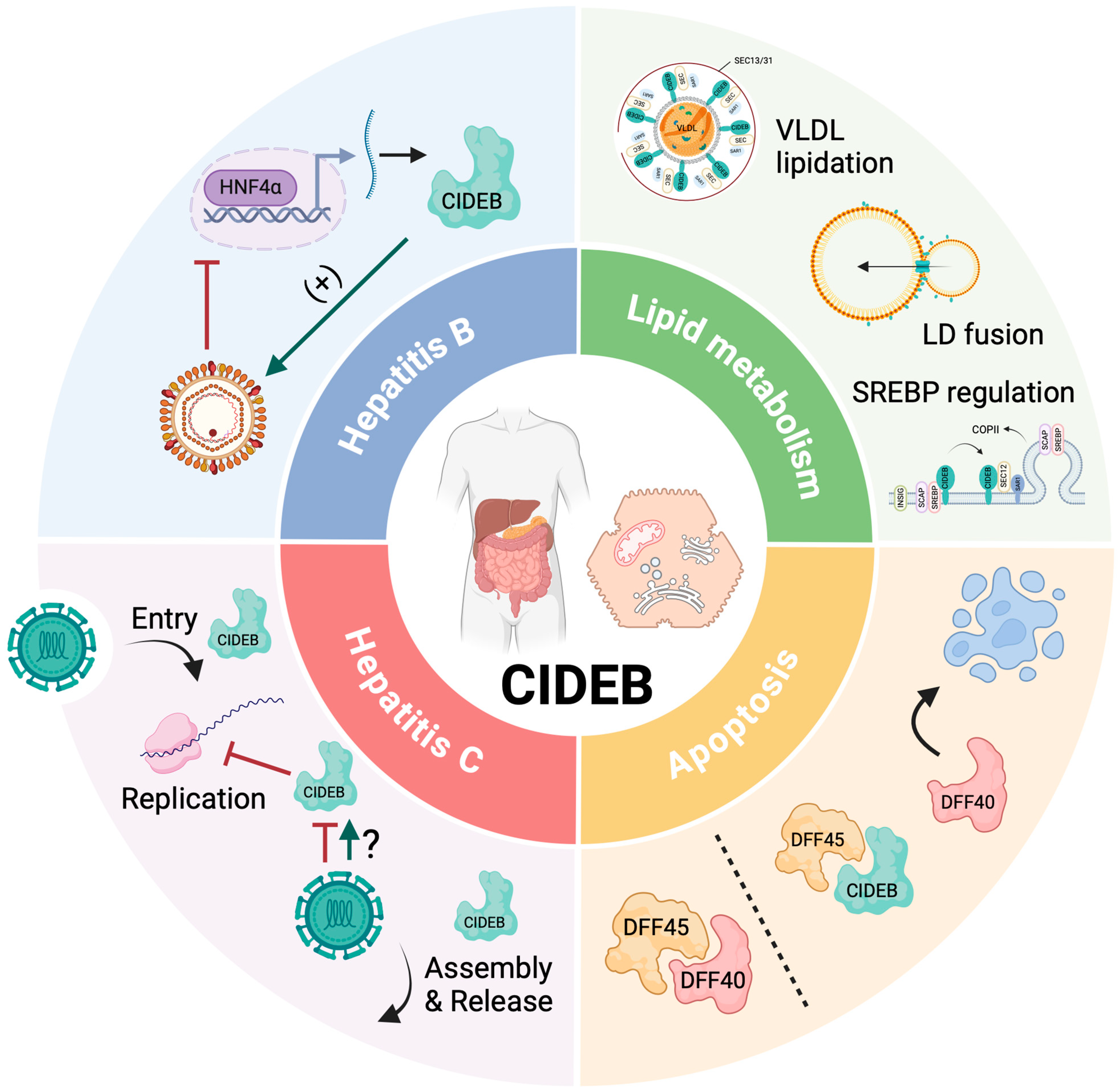
Unveiling the Multifaceted Role of CIDEB: From Apoptosis to Lipid Metabolism and Liver Health
Abstract
1. Introduction
2. CIDEB: Structure, Expression, and Phylogeny
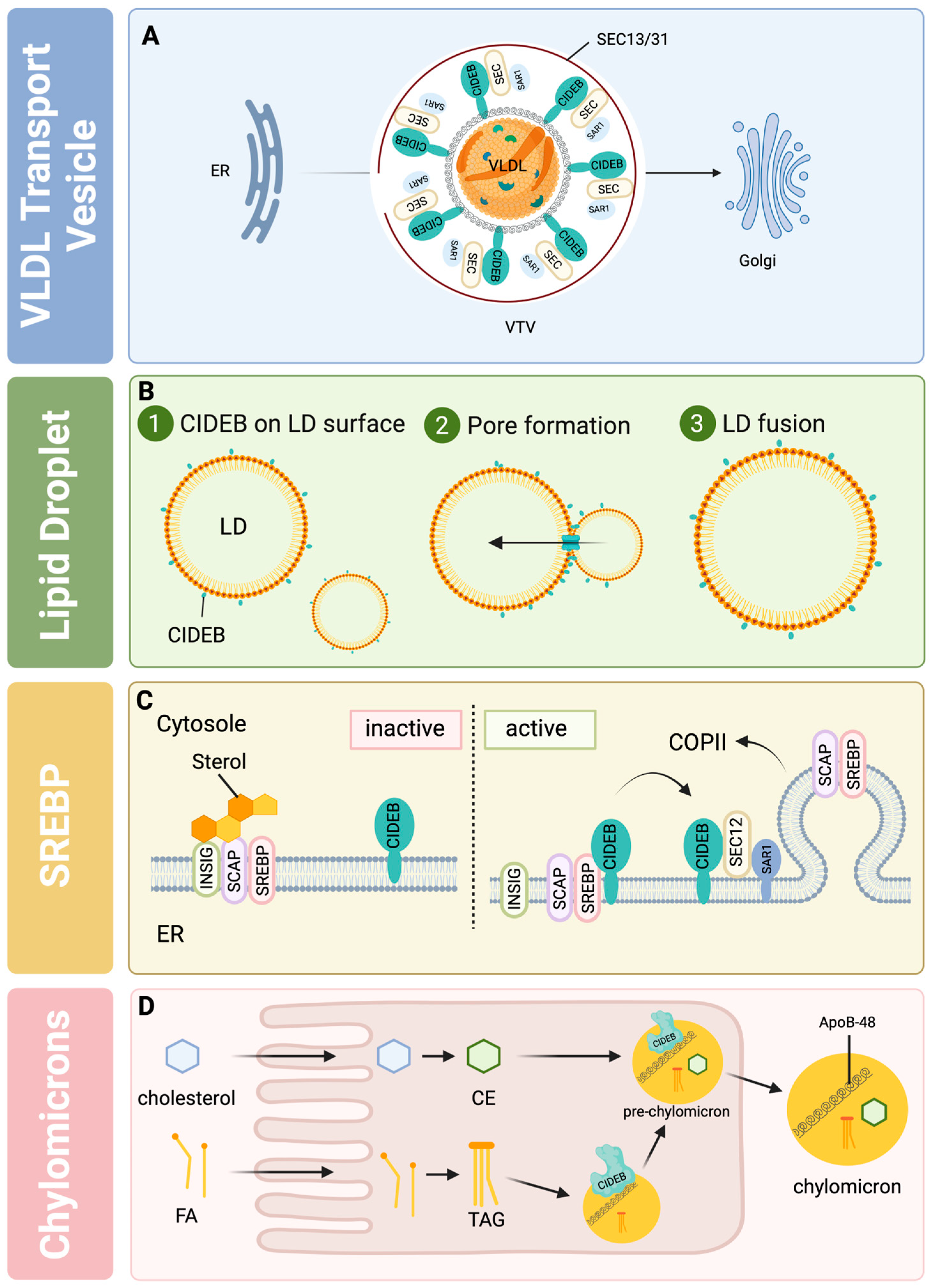
3. CIDEB in Lipid Metabolism
3.1. CIDEB in Diabetes and Metabolic Syndrome
3.2. CIDEB in MAFLD
3.3. The Protective Role of CIDEB Variants in Liver Disease
4. CIDEB in Apoptosis
5. CIDEB in Viral Hepatitis
5.1. CIDEB in Hepatitis C
5.2. CIDEB in Hepatitis B
6. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Naghavi, M.; Ong, K.L.; Aali, A.; Ababneh, H.S.; Abate, Y.H.; Abbafati, C.; Abbasgholizadeh, R.; Abbasian, M.; Abbasi-Kangevari, M.; Abbastabar, H.; et al. Global Burden of 288 Causes of Death and Life Expectancy Decomposition in 204 Countries and Territories and 811 Subnational Locations, 1990–2021: A Systematic Analysis for the Global Burden of Disease Study 2021. Lancet 2024, 403, 2100–2132. [Google Scholar] [CrossRef] [PubMed]
- Moon, A.M.; Singal, A.G.; Tapper, E.B. Contemporary Epidemiology of Chronic Liver Disease and Cirrhosis. Clin. Gastroenterol. Hepatol. 2020, 18, 2650–2666. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Wong, G.; Anstee, Q.M.; Henry, L. The Global Burden of Liver Disease. Clin. Gastroenterol. Hepatol. 2023, 21, 1978–1991. [Google Scholar] [CrossRef]
- Eslam, M.; Newsome, P.N.; Sarin, S.K.; Anstee, Q.M.; Targher, G.; Romero-Gomez, M.; Zelber-Sagi, S.; Wai-Sun Wong, V.; Dufour, J.-F.; Schattenberg, J.M.; et al. A New Definition for Metabolic Dysfunction-Associated Fatty Liver Disease: An International Expert Consensus Statement. J. Hepatol. 2020, 73, 202–209. [Google Scholar] [CrossRef] [PubMed]
- EASL–EASD–EASO Clinical Practice Guidelines for the Management of Non-Alcoholic Fatty Liver Disease. J. Hepatol. 2016, 64, 1388–1402. [CrossRef] [PubMed]
