Abstract
With inadequate water purification and sewage disposal systems, cholera poses a significant health threat in developing nations. Vibrio cholerae (V. cholerae) is one of the main microscopic organisms associated with cholera illness. If cholera is not treated, renal failure, shock, hypokalemia, and pulmonary edema can occur, resulting in death in a matter of hours. The machinery of bacterial virulence factors is what causes this disease. Among the various V. cholerae strains, V. cholerae O1 is the most prevalent and pathogenic strain. The total genome succession of V. cholerae unravels the presence of different genes and uncharacterized proteins whose capabilities are not yet perceived. Therefore, it is essential to comprehend V. cholerae by analyzing the structure and annotating the function of uncharacterized proteins. The NCBI sequence of uncharacterized V. cholerae O1 EET91795.1 proteins was annotated for this study. The domain family, protein solubility, ligand binding sites, and other parameters were all determined using a variety of databases and computational tools. The protein’s ligand-binding sites were found, and its three-dimensional structure was modeled. According to the analysis, the hotdog family protein may play metabolic roles like thioester hydrolysis in the metabolism of fatty acids and the breakdown of two products such as phenylacetic acid and the pollutant 4-chlorobenzoate. The structural prediction of this protein and detection of binding sites suggested a potential target to uncover promising inhibitors against the protein to treat infection caused by the target strain.
1. Introduction
V. cholerae is a Gram-negative, facultatively anaerobic, highly mobile bacteria with an arch or comma-shaped with a single polar flagellum. Gammaproteobacteria secrete enterotoxin, which induces severe diarrhea known as cholera [1]. V. cholerae is primarily found in uncooked foods and polluted water and is transmitted by the fecal-oral route. An enzymatically active factor that raises cyclic adenosine 5-monophosphate (cAMP) production is secreted when the toxin binds to a specific receptor, monosialosyl ganglioside GM1, in the intestinal epithelial cells of the plasma membrane. Due to the cell’s high cAMP levels, the intestinal lumen will overflow with electrolytes and water [2,3]. At an ever-increasing rate, structural genomics initiatives produce many uncharacterized protein structures. However, this enormous structural storage is only helpful with functional annotation for biologists interested in particular molecular systems [4].
We implement bioinformatic tools for the functional annotation for the EET91795.1 protein of V. cholerae, the causative agent of cholera, particularly in Southeast Asia [5]. This uncharacterized protein could be helpful as a pharmacological target and marker if annotated. According to the findings of various bioinformatics databases and studies, these proteins play a significant part in the pathogenesis of V. cholerae.
2. Materials and Methods
2.1. Protein Sequence Retrieval
The protein sequence was retrieved from the NCBI-protein database [6] with the accession number of EET91795 (version: EET91795.1).
2.2. Physicochemical Characterization
The amino acid sequence composition, the instability index, the aliphatic index, the GRAVY (the measurement of hydrophobicity or hydrophilicity of a protein), extinction coefficients, and the theoretical isoelectric point (pI) of the EET91795 protein were measured by the ExPaSy’s ProtParam program [7,8].
2.3. Functional Annotation Prediction
The NCBI platform’s CD search tool was applied to anticipate the conserved domain in the protein EET91795.1. Protein motif determination was implemented using the ScanProsite tool [9,10].
2.4. Subcellular Location Identification
The protein’s subcellular location was documented using the PSORTb v.3.0.3, SOSUI assessment tool, PSLpred server, HMMTOP v.2.0, and TMHMM server v.2.0 [11,12,13].
2.5. Secondary Structural Assessment
The SOPMA server exploited the secondary structural elements’ prediction following the default parameters of the protein EET91795.1 present in V. cholerae. Moreover, the PSIPRED v.4.0 tool was used to predict the secondary structure and the disordered areas [14].
