DNA-Binding Properties of Cytotoxic Naphtindolizinedione-Carboxamides Acting as Type II Topoisomerase Inhibitors. A Combined In Silico and Experimental Study †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Molecular Docking
2.1.1. Calculations for the Protonated Ligand/Oligomeric DNA Complexes
2.1.2. Docking Calculation on Topoisomerase II
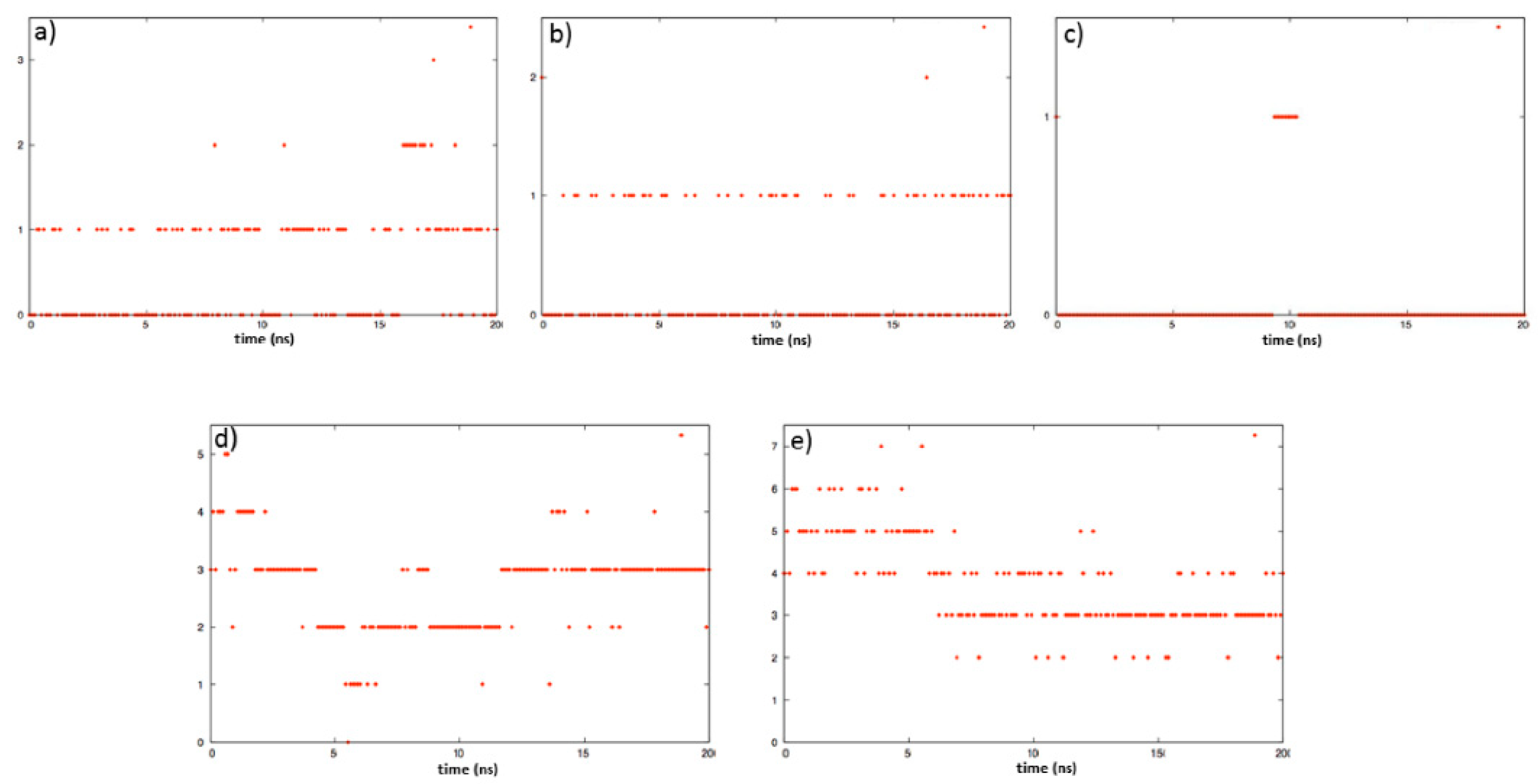
2.2. Molecular Dynamics
3. Results and Discussion
3.1. Computational Study
3.2. Spectroscopic Analyses
3.3. Topoisomerase IIα Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Deweese, J.E.; Osheroff, N. The DNA cleavage reaction of topoisomerase II: Wolf in sheep’s clothing. Nucleic Acids Res. 2009, 37, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Martinez, R.; Chacon-Garcia, L. The Search of DNA-Intercalators as Antitumoral Drugs: What it Worked and What did not Work. Curr. Med. Chem. 2005, 12, 127–151. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Li, T.K.; Farh, L.; Lin, L.Y.; Lin, T.S.; Yu, Y.J.; Yen, T.J.; Chiang, C.W.; Chan, N.L. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science 2011, 333, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Heck, M.M.; Earnshaw, W.C. Mitotic chromatin condensation in vitro using somatic cell extracts and nuclei with variable levels of endogenous topoisomerase II. J. Cell Biol. 1986, 103, 2569–2581. [Google Scholar] [CrossRef] [PubMed]
- Defant, A.; Guella, G.; Mancini, I. Synthesis and in vitro cytotoxicity evaluation of novel naphthindolizinedione derivatives. Arch. Pharm. Chem. Life Sci. 2007, 340, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Defant, A.; Guella, G.; Mancini, I. Synthesis and in-vitro cytotoxicity evaluation of novel naphtindolizinedionederivatives, part II: Improved activity for aza-analogues. Arch. Pharm. Chem. Life Sci. 2009, 342, 80–86. [Google Scholar] [CrossRef]
- Defant, A.; Guella, G.; Mancini, I. Microwave-assisted multicomponent synthesis of aza-, diaza-, benzo-, and dibenzofluorenedione derivatives. Synth. Commun. 2008, 38, 3003–3016. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Sanner, M.F. Python: A Programming Language for Software Integration and Development. J. Mol. Graph Model. 1999, 17, 57–61. [Google Scholar] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 16, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://code.google.com/archive/p/d2md/downloads (accessed on 20 October 2020).
- Strat, D.; Missailidis, S.; Drake, A.F. A novel methodological approach for the analysis of host-ligand interactions. ChemPhysChem 2007, 8, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Stootman, F.H.; Fisher, D.M.; Rodgerc, A.; Aldrich-Wright, J.R. Improved curve fitting procedures to determine equilibrium binding constants. Analyst 2006, 131, 1145–1151. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.-L.; Lacroix, L. Analysis of thermal melting curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef] [PubMed]
