Abstract
Nipah henipavirus is an emerging RNA virus that poses a danger to world safety due to its high fatality rate. The Nipah virus has caused several illness epidemics in South and Southeast Asia. The matrix protein of Nipah henipavirus plays a crucial function in linking the viral envelope to the virus core. Connecting the viral envelope to the virus core is critical for virus assembly. Through analyses of structural and functional protein explanations, bioinformatics tools can aid in our comprehension of the protein. This study intends to provide structural and functional annotations to proteins. Using in silico approaches, the analysis also assigns the protein’s physicochemical properties, three-dimensional structure, and functional annotation. The in silico research validated the protein’s hydrophilic nature and alpha (α) helix-dominated secondary structure. The protein’s tertiary structure model is generally consistent based on various quality evaluation approaches. The functional explanation claimed that the protein is a structural protein that connects the viral envelope to the virus core, a protein that is necessary for virus assembly. This study reveals the importance of the matrix protein as a functional protein needed by Nipah henipavirus.
1. Introduction
Nipah henipavirus is a bat-borne virus that can infect humans and other animals [1,2]. Nipah virus infection is a zoonotic disease that is spread from animals to humans. It can also be spread through contaminated food or from person to person. It causes a variety of symptoms in infected individuals, ranging from asymptomatic (subclinical) infection to acute respiratory sickness and deadly encephalitis [3,4,5]. The Nipah virus is a member of the Paramyxoviridae family and the genus Henipavirus along with the Hendra virus, which has also caused disease outbreaks [6,7]. The Nipah virus genome is a single (nonsegmented) negative-sense, single-stranded RNA with a length of over 18 kb, which is far longer than that of other paramyxoviruses [8,9]. The Nipah virus was initially detected in pigs and pig farmers in Peninsular Malaysia in 1998 [9]. Infection outbreaks of the Nipah virus have been documented in Malaysia, Singapore, Bangladesh, and India [6]. The highest rates of death from Nipah virus infection have been reported in Bangladesh, where outbreaks are most common in the winter [10,11]. The consumption of fruits or fruit products (such as raw date palm juice) contaminated with urine or saliva from infected fruit bats was the most likely cause of infection in later outbreaks in Bangladesh and India [11]. About 700 human cases of the Nipah virus had been reported as of May 2018, with 50 to 75 percent of those affected dying [12]. In the Indian state of Kerala, an epidemic of the disease led to 17 deaths in May 2018 [13].
A study of the proteins using bioinformatics tools allows researchers to assess their three-dimensional structural conformation, classify new domains, explore certain pathways to gain a better understanding of our evolutionary tree, uncover more clusters, and assign roles to the proteins [14,15,16]. This knowledge can also be used to develop successful pharmacological methods and aid in the development of new drugs to treat a wide range of diseases [17,18,19]. This study demonstrated the matrix protein secondary as well as tertiary characteristics that are associated with protein–structure relationships. The selected protein can be used as a potential target for protein-based drug and protein-based vaccine design candidates to minimize the viral infection.
2. Materials and Methods
2.1. Protein Selection and Sequence Retrieval
The amino acid (aa) sequence of the matrix protein found in Nipah henipavirus was obtained in FASTA format from the NCBI database [20].
2.2. Physicochemical Characterization of the Selected Protein
The amino acid sequence composition, instability index, aliphatic index, GRAVY (assessment of the hydrophobicity or hydrophilicity of a protein), and extinction coefficients were all measured using the ExPASy server ProtParam tool [21]. The theoretical isoelectric point (pI) of the QBQ56721.1 protein was also measured using SMS Suite (v.2.0) [22].
2.3. Functional Annotation of the Selected Protein
The conserved domain in the protein QBQ56721.1 was predicted using the NCBI platform’s CD-search tool [23,24]. The ExPASy software’s ScanProsite tool [25] and Pfam tool [26] were used to determine protein motifs. The evolutionary relationships of the protein QBQ56721.1 were assigned by the SuperFamily program [27].
2.4. Secondary Structural Properties and Assessment
The self-optimized prediction method with alignment (SOPMA) was used to predict secondary structure elements [28,29]. The secondary structure was predicted using the SPIPRED (v.4.0) [30] algorithm.
2.5. Three-Dimensional Structure Prediction and Validation of the Selected Protein
With Modeller [31], HHpred [32] predicted the three-dimensional (tertiary) structure. The most suitable template (HHpred ID: 6BK6 A) was chosen for creating the tertiary structure, with a probability, an E value, an aligned Cols, and goal lengths of 100, 2.4 × 10-116, 342, and 372, respectively. To predict the Ramachandran plot and validate the expected tertiary structure, the PROCHECK tool of the SAVES (v.6.0) program [33,34] was used.
3. Results and Discussion
3.1. Protein Sequence Retrieval
The amino acid (aa) sequence of the Nipah henipavirus protein (QBQ56721.1) was obtained from the NCBI database. The 352-amino-acid-long protein sequence was used to model the tertiary structure of the protein QBQ56721.1. Table 1 provides additional information on the protein (QBQ56721.1).