- Loomba, R.; Sanyal, A.J. The Global NAFLD Epidemic. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 686–690. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Golabi, P.; Paik, J.M.; Henry, A.; Van Dongen, C.; Henry, L. The Global Epidemiology of Nonalcoholic Fatty Liver Disease (NAFLD) and Nonalcoholic Steatohepatitis (NASH): A Systematic Review. Hepatology 2023, 77, 1335–1347. [Google Scholar] [CrossRef]
- Wong, M.C.S.; Huang, J.; Wang, J.; Chan, P.S.F.; Lok, V.; Chen, X.; Leung, C.; Wang, H.H.X.; Lao, X.Q.; Zheng, Z.-J. Global, Regional and Time-Trend Prevalence of Central Obesity: A Systematic Review and Meta-Analysis of 13.2 Million Subjects. Eur. J. Epidemiol. 2020, 35, 673–683. [Google Scholar] [CrossRef]
- Koh, J.H.; Ng, C.H.; Nah, B.; Tan, D.J.H.; Loomba, R.; Huang, D.Q. NASH Is the Leading Cause of Hepatocellular Carcinoma in Liver Transplant Candidates. Clin. Gastroenterol. Hepatol. 2024, 22, 197–199.e3. [Google Scholar] [CrossRef]
- Younossi, Z.; Stepanova, M.; Ong, J.P.; Jacobson, I.M.; Bugianesi, E.; Duseja, A.; Eguchi, Y.; Wong, V.W.; Negro, F.; Yilmaz, Y.; et al. Nonalcoholic Steatohepatitis Is the Fastest Growing Cause of Hepatocellular Carcinoma in Liver Transplant Candidates. Clin. Gastroenterol. Hepatol. 2018, 17, 748–755.e3. [Google Scholar] [CrossRef]
- Anstee, Q.M.; Day, C.P. The Genetics of NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Semmler, G.; Datz, C.; Reiberger, T.; Trauner, M. Diet and Exercise in NAFLD/NASH: Beyond the Obvious. Liver Int. 2021, 41, 2249–2268. [Google Scholar] [CrossRef]
- FDA Approves First Treatment for Patients with Liver Scarring Due to Fatty Liver Disease. FDA. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-patients-liver-scarring-due-fatty-liver-disease (accessed on 20 May 2024).