- Lerman, L.S. Structural considerations in the interaction of DNA and acridines. J. Mol. Biol. 1961, 3, 18–30. [Google Scholar] [CrossRef]
- Johnson, Β.B.; Dahl, Κ.S.; Tinoco, I.; Ivanov, V.; Zhurkin, V.B. Correlations between deoxyribonucleic acid structural parameters and calculated circular dichroism spectra. Biochemistry 1981, 20, 73–78. [Google Scholar] [CrossRef]
- Rajendran, B.U.A. Nair. Biochim. Biophys. Acta 2006, 1760, 1794–1801. [Google Scholar] [CrossRef] [PubMed]
- Garbett, N.C.; Ragazzon, P.A.; Chaires, J.B. Circular dichroism to determine binding mode and affinity of ligand–DNA interactions. Nat. Protoc. 2007, 2, 3166–3172. [Google Scholar] [CrossRef] [PubMed]
- Furlanetto, V.; Zagotto, G.; Pasquale, R.; Moro, S.; Gatto, B. Ellagic acid and polyhydroxylated urolithins are potent catalytic inhibitors of human topoisomerase II: An in vitro study. J. Agric. Food Chem. 2012, 60, 9162–9170. [Google Scholar] [CrossRef] [PubMed]

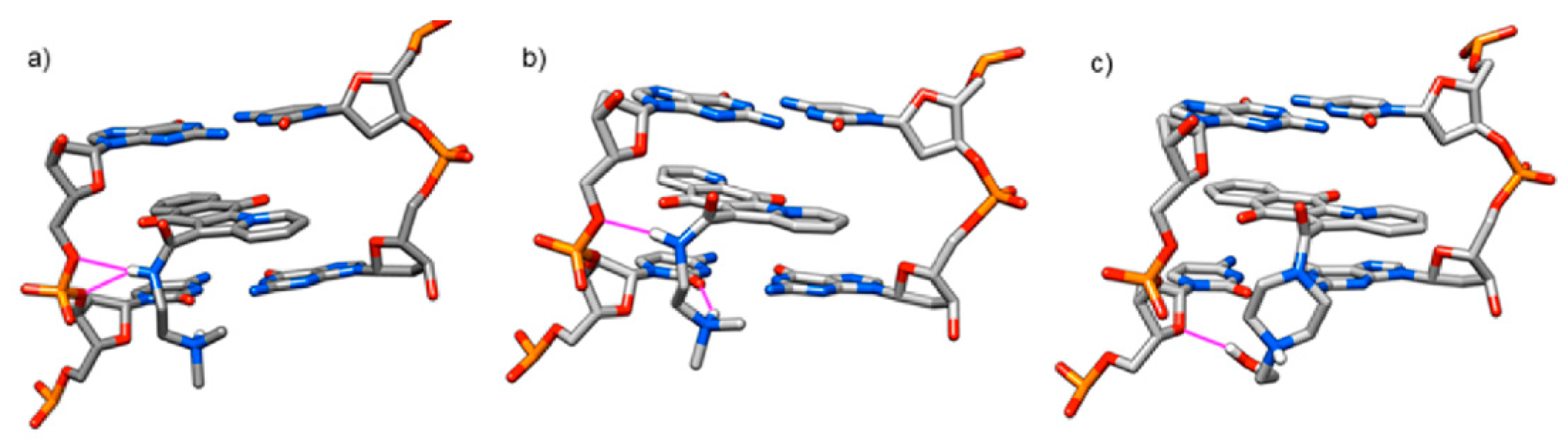
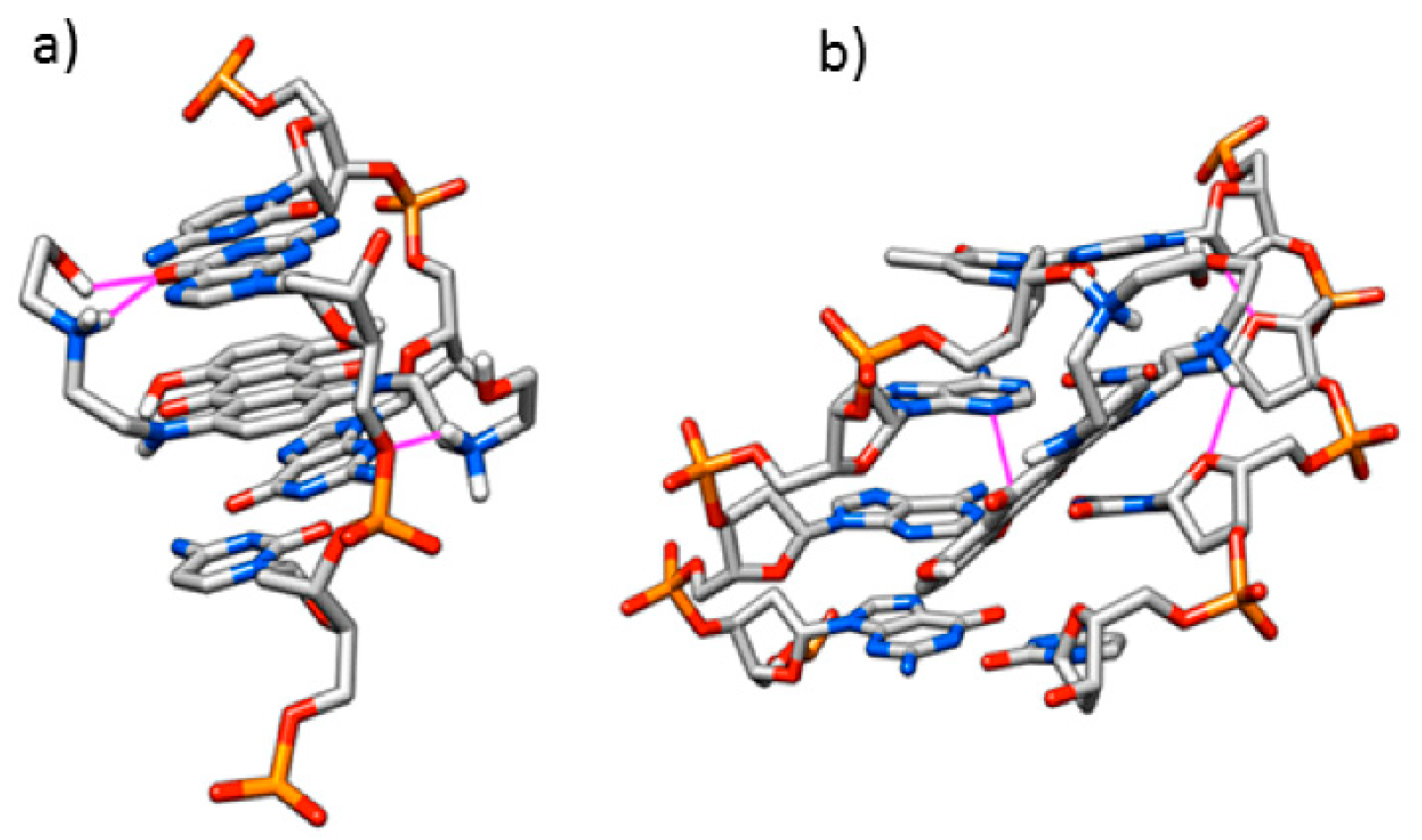
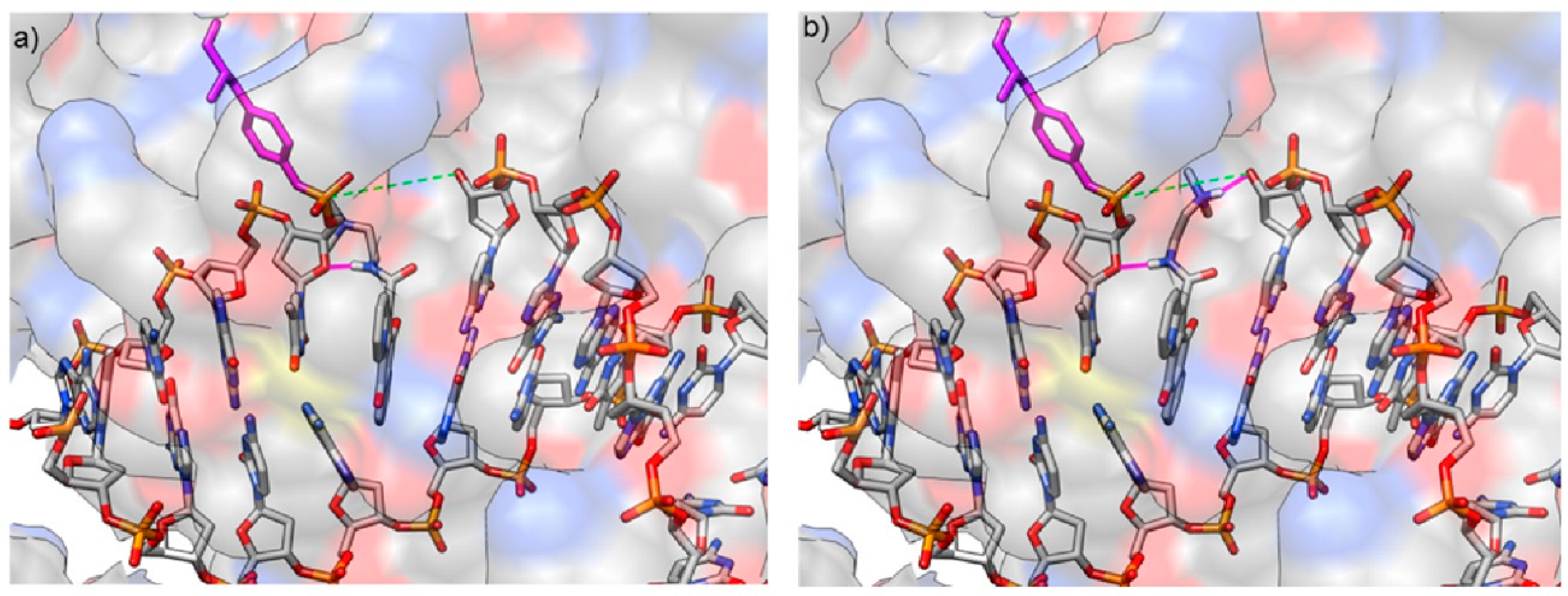
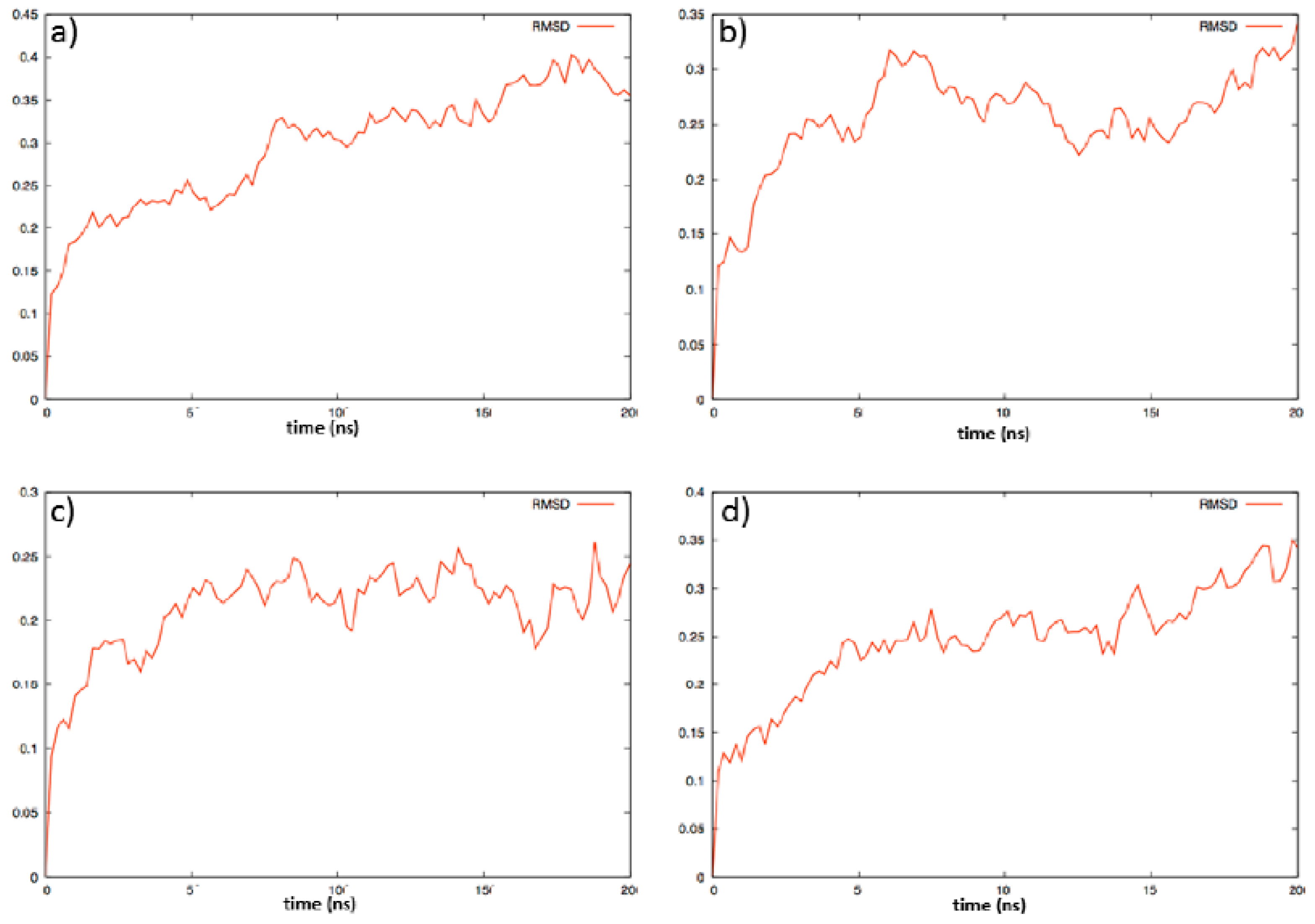
| Ligand | ΔG (Kcal/mol) | K |
|---|---|---|
| 1 | −8.4 | 1.4·10⁶ |
| 2 | −8.5 | 1.7·10⁶ |
| 3 | −9.1 | 5.0·10⁶ |
| Mitoxantrone | −7.8 | 1.5·105 |
| Ligand | Ligand RMSD [nm] | Oligomer RMSD [nm] | H-bond Number |
|---|---|---|---|
| 1 | 0.14 | 0.29 | 0.532 |
| 2 | 0.11 | 0.26 | 0.342 |
| 3 | 0.13 | 0.21 | 0.060 |
| Mitoxantrone (intercalation) | 0.09 | 0.25 | 2.672 |
| Mitoxantrone (minor groove interaction) | 0.11 | 0.22 | 3.756 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Defant, A.; Gatti, P.; Poli, A.; Serena, M.; Sosic, A.; Gatto, B.; Mancini, I. DNA-Binding Properties of Cytotoxic Naphtindolizinedione-Carboxamides Acting as Type II Topoisomerase Inhibitors. A Combined In Silico and Experimental Study. Chem. Proc. 2021, 3, 96. https://doi.org/10.3390/ecsoc-24-08103