Table 1.
Protein retrieval.
3.2. Identification of the Physicochemical Properties of the Protein
The amino acid sequence of QBQ56721.1, which is found in Nipah henipavirus, was obtained in FASTA format and utilized as a query sequence for physicochemical parameter measurement. The protein is stable because its instability index is 30.59 (less than 40.00) [35]. The theoretical isoelectric point (pI) of the protein (pI 9.31, 9.65 *) indicates that it is basic [36,37,38]. The molecular weight, aliphatic index, instability index, and GRAVY are 39,847.16 Dalton, 89.69, 30.59, and −0.212, respectively (Table 2) [39,40,41]. The protein’s higher aliphatic index value of 89.69 indicates increased thermos-stability over a wide temperature range, which is a favorable factor [42,43]. The GRAVY index value of −0.212 suggested the protein’s hydrophilic character and, hence, the prospect of more water interaction [44,45].

Table 2.
Physicochemical parameters.
3.3. Functional Annotation Anticipation of the Selected Protein
The NCBI CDD tool identifies the domain that appears in identical protein sequences. RPS-BLAST is used by CD-Search to compare a test sequence to position-specific rating datasets compiled from conserved domain (CD) alignments in the CD protein cluster. The CD search engine identified a conserved domain in the protein QBQ56721.1 as a viral matrix protein (matrix, accession no. pfam00661). Viral matrix proteins are structural proteins that connect the viral envelope and the virus core [46,47]. The matrix protein plays an important role in virus assembly and in linking the viral envelope with the virus core. It is possible that they are found in Morbillivirus, Paramyxovirus, and Pneumovirus [47,48]. A motif was also predicted by the Pfam software at locations 16–349 (Pfam ID: PF00661; Viral matrix protein; e value of 1:7 10146). Protein motifs are small regions of a three-dimensional protein structure or amino acid sequence that are shared by multiple proteins [48,49]. Motifs are distinct regions of a protein structure that may or may not be defined by a distinct chemical or biological function [48,50,51,52,53].
The CDD technique also confirmed the presence of the viral matrix protein at the 17–348 position. The lone member of the superfamily cl02918 is the viral matrix protein (CDD no. pfam00661). A protein superfamily is a group of proteins made up of one or more protein families [46,53,54]. The set of all superfamilies must be a partitioning of the set of all protein sequences or subsequences defined by the protein families’ relationship, and each superfamily must be closed under transitivity [54]. The protein QBQ56721.1 (Figure 1) was predicted to be closely related to the matrix superfamily by the SuperFamily tool (e value of 0.0). Main text paragraph (M_Text).
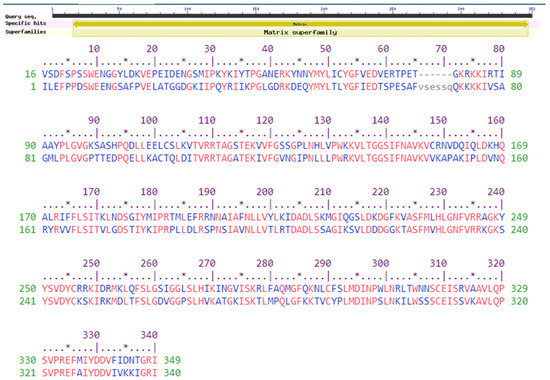
Figure 1.
Functional annotation of the selected protein QBQ56721.1. The graphical summary represents the conserved domains identified in the query sequence. The aligned sequences represent the conserved domains identified on the query sequence by comparing them with the conserved protein domain family matrix (CDD accession no. pfam00661). The Pfam software predicted a motif at 16–349 (accession no. PF00661) as a viral matrix protein. The SuperFamily program predicted the protein as a member of the matrix superfamily. An * (asterisk) indicates positions with a single, fully conserved residue.