- Armstrong, M.J.; Gaunt, P.; Aithal, G.P.; Barton, D.; Hull, D.; Parker, R.; Hazlehurst, J.M.; Guo, K.; Abouda, G.; Aldersley, M.A.; et al. Liraglutide Safety and Efficacy in Patients with Non-Alcoholic Steatohepatitis (LEAN): A Multicentre, Double-Blind, Randomised, Placebo-Controlled Phase 2 Study. Lancet 2016, 387, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Newsome, P.N.; Buchholtz, K.; Cusi, K.; Linder, M.; Okanoue, T.; Ratziu, V.; Sanyal, A.J.; Sejling, A.-S.; Harrison, S.A. A Placebo-Controlled Trial of Subcutaneous Semaglutide in Nonalcoholic Steatohepatitis. N. Engl. J. Med. 2021, 384, 1113–1124. [Google Scholar] [CrossRef]
- Aithal, G.P.; Thomas, J.A.; Kaye, P.V.; Lawson, A.; Ryder, S.D.; Spendlove, I.; Austin, A.S.; Freeman, J.G.; Morgan, L.; Webber, J. Randomized, Placebo-Controlled Trial of Pioglitazone in Nondiabetic Subjects with Nonalcoholic Steatohepatitis. Gastroenterology 2008, 135, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Tahara, A.; Kurosaki, E.; Yokono, M.; Yamajuku, D.; Kihara, R.; Hayashizaki, Y.; Takasu, T.; Imamura, M.; Li, Q.; Tomiyama, H.; et al. Effects of SGLT2 Selective Inhibitor Ipragliflozin on Hyperglycemia, Hyperlipidemia, Hepatic Steatosis, Oxidative Stress, Inflammation, and Obesity in Type 2 Diabetic Mice. Eur. J. Pharmacol. 2013, 715, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Emdin, C.A.; Haas, M.E.; Khera, A.V.; Aragam, K.; Chaffin, M.; Klarin, D.; Hindy, G.; Jiang, L.; Wei, W.-Q.; Feng, Q.; et al. A Missense Variant in Mitochondrial Amidoxime Reducing Component 1 Gene and Protection against Liver Disease. PLoS Genet. 2020, 16, e1008629. [Google Scholar] [CrossRef]
- Verweij, N.; Haas, M.E.; Nielsen, J.B.; Sosina, O.A.; Kim, M.; Akbari, P.; De, T.; Hindy, G.; Bovijn, J.; Persaud, T.; et al. Germline Mutations in CIDEB and Protection against Liver Disease. N. Engl. J. Med. 2022, 387, 332–344. [Google Scholar] [CrossRef]
- Abul-Husn, N.S.; Cheng, X.; Li, A.H.; Xin, Y.; Schurmann, C.; Stevis, P.; Liu, Y.; Kozlitina, J.; Stender, S.; Wood, G.C.; et al. A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N. Engl. J. Med. 2018, 378, 1096–1106. [Google Scholar] [CrossRef]
- Motomura, T.; Amirneni, S.; Diaz-Aragon, R.; Faccioli, L.A.P.; Malizio, M.R.; Coard, M.C.; Kocas-Kilicarslan, Z.N.; Frau, C.; Haep, N.; Ostrowska, A.; et al. Is HSD17B13 Genetic Variant a Protector for Liver Dysfunction? Future Perspective as a Potential Therapeutic Target. J. Pers. Med. 2021, 11, 619. [Google Scholar] [CrossRef]
- Wu, C.; Zhang, Y.; Sun, Z.; Li, P. Molecular Evolution of Cide Family Proteins: Novel Domain Formation in Early Vertebrates and the Subsequent Divergence. BMC Evol. Biol. 2008, 8, 159. [Google Scholar] [CrossRef]
- Li, Q.-L.; Zheng, H.; Luo, Z.; Wu, L.-X.; Xu, P.-C.; Guo, J.-C.; Song, Y.-F.; Tan, X.-Y. Characterization and Expression Analysis of Seven Lipid Metabolism-Related Genes in Yellow Catfish Pelteobagrus Fulvidraco Fed High Fat and Bile Acid Diet. Gene 2024, 894, 147972. [Google Scholar] [CrossRef]
- Inohara, N.; Koseki, T.; Chen, S.; Wu, X.; Núñez, G. CIDE, a Novel Family of Cell Death Activators with Homology to the 45 kDa Subunit of the DNA Fragmentation Factor. EMBO J. 1998, 17, 2526–2533. [Google Scholar] [CrossRef]
- Lugovskoy, A.A.; Zhou, P.; Chou, J.J.; McCarty, J.S.; Li, P.; Wagner, G. Solution Structure of the CIDE-N Domain of CIDE-B and a Model for CIDE-N/CIDE-N Interactions in the DNA Fragmentation Pathway of Apoptosis. Cell 1999, 99, 747–755. [Google Scholar] [CrossRef]
- Zou, H.; Henzel, W.J.; Liu, X.; Lutschg, A.; Wang, X. Apaf-1, a Human Protein Homologous to C. Elegans CED-4, Participates in Cytochrome c–Dependent Activation of Caspase-3. Cell 1997, 90, 405–413. [Google Scholar] [CrossRef] [PubMed]
- McCarty, J.S.; Toh, S.Y.; Li, P. Study of DFF45 in Its Role of Chaperone and Inhibitor: Two Independent Inhibitory Domains of DFF40 Nuclease Activity. Biochem. Biophys. Res. Commun. 1999, 264, 176–180. [Google Scholar] [CrossRef]