2.6. Tertiary Structure Modeling and Validation
The three-dimensional structure of the protein was modeled by utilizing the Swiss-Model server. The anticipated tertiary structure obtained from the Swiss-Model server was subjected to structural quality evaluation experiments. The PROCHECK of the SAVES v.6.0 program was performed for Ramachandran plot calculation [15].
2.7. Active Site Determination
The CASTp v.3.0 server was used to predict the active sites of the modeled protein [16].
3. Results and Discussion
3.1. Protein Sequence Retrieval
Protein sequencing identifies a protein or peptide’s amino-acid sequence, either as a whole or a segment. The NCBI Protein data bank is a big store of sequences from multiple sources, including GenBank, RefSeq SwissProt, PDB, etc. The protein obtained from the NCBI database with the accession number EET91795 (version: EET91795.1) was present in the locus EET91795 containing 161 amino acids (aa).
3.2. Physicochemical Characterization
By examining the properties of each amino acid in a protein, we can understand how proteins’ physical and chemical properties are defined [17]. The ProtParam program on the ExPASy server was used to determine the physicochemical characteristics of the protein (EET91795.1). The protein consists of 161 amino acids, where Val (17) represented the amino acid of the greatest quantity, followed by Ala (14), Arg (7), Asn (9), Asp (3), Cys (3), Gln (6), Glu (10), Gly (10), His (7), Ile (12), Leu (13), Lys (5), Met (5), Phe (4), Pro (6), Ser (13), Thr (12), Trp (2), and Tyr (3). The time it takes for the radiolabeled target protein concentration to decrease by 50% from the level at the beginning of the chase is known as the protein half-life. The protein (EET91795.1) V. Cholerae has an estimated half-life of about 30 h (mammalian reticulocytes, in vitro), >20 h (yeast, in vivo), and >10 h (Escherichia coli, in vivo). The calculated isoelectric point (pI), molecular weight (MW), and the number of total atoms were 6.64, 17,568.20 Dalton, and 2476, respectively.
3.3. Functional Annotation Prediction
The CD search tool of NCBI predicted a conserved domain as a hot-dog superfamily protein (accession: cl00509) of the protein. The structure of E. coli’s FabA (beta-hydroxydecanoyl-acyl carrier protein (ACP)-dehydratase) and Pseudomonas’ 4HBT (4-hydroxy benzoyl-CoA thioesterase) both contained the hot-dog fold. Various other inconsequential proteins additionally share the hot dog-fold. The metabolic functions of these proteins include the thioester hydrolysis of fatty acids, degradation of phenylacetic acid, and the environmental pollutant 4-chlorobenzoate, among other related but distinct catalytic activities. FapR is a member of this superfamily involved in the transcriptional regulation of fatty acid biosynthesis. Moreover, the ScanProsite tool predicts four pattern sites of the protein EET91795.1. These are the Protein kinase C phosphorylation site (accession no.PS00005), N-glycosylation site (accession no. PS00001), Casein kinase II phosphorylation site (accession: PS00006), and N-myristoylation site (accession: PS00008).
3.4. Subcellular Location Determination
The PSORTb (v.3.0.2), SOSUIGramN, and PSLpred tools were used for subcellular location analysis of the protein (EET91795.1). The tools predicted the subcellular location of the protein as a cytoplasmic protein. The HMMTOP (v.2.0) and TMHMM (v.2.0) programs anticipated that there were no transmembrane helices in the protein (EET91795.1) and highlighted the cytoplasmic location of the protein present in V. cholerae (Table 1).

Table 1.
Subcellular localization assessment.
3.5. Secondary Structure Inquiry
Protein function and structure are vigorously connected. The secondary structural elements, e.g., helix, coil, sheet, and turn, have an excellent relationship with protein function, construction, and engagement. The SOPMA program predicted the secondary-structural component of the protein (EET91795.1) where the alpha-helix (Hh), extended-strand (Ee), and random-coils (Cc) were 49 (30.43%), 44 (27.33%), 68 (42.24%), respectively. The PSIPRED v.4.0 tool predicted the sequence plot and secondary structure. The sequence plot from the two-dimensional structure of the protein indicates that most of the protein is cytoplasmic.