3.4. Secondary Structural Inquiry
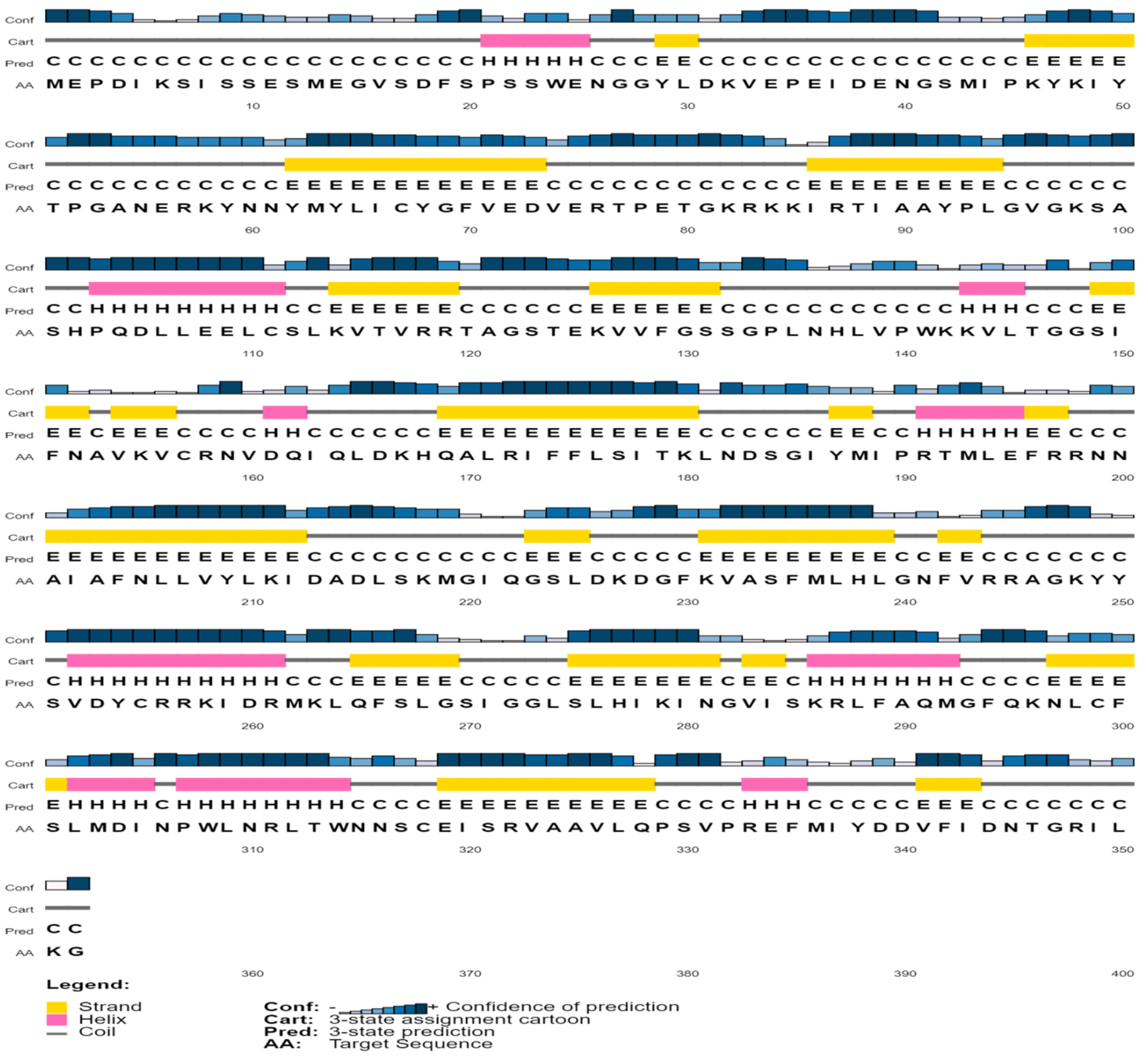
Protein structure and function are inextricably linked. Helix, coil, sheet, and turn are secondary structural elements that have a fantastic association with protein function, structure, and engagement [55,56,57,58]. The secondary-structural element of the protein (QBQ56721.1) was predicted by the SOPMA software when the alpha helix (Hh), extended strand (Ee), beta turn (Tt), and random coil (Cc) were 60 (17.05%), 87 (24.72%), 20 (5.68%), and 185 (52.56%), respectively (Table 3 and Table 4). The PSIPRED software predicts the helix, strand, and coil of the matrix protein (QBQ56721.1) with more confidence (Figure 2). Table 3 shows the amino acid composition obtained from the ExPASy service’s ProtParam Tool.

Table 3.
Amino acid composition.

Table 4.
Secondary structural elements.
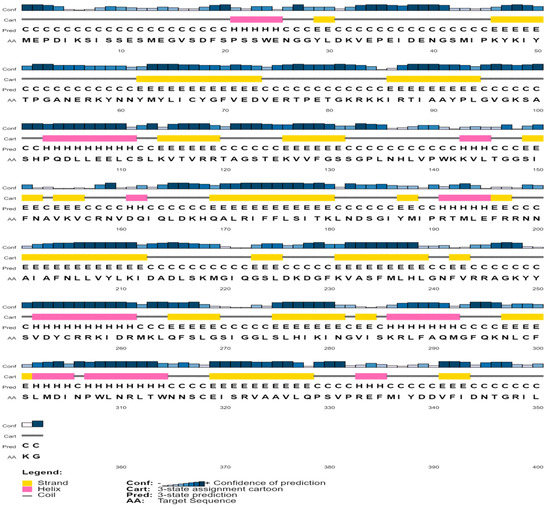
Figure 2.
Predicted secondary structure of the selected protein.
