singh AV. Perspective on Quantitative Structure–Toxicity Relationship (QSTR) Models to Predict Hepatic Biotransformation of Xenobiotics. Livers. 2023; 3(3):448-462. https://doi.org/10.3390/livers3030032
Chicago/Turabian StyleRai, Mansi, Namuna Paudel, Mesevilhou Sakhrie, Donato Gemmati, Inshad Ali Khan, Veronica Tisato, Anurag Kanase, Armin Schulz, and Ajay Vikram Singh. 2023. "Perspective on Quantitative Structure–Toxicity Relationship (QSTR) Models to Predict Hepatic Biotransformation of Xenobiotics" Livers 3, no. 3: 448-462. https://doi.org/10.3390/livers3030032
APA StyleRai, M., Paudel, N., Sakhrie, M., Gemmati, D., Khan, I. A., Tisato, V., Kanase, A., Schulz, A., & Singh, A. V. (2023). Perspective on Quantitative Structure–Toxicity Relationship (QSTR) Models to Predict Hepatic Biotransformation of Xenobiotics. Livers, 3(3), 448-462. https://doi.org/10.3390/livers3030032