- Chen, Z.; Guo, K.; Toh, S.Y.; Zhou, Z.; Li, P. Mitochondria Localization and Dimerization Are Required for CIDE-B to Induce Apoptosis. J. Biol. Chem. 2000, 275, 22619–22622. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Ye, J.; Xue, B.; Qi, J.; Zhang, J.; Zhou, Z.; Li, Q.; Wen, Z.; Li, P. Cideb Regulates Diet-Induced Obesity, Liver Steatosis, and Insulin Sensitivity by Controlling Lipogenesis and Fatty Acid Oxidation. Diabetes 2007, 56, 2523–2532. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-J.; Wang, C.; Yuan, Y.; Wang, H.; Wu, J.; Liu, F.; Li, L.; Gao, X.; Zhao, Y.-L.; Hu, P.-Z.; et al. Cideb Facilitates the Lipidation of Chylomicrons in the Small Intestine. J. Lipid Res. 2014, 55, 1279–1287. [Google Scholar] [CrossRef]
- Li, H.; Song, Y.; Zhang, L.J.; Li, F.F.; Gu, Y.; Zhang, J.; Dong, W.P.; Xue, L.; Zhang, L.Y.; Liu, F.; et al. Cell Death-Inducing DFF45-like Effector b (Cideb) Is Present in Pancreatic Beta-Cells and Involved in Palmitate Induced Beta-Cell Apoptosis. Diabetes/Metab. Res. Rev. 2012, 28, 145–155. [Google Scholar] [CrossRef]
- Ye, J.; Li, J.Z.; Liu, Y.; Li, X.; Yang, T.; Ma, X.; Li, Q.; Yao, Z.; Li, P. Cideb, an ER- and Lipid Droplet-Associated Protein, Mediates VLDL Lipidation and Maturation by Interacting with Apolipoprotein B. Cell Metab. 2009, 9, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ye, J.; Zhou, L.; Gu, W.; Fisher, E.A.; Li, P. eroxisome Proliferator-Activated Receptor-γ Coactivator-1α (PGC-1α) Stimulates VLDL Assembly through Activ VLDL Lipidation. J. Lipid Res. 2012, 53, 1877–1889. [Google Scholar] [CrossRef]
- Olofsson, S.-O.; Stillemark-Billton, P.; Asp, L. Intracellular Assembly of VLDL: Two Major Steps in Separate Cell Compartments. Trends Cardiovasc. Med. 2000, 10, 338–345. [Google Scholar] [CrossRef]
- Wetterau, J.R.; Aggerbeck, L.P.; Bouma, M.-E.; Eisenberg, C.; Munck, A.; Hermier, M.; Schmitz, J.; Gay, G.; Rader, D.J.; Gregg, R.E. Absence of Microsomal Triglyceride Transfer Protein in Individuals with Abetalipoproteinemia. Science 1992, 258, 999–1001. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Siddiqi, S.A. Intracellular Trafficking and Secretion of Very Low Density Lipoproteins. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 1079–1086. [Google Scholar] [CrossRef]
- Yamaguchi, J.; Gamble, M.V.; Conlon, D.; Liang, J.; Ginsberg, H.N. The Conversion of ApoB100 Low Density Lipoprotein/High Density Lipoprotein Particles to ApoB100 Very Low Density Lipoproteins in Response to Oleic Acid Occurs in the Endoplasmic Reticulum and Not in the Golgi in McA RH7777 Cells. J. Biol. Chem. 2003, 278, 42643–42651. [Google Scholar] [CrossRef] [PubMed]
- Gusarova, V.; Brodsky, J.L.; Fisher, E.A. Apolipoprotein B100 Exit from the Endoplasmic Reticulum (ER) Is COPII-Dependent, and Its Lipidation to Very Low Density Lipoprotein Occurs Post-ER. J. Biol. Chem. 2003, 278, 48051–48058. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, S.A. VLDL Exits from the Endoplasmic Reticulum in a Specialized Vesicle, the VLDL Transport Vesicle, in Rat Primary Hepatocytes. Biochem. J. 2008, 413, 333–342. [Google Scholar] [CrossRef]
- Yuan, L.; Kenny, S.J.; Hemmati, J.; Xu, K.; Schekman, R. TANGO1 and SEC12 Are Copackaged with Procollagen I to Facilitate the Generation of Large COPII Carriers. Proc. Natl. Acad. Sci. USA 2018, 115, E12255–E12264. [Google Scholar] [CrossRef]
- Zanetti, G.; Pahuja, K.B.; Studer, S.; Shim, S.; Schekman, R. COPII and the Regulation of Protein Sorting in Mammals. Nat. Cell Biol. 2012, 14, 20–28. [Google Scholar] [CrossRef]
- Fromme, J.C.; Schekman, R. COPII-Coated Vesicles: Flexible Enough for Large Cargo? Curr. Opin. Cell Biol. 2005, 17, 345–352. [Google Scholar] [CrossRef]
- Tiwari, S.; Siddiqi, S.; Siddiqi, S.A. CideB Protein Is Required for the Biogenesis of Very Low Density Lipoprotein (VLDL) Transport Vesicle. J. Biol. Chem. 2013, 288, 5157–5165. [Google Scholar] [CrossRef]
- Su, L.; Zhou, L.; Chen, F.-J.; Wang, H.; Qian, H.; Sheng, Y.; Zhu, Y.; Yu, H.; Gong, X.; Cai, L.; et al. Cideb Controls Sterol-Regulated ER Export of SREBP/SCAP by Promoting Cargo Loading at ER Exit Sites. EMBO J. 2019, 38, e100156. [Google Scholar] [CrossRef]