3.5. Tertiary-Structure Anticipation and Validation of the Protein
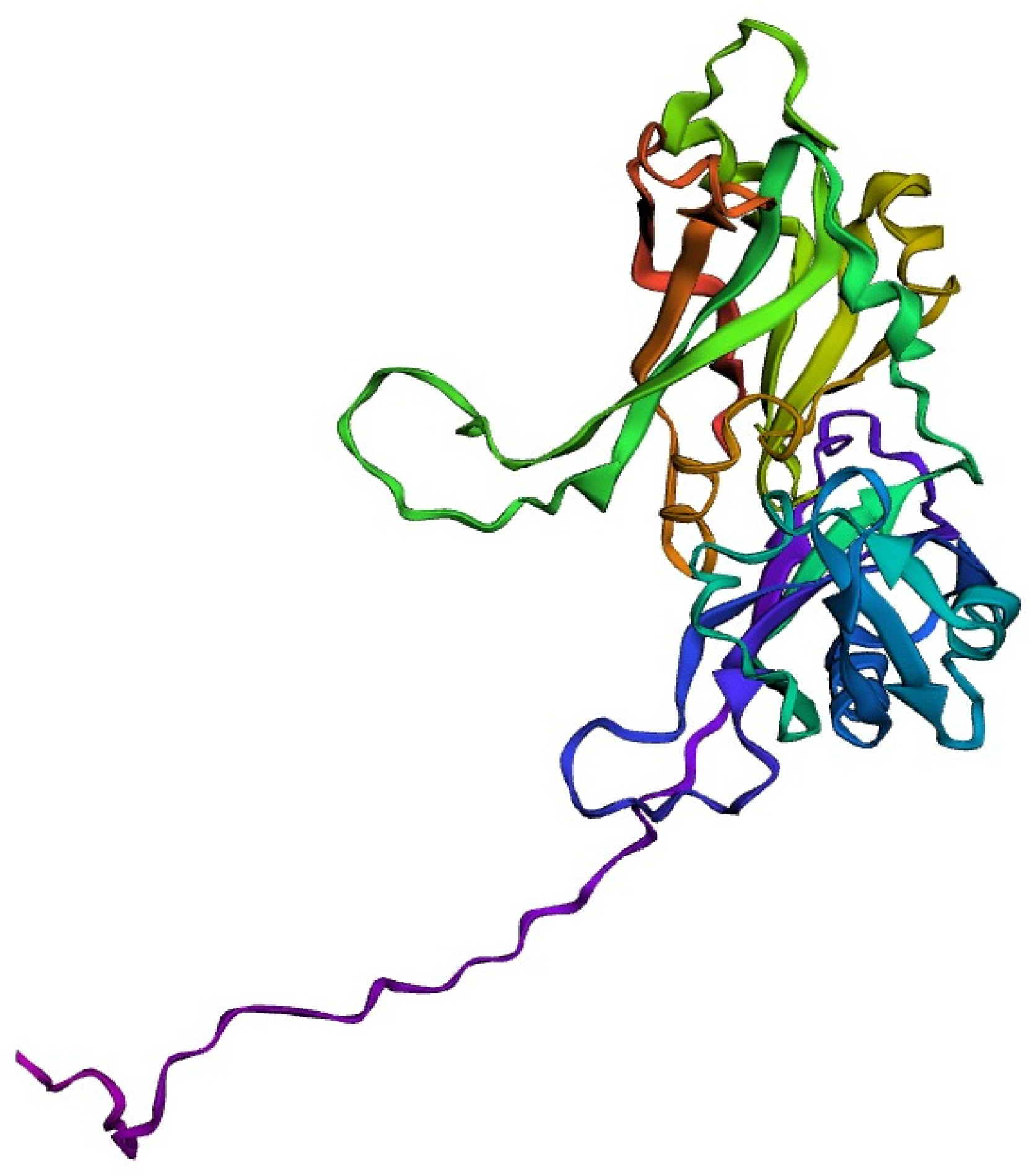
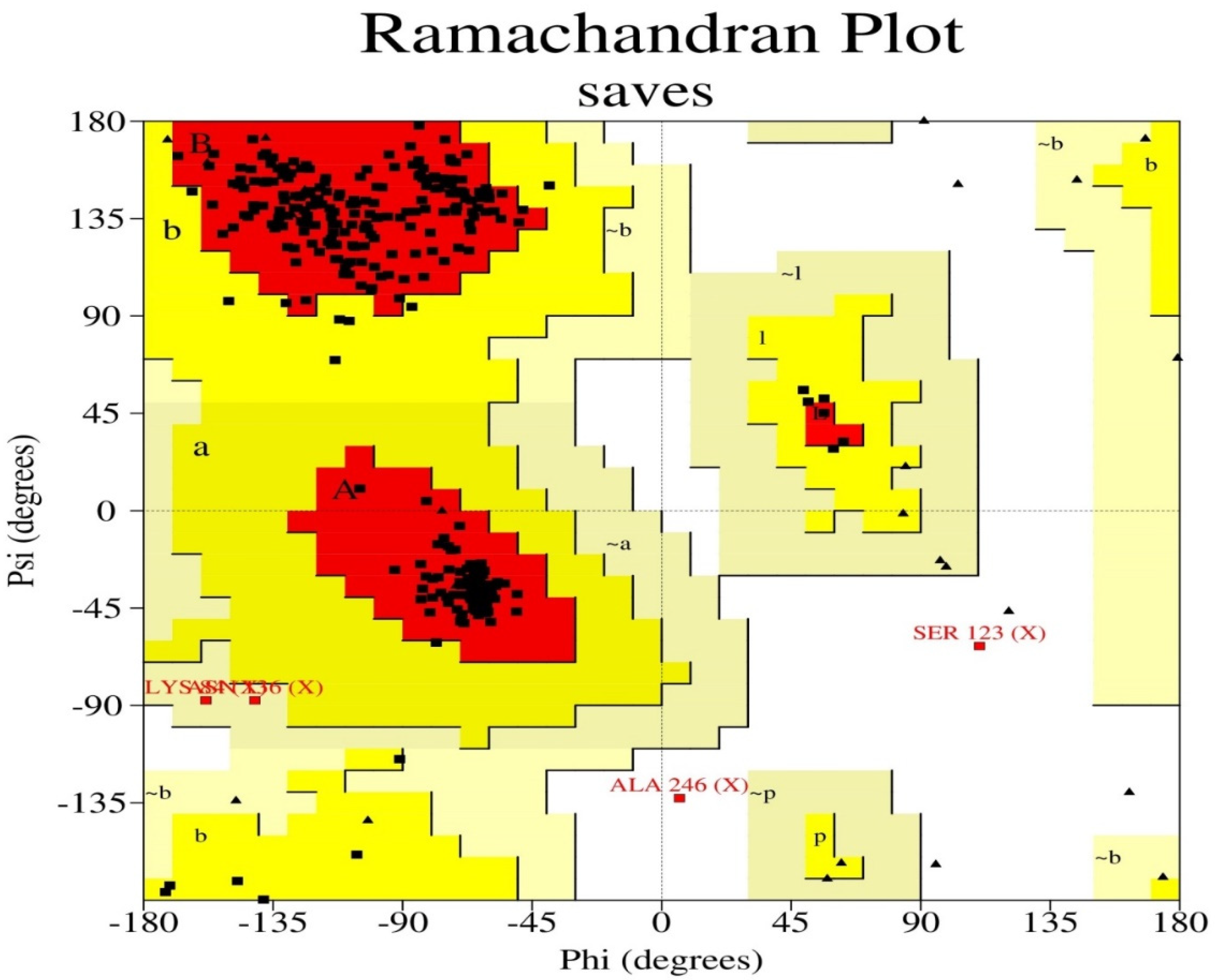
The target sequence of QBQ56721.1 in FASTA format was inserted into the HHpred Template Selection tool as the input, and the most suitable template (6BK6 A) was selected with a probability rate of 100%, an E-Value of 2.4 × 10-116, a Cols of 342, and a target length of 372 and finally stored the tertiary modeled protein structure in PDB format, as predicted by Modeller (Figure 3). The Ramachandran plot by PROCHECK (Figure 4) was used to assess the matrix protein’s tertiary structure, which revealed that 92.4 percent of the total residues (342) were found in the core (A, B, L); 6.3 percent of residues were in the additional allowed regions (a, b, l, p); and 0.7 percent of residues were in the generously allowed regions (a, b, l, p). The total number of non-glycine and non-proline residues was 301; that of the the end-residues (excluding Gly and Pro) was 1.0; that of the glycine and proline residues was 27 and 13, respectively, out of 473 total residues (Table 5). Verify 3D: a tertiary structure evaluation tool was used to demonstrate that the anticipated tertiary structure passed the evaluation.
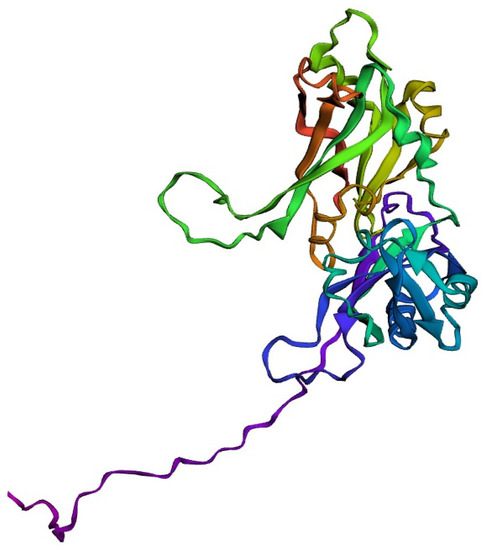
Figure 3.
Tertiary structure predicted by the HHpred tool employing the Modeller application.
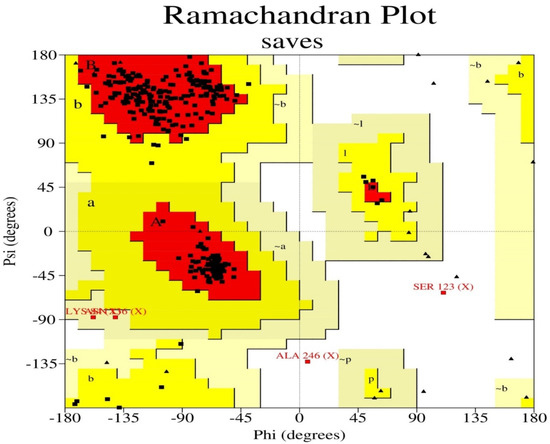
Figure 4.