3.6. Tertiary-Structure Modeling and Validation
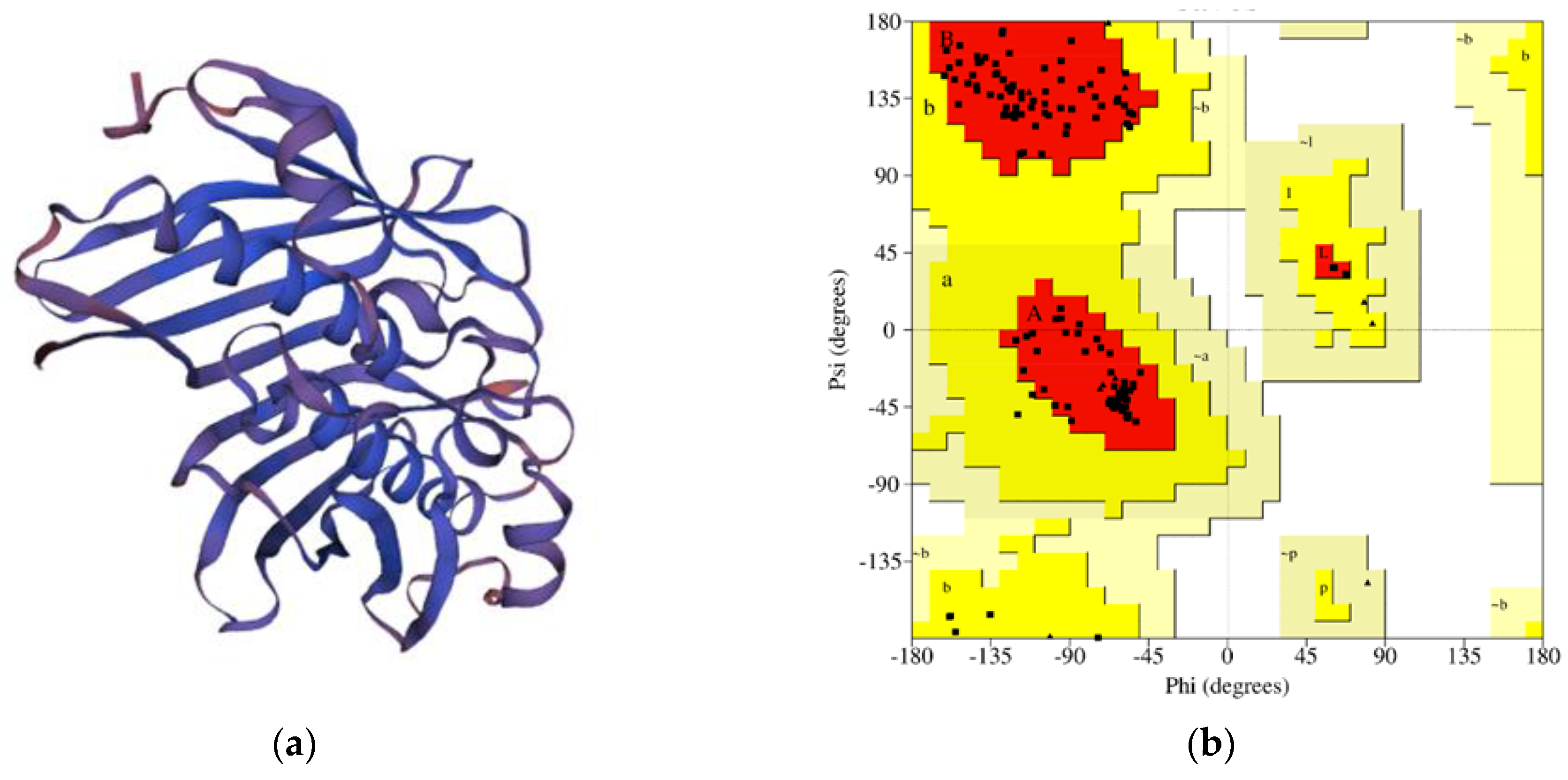
The Swiss-Model server was utilized for tertiary-structure prediction of the protein EET91795.1. The Swiss-Model server predicted the tertiary structure of the protein based on the most favored template (4qdb.1.A). This template has a the Global Model Quality Estimation (GMQE) value, QMEANDisCo Global value, and sequence identity score of 0.72, 0.80, and 46.48%, respectively. The Ramachandran plot analysis by PROCHECK was applied for the structural assessment of the modeled tertiary structure obtained from the Swiss-Model server [18,19,20,21].