- Xu, W.; Wu, L.; Yu, M.; Chen, F.-J.; Arshad, M.; Xia, X.; Ren, H.; Yu, J.; Xu, L.; Xu, D.; et al. Differential Roles of Cell Death-Inducing DNA Fragmentation Factor-α-like Effector (CIDE) Proteins in Promoting Lipid Droplet Fusion and olecular Cloning, Genomic Organization, Chromosome. J. Biol. Chem. 2016, 291, 4282–4293. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Lee, E.M.; Hammack, C.; Robotham, J.M.; Basu, M.; Lang, J.; Brinton, M.A.; Tang, H. Cell Death-Inducing DFFA-Like Effector b Is Required for Hepatitis C Virus Entry into Hepatocytes. J. Virol. 2014, 88, 8433–8444. [Google Scholar] [CrossRef] [PubMed]
- Singaravelu, R.; Delcorde, J.; Lyn, R.K.; Steenbergen, R.H.; Jones, D.M.; Tyrrell, D.L.; Russell, R.S.; Pezacki, J.P. Investigating the Antiviral Role of Cell Death-Inducing DFF45-like Effector B in HCV Replication. FEBS J. 2014, 281, 3751–3765. [Google Scholar] [CrossRef]
- Singaravelu, R.; Lyn, R.K.; Srinivasan, P.; Delcorde, J.; Steenbergen, R.H.; Tyrrell, D.L.; Pezacki, J.P. Human Serum Activates CIDEB-Mediated Lipid Droplet Enlargement in Hepatoma Cells. Biochem. Biophys. Res. Commun. 2013, 441, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Sun, Z.; Wu, L.; Xu, W.; Schieber, N.; Xu, D.; Shui, G.; Yang, H.; Parton, R.G.; Li, P. Fsp27 Promotes Lipid Droplet Growth by Lipid Exchange and Transfer at Lipid Droplet Contact Sites. J. Cell Biol. 2011, 195, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Lei, Y.; Wang, Y.; Zhang, Y.; Ye, J.; Xia, X.; Pan, X.; Li, P. Control of Cholesterol Biosynthesis, Uptake and Storage in Hepatocytes by Cideb. Biochim. Biophys. Acta 2010, 1801, 577–586. [Google Scholar] [CrossRef]
- Matsuda, M.; Korn, B.S.; Hammer, R.E.; Moon, Y.-A.; Komuro, R.; Horton, J.D.; Goldstein, J.L.; Brown, M.S.; Shimomura, I. SREBP Cleavage-Activating Protein (SCAP) Is Required for Increased Lipid Synthesis in Liver Induced by Cholesterol Deprivation and Insulin Elevation. Genes Dev. 2001, 15, 1206–1216. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, P.; Weiskirchen, R. The Role of SCAP/SREBP as Central Regulators of Lipid Metabolism in Hepatic Steatosis. Int. J. Mol. Sci. 2024, 25, 1109. [Google Scholar] [CrossRef]
- Ko, C.-W.; Qu, J.; Black, D.D.; Tso, P. Regulation of Intestinal Lipid Metabolism: Current Concepts and Relevance to Disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 169–183. [Google Scholar] [CrossRef]
- Da, L.; Li, D.; Yokoyama, K.K.; Li, T.; Zhao, M. Dual Promoters Control the Cell-Specific Expression of the Human Cell Death-Inducing DFF45-like Effector B Gene. Biochem. J. 2006, 393 Pt 3, 779–788. [Google Scholar] [CrossRef][Green Version]
- Chen, Z.; Norris, J.Y.; Finck, B.N. Peroxisome Proliferator-Activated Receptor-γ Coactivator-1α (PGC-1α) Stimulates VLDL Assembly through Activation of Cell Death-Inducing DFFA-like Effector B (CideB). J. Biol. Chem. 2010, 285, 25996. [Google Scholar] [CrossRef]
- Daigo, K.; Kawamura, T.; Ohta, Y.; Ohashi, R.; Katayose, S.; Tanaka, T.; Aburatani, H.; Naito, M.; Kodama, T.; Ihara, S.; et al. Proteomic Analysis of Native Hepatocyte Nuclear Factor-4α (HNF4α) Isoforms, Phosphorylation Status, and Interactive Cofactors. J. Biol. Chem. 2011, 286, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Collison, K.S.; Inglis, A.; Shibin, S.; Saleh, S.; Andres, B.; Ubungen, R.; Thiam, J.; Mata, P.; Al-Mohanna, F.A. Effect of Developmental NMDAR Antagonism with CGP 39551 on Aspartame-Induced Hypothalamic and Adrenal Gene Expression. PLoS ONE 2018, 13, e0194416. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.M.; Alsagheir, A.; Wu, X.; Hammack, C.; McLauchlan, J.; Watanabe, N.; Wakita, T.; Kneteman, N.M.; Douglas, D.N.; Tang, H. Hepatitis C Virus-Induced Degradation of Cell Death-Inducing DFFA-Like Effector B Leads to Hepatic Lipid Dysregulation. J. Virol. 2016, 90, 4174–4185. [Google Scholar] [CrossRef]
- Aguilar, M.; Bhuket, T.; Torres, S.; Liu, B.; Wong, R.J. Prevalence of the Metabolic Syndrome in the United States, 2003–2012. JAMA 2015, 313, 1973–1974. [Google Scholar] [CrossRef] [PubMed]