The Ramachandran plot statistics of the Modeller-predicted three-dimensional protein structure validated by the PROCHECK program. The red regions in the graph indicate the most allowed areas. Additional allowed, generously allowed, and disallowed regions are receptively marked as yellow, light yellow, and white fields.

Table 5.
Ramachandran plot statistics of the modeled protein.
4. Conclusions
Understanding how proteins function is vital for describing how they work, and this protein is critical for virus assembly. With the virus core, the matrix protein binds to the viral envelope. This research reveals the protein’s fundamental features, such as its hydrophilic nature and functional annotation, in relation to its tertiary structure. As a result, the outcomes of this study demonstrate the efficacy and scope of future research on the matrix protein using the bioinformatics methodologies used in this investigation. The selected protein’s secondary and tertiary structures demonstrated the protein–function relationships of the matrix protein. This research will strengthen and sharpen our understanding of pathophysiology, allowing for the development of promising protein-based drugs and vaccine candidates to combat Nipah virus infection.
Author Contributions
Conceptualization, A.S.M.S.; methodology, A.S.M.S. and A.K.P.; software, A.S.M.S.; validation, A.S.M.S. and D.D.; formal analysis, A.S.M.S.; investigation, A.S.M.S.; resources, A.S.M.S. and A.K.P.; data curation, A.S.M.S., A.K.P., and D.D.; writing—original draft preparation, A.S.M.S., A.K.P. and D.D.; writing—review and editing, A.S.M.S., R.C.D. and M.C.D.; visualization, A.S.M.S., A.K.P. and D.D.; supervision, A.S.M.S.; project administration, A.S.M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ang, B.S.P.; Lim, T.C.C.; Wang, L. Nipah Virus Infection. J. Clin. Microbiol. 2018, 56, 1–10. [Google Scholar] [CrossRef]
- Paul, L. Nipah virus in Kerala: A deadly Zoonosis. Clin. Microbiol. Infect. 2018, 24, 1113–1114. [Google Scholar] [CrossRef] [PubMed]
- Aditi; Shariff, M. Nipah virus infection: A review. Epidemiol. Infect. 2019, 147, e95. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Kaushik, S.; Kumar, R.; Yadav, J.P.; Kaushik, S. Emerging trends of Nipah virus: A review. Rev. Med. Virol. 2019, 29, e2010. [Google Scholar] [CrossRef] [PubMed]
- Soman Pillai, V.; Krishna, G.; Veettil, M.V. Nipah Virus: Past Outbreaks and Future Containment. Viruses 2020, 12, 465. [Google Scholar] [CrossRef]
- Lo, M.K.; Rota, P.A. The emergence of Nipah virus, a highly pathogenic paramyxovirus. J. Clin. Virol. 2008, 43, 396–400. [Google Scholar] [CrossRef]
- Ternhag, A.; Penttinen, P. Nipah virus--another product from the Asian “virus factory”. Lakartidningen 2005, 102, 1046–1047. [Google Scholar]
- Choi, C. Nipah’s return. The lethal “flying fox” virus may spread between people. Sci. Am. 2004, 291, 21A–22A. [Google Scholar] [CrossRef]
- Singh, R.K.; Dhama, K.; Chakraborty, S.; Tiwari, R.; Natesan, S.; Khandia, R.; Munjal, A.; Vora, K.S.; Latheef, S.K.; Karthik, K.; et al. Nipah virus: Epidemiology, pathology, immunobiology and advances in diagnosis, vaccine designing and control strategies—A comprehensive review. Vet. Q. 2019, 39, 26–55. [Google Scholar] [CrossRef]
- Epstein, J.H.; Anthony, S.J.; Islam, A.; Kilpatrick, A.M.; Ali Khan, S.; Balkey, M.D.; Ross, N.; Smith, I.; Zambrana-Torrelio, C.; Tao, Y.; et al. Nipah virus dynamics in bats and implications for spillover to humans. Proc. Natl. Acad. Sci. USA 2020, 117, 29190–29201. [Google Scholar] [CrossRef]
- Yadav, P.D.; Shete, A.M.; Kumar, G.A.; Sarkale, P.; Sahay, R.R.; Radhakrishnan, C.; Lakra, R.; Pardeshi, P.; Gupta, N.; Gangakhedkar, R.R.; et al. Nipah Virus Sequences from Humans and Bats during Nipah Outbreak, Kerala, India, 2018. Emerg. Infect. Dis. 2019, 25, 1003–1006. [Google Scholar] [CrossRef] [PubMed]
- Sudeep, A.B.; Yadav, P.D.; Gokhale, M.D.; Balasubramanian, R.; Gupta, N.; Shete, A.; Jain, R.; Patil, S.; Sahay, R.R.; Nyayanit, D.A.; et al. Detection of Nipah virus in Pteropus medius in 2019 outbreak from Ernakulam district, Kerala, India. BMC Infect. Dis. 2021, 21, 162. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.D.; Raut, C.G.; Shete, A.M.; Mishra, A.C.; Towner, J.S.; Nichol, S.T.; Mourya, D.T. (Detection of Nipah virus RNA in fruit bat (Pteropus giganteus) from India. Am. J. Trop. Med. Hyg. 2012, 87, 576–578. [Google Scholar] [CrossRef] [PubMed]