Defant A, Gatti P, Poli A, Serena M, Sosic A, Gatto B, Mancini I. DNA-Binding Properties of Cytotoxic Naphtindolizinedione-Carboxamides Acting as Type II Topoisomerase Inhibitors. A Combined In Silico and Experimental Study. Chemistry Proceedings. 2021; 3(1):96. https://doi.org/10.3390/ecsoc-24-08103
Chicago/Turabian StyleDefant, Andrea, Paolo Gatti, Alessandro Poli, Marta Serena, Alice Sosic, Barbara Gatto, and Ines Mancini. 2021. "DNA-Binding Properties of Cytotoxic Naphtindolizinedione-Carboxamides Acting as Type II Topoisomerase Inhibitors. A Combined In Silico and Experimental Study" Chemistry Proceedings 3, no. 1: 96. https://doi.org/10.3390/ecsoc-24-08103
APA StyleDefant, A., Gatti, P., Poli, A., Serena, M., Sosic, A., Gatto, B., & Mancini, I. (2021). DNA-Binding Properties of Cytotoxic Naphtindolizinedione-Carboxamides Acting as Type II Topoisomerase Inhibitors. A Combined In Silico and Experimental Study. Chemistry Proceedings, 3(1), 96. https://doi.org/10.3390/ecsoc-24-08103