- Noubiap, J.J.; Nansseu, J.R.; Lontchi-Yimagou, E.; Nkeck, J.R.; Nyaga, U.F.; Ngouo, A.T.; Tounouga, D.N.; Tianyi, F.-L.; Foka, A.J.; Ndoadoumgue, A.L.; et al. Geographic Distribution of Metabolic Syndrome and Its Components in the General Adult Population: A Meta-Analysis of Global Data from 28 Million Individuals. Diabetes Res. Clin. Pract. 2022, 188, 109924. [Google Scholar] [CrossRef]
- Zhou, Z.; Yon Toh, S.; Chen, Z.; Guo, K.; Peng Ng, C.; Ponniah, S.; Lin, S.-C.; Hong, W.; Li, P. Cidea-Deficient Mice Have Lean Phenotype and Are Resistant to Obesity. Nat. Genet. 2003, 35, 49–56. [Google Scholar] [CrossRef]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD Development and Therapeutic Strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, A.J. Past, Present and Future Perspectives in Nonalcoholic Fatty Liver Disease. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Romeo, S.; Kozlitina, J.; Xing, C.; Pertsemlidis, A.; Cox, D.; Pennacchio, L.A.; Boerwinkle, E.; Cohen, J.C.; Hobbs, H.H. Genetic Variation in PNPLA3 Confers Susceptibility to Nonalcoholic Fatty Liver Disease. Nat. Genet. 2008, 40, 1461–1465. [Google Scholar] [CrossRef]
- Meffert, P.J.; Repp, K.D.; Völzke, H.; Weiss, F.U.; Homuth, G.; Kühn, J.P.; Lerch, M.M.; Aghdassi, A.A. The PNPLA3 SNP Rs738409:G Allele Is Associated with Increased Liver Disease-Associated Mortality but Reduced Overall Mortality in a Population-Based Cohort. J. Hepatol. 2018, 68, 858–860. [Google Scholar] [CrossRef]
- Stender, S.; Kozlitina, J.; Nordestgaard, B.G.; Tybjærg-Hansen, A.; Hobbs, H.H.; Cohen, J.C. Adiposity Amplifies the Genetic Risk of Fatty Liver Disease Conferred by Multiple Loci. Nat. Genet. 2017, 49, 842–847. [Google Scholar] [CrossRef]
- Eslam, M.; George, J. Genetic Contributions to NAFLD: Leveraging Shared Genetics to Uncover Systems Biology. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 40–52. [Google Scholar] [CrossRef]
- Aron-Wisnewsky, J.; Warmbrunn, M.V.; Nieuwdorp, M.; Clément, K. Nonalcoholic Fatty Liver Disease: Modulating Gut Microbiota to Improve Severity? Gastroenterology 2020, 158, 1881–1898. [Google Scholar] [CrossRef]
- Caussy, C.; Tripathi, A.; Humphrey, G.; Bassirian, S.; Singh, S.; Faulkner, C.; Bettencourt, R.; Rizo, E.; Richards, L.; Xu, Z.Z.; et al. A Gut Microbiome Signature for Cirrhosis Due to Nonalcoholic Fatty Liver Disease. Nat. Commun. 2019, 10, 1406. [Google Scholar] [CrossRef] [PubMed]
- Loomba, R.; Seguritan, V.; Li, W.; Long, T.; Klitgord, N.; Bhatt, A.; Dulai, P.S.; Caussy, C.; Bettencourt, R.; Highlander, S.K.; et al. Gut Microbiome Based Metagenomic Signature for Non-Invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017, 25, 1054–1062.e5. [Google Scholar] [CrossRef]
- Erdtmann, L.; Franck, N.; Lerat, H.; Le Seyec, J.; Gilot, D.; Cannie, I.; Gripon, P.; Hibner, U.; Guguen-Guillouzo, C. The Hepatitis C Virus NS2 Protein Is an Inhibitor of CIDE-B-Induced Apoptosis. J. Biol. Chem. 2003, 278, 18256–18264. [Google Scholar] [CrossRef]
- Ha, H.J.; Park, H.H. Molecular Basis of Apoptotic DNA Fragmentation by DFF40. Cell Death Dis. 2022, 13, 198. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Yao, W.; Li, L.; Li, X.; Hu, L.; Mai, R.; Peng, T. Cell-Death-Inducing DFFA-like Effector B Contributes to the Assembly of Hepatitis C Virus (HCV) Particles and Interacts with HCV NS5A. Sci. Rep. 2016, 6, 27778. [Google Scholar] [CrossRef]
- Boson, B.; Granio, O.; Bartenschlager, R.; Cosset, F.-L. A Concerted Action of Hepatitis C Virus P7 and Nonstructural Protein 2 Regulates Core Localization at the Endoplasmic Reticulum and Virus Assembly. PLoS Pathog. 2011, 7, e1002144. [Google Scholar] [CrossRef]
- Li, Q.; Pène, V.; Krishnamurthy, S.; Cha, H.; Liang, T.J. Hepatitis C Virus Infection Activates a Novel Innate Pathway Involving IKKα in Lipogenesis and Viral Assembly. Nat. Med. 2013, 19, 722–729. [Google Scholar] [CrossRef]
- Bassendine, M.F.; Sheridan, D.A.; Felmlee, D.J.; Bridge, S.H.; Toms, G.L.; Neely, R.D.G. HCV and the Hepatic Lipid Pathway as a Potential Treatment Target. J. Hepatol. 2011, 55, 1428–1440. [Google Scholar] [CrossRef][Green Version]