In the case of the anticipated tertiary structure by the Swiss Model, the assessment experiment executed by the Ramachandran Map (PROCHECK) indicated that 93% of the total residues (227) were found in the core, 7.0% of residues were found in the additional allowed regions, and there was no residue found in the abundantly authorized parts and the disallowed areas [22,23]. The number of non-glycine and non-proline residues was 244, which was 100%, and the end residues were four; the glycine and proline residues were 20 and 8, respectively, among the 276 total residues (Figure 1).
Figure 1.
(a) Structure of EET91795.1 protein predicted by Swiss Model, and (b) the Ramachandran plot statistics of the modeled three-dimensional structure validated by the PROCHECK program. The 3D structure of the protein was anticipated using the Swiss-Model server (https://swissmodel.expasy.org/, accessed on 1 November 2022). Triangles represent the number of glycine residues. Residues are in the most favorable regions in the A and B areas. Moreover, the residues in the additional allowed regions are found in the a and b areas. The red color represents the most favored regions, whereas the yellow color denotes the additional allowed regions.
3.7. Active Site Determination
The CASTp version 3.0 program predicted 35 different active sites of the modeled protein. This server recognized all surface pockets, internal cavities, and transversal channels in the protein structure and described all atoms involved in their formation. It also calculated their exact size and area and the size of the mouth openings, if any. These dimensions were calculated analytically using a solvent-accessible surface model (Richards surface) and a molecular surface model (Connolly surface). CASTp could be utilized to investigate surface properties and protein operational zones. CASTp also helped study a protein’s surface properties and functional parts. CASTp provided a graphical, versatile, dynamic user interface and user-provided constructs for rapid measurements. The top active sites of the modeled protein were identified between the area of 256.436 and the volume of 94.190.
4. Conclusions
In this study, the protein EET91795.1 of V. Cholerae was retrieved from the NCBI database, and we determined their physicochemical properties and identified domains and families using various bioinformatics tools and databases. This protein is hydrophobic and localized in the cytoplasm of the cell. In addition, it is a hot-dog superfamily protein that may act in metabolic roles. This protein has a coenzyme binding site and four more enzymatic pattern sites. The secondary structure verified that alpha-helix, random spiral, extended strand, and beta-turns were uppermost in most sequences. The Swiss-Model server was used to evaluate tertiary systems, and PROCHECK for Ramachandran Map Analysis showed that the protein residues in most favored regions were 93%. The CASTp program predicted 35 distinct functional areas of this modeled protein. The findings of this study may contribute to the modulation of new target identification and drug discovery for the control of cholera, thereby reducing this deadly global epidemic.
Author Contributions
Conceptualization, A.S.M.S.; methodology, A.S.M.S.; software, M.Y. and A.S.M.S.; validation, A.S.M.S.; formal analysis, A.S.M.S.; investigation, A.S.M.S.; resources, A.S.M.S.; data curation, M.Y. and A.S.M.S.; writing—original draft preparation, M.Y.; writing—review and editing, A.S.M.S. and M.E.U.; visualization, M.Y. and A.S.M.S.; supervision, A.S.M.S.; project administration, A.S.M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Upon request to the corresponding author.