- Gaudino, M.; Aurine, N.; Dumont, C.; Fouret, J.; Ferren, M.; Mathieu, C.; Reynard, O.; Volchkov, V.E.; Legras-Lachuer, C.; Georges-Courbot, M.C.; et al. High Pathogenicity of Nipah Virus from Pteropus lylei Fruit Bats, Cambodia. Emerg. Infect. Dis. 2020, 26, 104–113. [Google Scholar] [CrossRef]
- Rathish, B.; Vaishnani, K. Nipah Virus. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2022. [Google Scholar]
- Looi, L.M.; Chua, K.B. Lessons from the Nipah virus outbreak in Malaysia. Malays. J. Pathol. 2007, 29, 63–67. [Google Scholar]
- Lam, S.K.; Chua, K.B. Nipah virus encephalitis outbreak in Malaysia. Clin. Infect. Dis. 2002, 34 (Suppl. 2), S48–S51. [Google Scholar] [CrossRef]
- Singhai, M.; Jain, R.; Jain, S.; Bala, M.; Singh, S.; Goyal, R. Nipah Virus Disease: Recent Perspective and One Health Approach. Ann. Glob. Health 2021, 87, 102. [Google Scholar] [CrossRef]
- Gómez Román, R.; Wang, L.F.; Lee, B.; Halpin, K.; de Wit, E.; Broder, C.C.; Rahman, M.; Kristiansen, P.; Saville, M. Nipah@20: Lessons Learned from Another Virus with Pandemic Potential. mSphere 2020, 5, e00602-20. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021, 49, D10–D17. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Sigrist, C.J.; de Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013, 41, D344–D347. [Google Scholar] [CrossRef] [PubMed]
- Sigrist, C.J.; Cerutti, L.; Hulo, N.; Gattiker, A.; Falquet, L.; Pagni, M.; Bairoch, A.; Bucher, P. PROSITE: A documented database using patterns and profiles as motif descriptors. Brief. Bioinform. 2002, 3, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.; Madera, M.; Vogel, C.; Chothia, C.; Gough, J. The SUPERFAMILY database in 2007: Families and functions. Nucleic Acids Res. 2007, 35, D308–D313. [Google Scholar] [CrossRef]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Deléage, G. ALIGNSEC: Viewing protein secondary structure predictions within large multiple sequence alignments. Bioinformatics 2017, 33, 3991–3992. [Google Scholar] [CrossRef]
- Moffat, L.; Jones, D.T. Increasing the accuracy of single sequence prediction methods using a deep semi-supervised learning framework. Bioinformatics 2021, 37, 3744–3751. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef]
- Gabler, F.; Nam, S.Z.; Till, S.; Mirdita, M.; Steinegger, M.; Söding, J.; Lupas, A.N.; Alva, V. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinform. 2020, 72, e108. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Rullmannn, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Bowie, J.U.; Lüthy, R.; Eisenberg, D. A method to identify protein sequences that fold into a known three-dimensional structure. Science 1991, 253, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Gamage, D.G.; Gunaratne, A.; Periyannan, G.R.; Russell, T.G. Applicability of Instability Index for In vitro Protein Stability Prediction. Protein Pept. Lett. 2019, 26, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Pihlasalo, S.; Auranen, L.; Hänninen, P.; Härmä, H. Method for estimation of protein isoelectric point. Anal. Chem. 2012, 84, 8253–8258. [Google Scholar] [CrossRef]
- Audain, E.; Ramos, Y.; Hermjakob, H.; Flower, D.R.; Perez-Riverol, Y. Accurate estimation of isoelectric point of protein and peptide based on amino acid sequences. Bioinformatics 2016, 32, 821–827. [Google Scholar] [CrossRef]
- Saikat, A.S.M. An In Silico Approach for Potential Natural Compounds as Inhibitors of Protein CDK1/Cks2. Chem. Proc. 2022, 8, 5. [Google Scholar]
- Wilkins, M.R.; Williams, K.L. Cross-species protein identification using amino acid composition, peptide mass fingerprinting, isoelectric point and molecular mass: A theoretical evaluation. J. Theor. Biol. 1997, 186, 7–15. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Khan, R.A.; Hossain, R.; Siyadatpanah, A.; Al-Khafaji, K.; Khalipha, A.B.R.; Dey, D.; Asha, U.H.; Biswas, P.; Saikat, A.S.M.; Chenari, H.A.; et al. Diterpenes/Diterpenoids and Their Derivatives as Potential Bioactive Leads against Dengue Virus: A Computational and Network Pharmacology Study. Molecules 2021, 26, 6821. [Google Scholar] [CrossRef]