- Bassendine, M.F.; Sheridan, D.A.; Bridge, S.H.; Felmlee, D.J.; Neely, R.D.G. Lipids and HCV. Semin. Immunopathol. 2013, 35, 87–100. [Google Scholar] [CrossRef]
- Lambert, J.E.; Bain, V.G.; Ryan, E.A.; Thomson, A.B.R.; Clandinin, M.T. Elevated Lipogenesis and Diminished Cholesterol Synthesis in Patients with Hepatitis C Viral Infection Compared to Healthy Humans. Hepatology 2013, 57, 1697–1704. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Hood, B.L.; Chadwick, S.L.; Liu, S.; Watkins, S.C.; Luo, G.; Conrads, T.P.; Wang, T. Fatty Acid Synthase Is Upregulated during HCV Infection and Regulates HCV Entry and Production. Hepatology 2008, 48, 1396–1403. [Google Scholar] [CrossRef] [PubMed]
- Lerat, H.; Honda, M.; Beard, M.R.; Loesch, K.; Sun, J.; Yang, Y.; Okuda, M.; Gosert, R.; Xiao, S.-Y.; Weinman, S.A.; et al. Steatosis and Liver Cancer in Transgenic Mice Expressing the Structural and Nonstructural Proteins of Hepatitis C Virus. Gastroenterology 2002, 122, 352–365. [Google Scholar] [CrossRef]
- Harris, C.; Herker, E.; Farese, R.V.; Ott, M. Hepatitis C Virus Core Protein Decreases Lipid Droplet Turnover. J. Biol. Chem. 2011, 286, 42615–42625. [Google Scholar] [CrossRef]
- Camus, G.; Schweiger, M.; Herker, E.; Harris, C.; Kondratowicz, A.S.; Tsou, C.-L.; Farese, R.V.; Herath, K.; Previs, S.F.; Roddy, T.P.; et al. The Hepatitis C Virus Core Protein Inhibits Adipose Triglyceride Lipase (ATGL)-Mediated Lipid Mobilization and Enhances the ATGL Interaction with Comparative Gene Identification 58 (CGI-58) and Lipid Droplets. J. Biol. Chem. 2014, 289, 35770–35780. [Google Scholar] [CrossRef]
- Kandangwa, M.; Liu, Q. HCV-2a NS5A Downregulates Viral Translation Predominantly through Domain I. Biochem. Biophys. Res. Commun. 2020, 529, 77–84. [Google Scholar] [CrossRef]
- Benga, W.J.A.; Krieger, S.E.; Dimitrova, M.; Zeisel, M.B.; Parnot, M.; Lupberger, J.; Hildt, E.; Luo, G.; Mclauchlan, J.; Baumert, T.F.; et al. Apolipoprotein E Interacts with Hepatitis C Virus Nonstructural Protein 5A and Determines Assembly of Infectious Particles. Hepatology 2010, 51, 43. [Google Scholar] [CrossRef] [PubMed]
- Santolini, E.; Pacini, L.; Fipaldini, C.; Migliaccio, G.; Monica, N. The NS2 Protein of Hepatitis C Virus Is a Transmembrane Polypeptide. J. Virol. 1995, 69, 7461–7471. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.; Beran, R.K.F.; Peters, C.; Lorenz, I.C.; Lindenbach, B.D. Hepatitis C Virus NS2 Protein Contributes to Virus Particle Assembly via Opposing Epistatic Interactions with the E1-E2 Glycoprotein and NS3-NS4A Enzyme Complexes. J. Virol. 2009, 83, 8379–8395. [Google Scholar] [CrossRef]
- Stapleford, K.A.; Lindenbach, B.D. Hepatitis C Virus NS2 Coordinates Virus Particle Assembly through Physical Interactions with the E1-E2 Glycoprotein and NS3-NS4A Enzyme Complexes. J. Virol. 2011, 85, 1706–1717. [Google Scholar] [CrossRef]
- Yasumoto, J.; Kasai, H.; Yoshimura, K.; Otoguro, T.; Watashi, K.; Wakita, T.; Yamashita, A.; Tanaka, T.; Takeda, S.; Moriishi, K. Hepatitis B Virus Prevents Excessive Viral Production via Reduction of Cell Death-Inducing DFF45-Like Effectors. J Gen Virol. 2017, 98, 1762–1773. [Google Scholar] [CrossRef]
- Nissim, O.; Melis, M.; Diaz, G.; Kleiner, D.E.; Tice, A.; Fantola, G.; Zamboni, F.; Mishra, L.; Farci, P. Liver Regeneration Signature in Hepatitis B Virus (HBV)-Associated Acute Liver Failure Identified by Gene Expression Profiling. PLoS ONE 2012, 7, e49611. [Google Scholar] [CrossRef]
- Wu, Q.; Liu, H.-O.; Liu, Y.-D.; Liu, W.-S.; Pan, D.; Zhang, W.-J.; Yang, L.; Fu, Q.; Xu, J.-J.; Gu, J.-X. Decreased Expression of Hepatocyte Nuclear Factor 4α (Hnf4α)/microRNA-122 (miR-122) Axis in Hepatitis B Virus-Associated Hepatocellular Carcinoma Enhances Potential Oncogenic GALNT10 Protein Activity. J. Biol. Chem. 2015, 290, 1170–1185. [Google Scholar] [CrossRef]
- Yi, S.; Ren, G.; Zhu, Y.; Cong, Q. Correlation Analysis of Hepatic Steatosis and Hepatitis B Virus: A Cross-Sectional Study. Virol. J. 2024, 21, 22. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.W.-S.; Wong, G.L.-H.; Chu, W.C.-W.; Chim, A.M.-L.; Ong, A.; Yeung, D.K.-W.; Yiu, K.K.-L.; Chu, S.H.-T.; Chan, H.-Y.; Woo, J.; et al. Hepatitis B Virus Infection and Fatty Liver in the General Population. J. Hepatol. 2012, 56, 533–540. [Google Scholar] [CrossRef]
- Jinjuvadia, R.; Liangpunsakul, S. Association between Metabolic Syndrome and Its Individual Components with Viral Hepatitis B. Am. J. Med. Sci. 2014, 347, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Hou, B.; Qin, L.; Huang, L. Liver Cancer Cells as the Model for Developing Liver-Targeted RNAi Therapeutics. Biochem. Biophys. Res. Commun. 2023, 644, 85–94. [Google Scholar] [CrossRef]
- Chen, Q.; Fang, W.; Shen, Y.; Xu, D.; Chen, Q.; Cui, K.; Mai, K.; Ai, Q. Suppression of Cideb under Endoplasmic Reticulum Stress Exacerbated Hepatic Inflammation by Inducing Hepatic Steatosis and Oxidative Stress. Free Radic. Biol. Med. 2022, 185, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, A.; Yang, Z.Q. Molecular Cloning, Genomic Organization, Chromosome Mapping, Tissues Expression Pattern and Identification of a Novel Splicing Variant of Porcine CIDEb Gene. Biochem. Biophys. Res. Commun. 2016, 478, 486–493. [Google Scholar] [CrossRef]
- Qiu, Y.; Yang, X.; Ma, X.; Xiong, Y.; Tian, Z.; Fan, Q.; Wang, L.; Jiang, Z. CIDE Gene Expression in Adipose Tissue, Liver, and Skeletal Muscle from Obese and Lean Pigs. J. Zhejiang Univ. Sci. B 2017, 18, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Ye, P.; Cheah, I.K.; Halliwell, B. High Fat Diets and Pathology in the Guinea Pig. Atherosclerosis or Liver Damage? Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2013, 1832, 355–364. [Google Scholar] [CrossRef]
- Fu, J.-F.; Fang, Y.-L.; Liang, L.; Wang, C.-L.; Hong, F.; Dong, G.-P. A Rabbit Model of Pediatric Nonalcoholic Steatohepatitis: The Role of Adiponectin. World J. Gastroenterol. 2009, 15, 912–918. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, X.; Liao, S.; Li, Y.; Zhang, Z.; Chang, Q.; Xiao, R.; Liang, B. Tree Shrew (Tupaia Belangeri Chinensis), a Novel Non-Obese Animal Model of Non-Alcoholic Fatty Liver Disease. Biol. Open 2016, 5, 1545–1552. [Google Scholar] [CrossRef]
- Hammoudeh, S.M.; Hammoudeh, A.M.; Bhamidimarri, P.M.; Mahboub, B.; Halwani, R.; Hamid, Q.; Rahmani, M.; Hamoudi, R. Insight into Molecular Mechanisms Underlying Hepatic Dysfunction in Severe COVID-19 Patients Using Systems Biology. World J. Gastroenterol. 2021, 27, 2850–2870. [Google Scholar] [CrossRef]
- Ng, S.W.K.; Rouhani, F.J.; Brunner, S.F.; Brzozowska, N.; Aitken, S.J.; Yang, M.; Abascal, F.; Moore, L.; Nikitopoulou, E.; Chappell, L.; et al. Convergent Somatic Mutations in Metabolism Genes in Chronic Liver Disease. Nature 2021, 598, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Wang, H.; Zhao, J.; Yuan, Y.; Wang, C.; Li, J.; Zhang, L.; Zhang, L.; Li, Q.; Ye, J. Expression of CIDE Proteins in Clear Cell Renal Cell Carcinoma and Their Prognostic Significance. Mol. Cell. Biochem. 2013, 378, 145–151. [Google Scholar] [CrossRef] [PubMed][Green Version]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wutsdorff, L.; Mougnekabol, J.; Tang, P.; Reutzel-Selke, A.; Sauer, I.M.; Haep, N. Unveiling the Multifaceted Role of CIDEB: From Apoptosis to Lipid Metabolism and Liver Health. Livers 2024, 4, 406-419. https://doi.org/10.3390/livers4030030
Wutsdorff L, Mougnekabol J, Tang P, Reutzel-Selke A, Sauer IM, Haep N. Unveiling the Multifaceted Role of CIDEB: From Apoptosis to Lipid Metabolism and Liver Health. Livers. 2024; 4(3):406-419. https://doi.org/10.3390/livers4030030
Chicago/Turabian StyleWutsdorff, Louise, Julienne Mougnekabol, Peter Tang, Anja Reutzel-Selke, Igor M. Sauer, and Nils Haep. 2024. "Unveiling the Multifaceted Role of CIDEB: From Apoptosis to Lipid Metabolism and Liver Health" Livers 4, no. 3: 406-419. https://doi.org/10.3390/livers4030030
APA StyleWutsdorff, L., Mougnekabol, J., Tang, P., Reutzel-Selke, A., Sauer, I. M., & Haep, N. (2024). Unveiling the Multifaceted Role of CIDEB: From Apoptosis to Lipid Metabolism and Liver Health. Livers, 4(3), 406-419. https://doi.org/10.3390/livers4030030