- Ikai, A. Thermostability and aliphatic index of globular proteins. J. Biochem. 1980, 88, 1895–1898. [Google Scholar] [PubMed]
- Dey, D.; Biswas, P.; Paul, P.; Mahmud, S.; Ema, T.I.; Khan, A.A.; Ahmed, S.Z.; Hasan, M.M.; Saikat, A.S.M.; Fatema, B.; et al. Natural flavonoids effectively block the CD81 receptor of hepatocytes and inhibit HCV infection: A computational drug development approach. Mol. Divers. 2022. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.T.; Jin, T.Y.; Zhang, Z.L.; Ye, Y.N.; Deng, Z.; Wang, J.; Guo, F.B. Quantitative elucidation of associations between nucleotide identity and physicochemical properties of amino acids and the functional insight. Comput. Struct. Biotechnol. J. 2021, 19, 4042–4048. [Google Scholar] [CrossRef] [PubMed]
- Saikat, A.S.M.; Islam, R.; Mahmud, S.; Imran, M.A.S.; Alam, M.S.; Masud, M.H.; Uddin, M.E. Structural and Functional Annotation of Uncharacterized Protein NCGM946K2_146 of Mycobacterium Tuberculosis: An In-Silico Approach. Proceedings 2020, 66, 13. [Google Scholar]
- Saikat, A.S.M.; Uddin, M.E.; Ahmad, T.; Mahmud, S.; Imran, M.A.S.; Ahmed, S.; Alyami, S.A.; Moni, M.A. Structural and Functional Elucidation of IF-3 Protein of Chloroflexus aurantiacus Involved in Protein Biosynthesis: An In Silico Approach. BioMed Res. Int. 2021, 2021, 9050026. [Google Scholar] [CrossRef] [PubMed]
- Battisti, A.J.; Meng, G.; Winkler, D.C.; McGinnes, L.W.; Plevka, P.; Steven, A.C.; Morrison, T.G.; Rossmann, M.G. Structure and assembly of a paramyxovirus matrix protein. Proc. Natl. Acad. Sci. USA 2012, 109, 13996–14000. [Google Scholar] [CrossRef]
- Shtykova, E.V.; Petoukhov, M.V.; Dadinova, L.A.; Fedorova, N.V.; Tashkin, V.Y.; Timofeeva, T.A.; Ksenofontov, A.L.; Loshkarev, N.A.; Baratova, L.A.; Jeffries, C.M.; et al. Solution Structure, Self-Assembly, and Membrane Interactions of the Matrix Protein from Newcastle Disease Virus at Neutral and Acidic pH. J. Virol. 2019, 93, e01450-18. [Google Scholar] [CrossRef]
- Stollar, E.J.; Smith, D.P. Uncovering protein structure. Essays Biochem. 2020, 64, 649–680. [Google Scholar] [CrossRef]
- Heizinger, L.; Merkl, R. Evidence for the preferential reuse of sub-domain motifs in primordial protein folds. Proteins 2021, 89, 1167–1179. [Google Scholar] [CrossRef]
- Xie, J.; Lai, L. Protein topology and allostery. Curr. Opin. Struct. Biol. 2020, 62, 158–165. [Google Scholar] [CrossRef]
- Santhouse, J.R.; Rao, S.R.; Horne, W.S. Analysis of folded structure and folding thermodynamics in heterogeneous-backbone proteomimetics. Methods Enzymol. 2021, 656, 93–122. [Google Scholar] [PubMed]
- Vishwanath, S.; de Brevern, A.G.; Srinivasan, N. Same but not alike: Structure, flexibility and energetics of domains in multi-domain proteins are influenced by the presence of other domains. PLoS Comput. Biol. 2018, 14, e1006008. [Google Scholar] [CrossRef] [PubMed]
- Berezovsky, I.N.; Guarnera, E.; Zheng, Z. Basic units of protein structure, folding, and function. Prog. Biophys. Mol. Biol. 2017, 128, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Padjasek, M.; Kocyła, A.; Kluska, K.; Kerber, O.; Tran, J.B.; Krężel, A. Structural zinc binding sites shaped for greater works: Structure-function relations in classical zinc finger, hook and clasp domains. J. Inorg. Biochem. 2020, 204, 110955. [Google Scholar] [CrossRef]
- Zhang, G.J.; Ma, L.F.; Wang, X.Q.; Zhou, X.G. Secondary Structure and Contact Guided Differential Evolution for Protein Structure Prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 1068–1081. [Google Scholar] [CrossRef] [PubMed]
- Rademaker, D.; van Dijk, J.; Titulaer, W.; Lange, J.; Vriend, G.; Xue, L. The Future of Protein Secondary Structure Prediction Was Invented by Oleg Ptitsyn. Biomolecules 2020, 10, 910. [Google Scholar] [CrossRef]
- Wardah, W.; Khan, M.G.M.; Sharma, A.; Rashid, M.A. Protein secondary structure prediction using neural networks and deep learning: A review. Comput. Biol. Chem. 2019, 81, 1–8. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).