Acknowledgments
The authors want to acknowledge the Department of Computational Biology and Bioinformatics, Advanced Bioscience Center for Collaborative Research (ABSCCR), Rajshahi 6250, Bangladesh, for supporting this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Weil, A.A.; Becker, R.L.; Harris, J.B. Vibrio cholerae at the Intersection of Immunity and the Microbiome. mSphere 2019, 4, e00597-19. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.Y.; Liu, R.; Macbeth, J.C.; Hsiao, A. The Interface of Vibrio cholerae and the Gut Microbiome. Gut Microbes 2021, 13, 1937015. [Google Scholar] [CrossRef] [PubMed]
- Conner, J.G.; Teschler, J.K.; Jones, C.J.; Yildiz, F.H. Staying Alive: Vibrio cholerae’s Cycle of Environmental Survival, Transmission, and Dissemination. Microbiol. Spectr. 2016, 4, 593–633. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio spp. infections. Nature reviews. Dis. Prim. 2018, 4, 8. [Google Scholar] [CrossRef]
- Deen, J.; Mengel, M.A.; Clemens, J.D. Epidemiology of cholera. Vaccine 2020, 38 (Suppl. S1), A31–A40. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021, 49, D10–D17. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Saikat, A.S.M.; Paul, A.K.; Dey, D.; Das, R.C.; Das, M.C. In-Silico Approaches for Molecular Characterization and Structure-Based Functional Annotation of the Matrix Protein from Nipah henipavirus. Chem. Proc. 2022, 12, 21. [Google Scholar] [CrossRef]
- de Castro, E.; Sigrist, C.J.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef]
- Saikat, A.S.M.; Islam, R.; Mahmud, S.; Imran, A.S.; Alam, M.S.; Masud, M.H.; Uddin, E. Structural and Functional Annotation of Uncharacterized Protein NCGM946K2_146 of Mycobacterium Tuberculosis: An In-Silico Approach. Proceedings 2020, 66, 13. [Google Scholar] [CrossRef]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- Tusnády, G.E.; Simon, I. The HMMTOP transmembrane topology prediction server. Bioinformatics 2001, 17, 849–850. [Google Scholar] [CrossRef] [PubMed]
- Saikat, A.S.M.; Uddin, M.E.; Ahmad, T.; Mahmud, S.; Imran, M.A.S.; Ahmed, S.; Alyami, S.A.; Moni, M.A. Structural and Functional Elucidation of IF-3 Protein of Chloroflexus aurantiacus Involved in Protein Biosynthesis: An In Silico Approach. BioMed Res. Int. 2021, 2021, 9050026. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef] [PubMed]
- Saikat, A.S.M. Computational approaches for molecular characterization and structure-based functional elucidation of a hypothetical protein from Mycobacterium tuberculosis. Genom. Inform. 2023, 21, e25. [Google Scholar] [CrossRef] [PubMed]
- Jisna, V.A.; Jayaraj, P.B. Protein Structure Prediction: Conventional and Deep Learning Perspectives. Protein J. 2021, 40, 522–544. [Google Scholar] [CrossRef] [PubMed]
- Saikat, A.S.M.; Ripon, A. Structure Prediction, Characterization, and Functional Annotation of Uncharacterized Protein BCRIVMBC126_02492 of Bacillus cereus: An In Silico Approach. Am. J. Pure Appl. Biosci. 2020, 2, 104–111. [Google Scholar] [CrossRef]
- Eisenhaber, F.; Persson, B.; Argos, P. Protein structure prediction: Recognition of primary, secondary, and tertiary structural features from amino acid sequence. Crit. Rev. Biochem. Mol. Biol. 1995, 30, 1–94. [Google Scholar] [CrossRef]
- Saikat, A.S.M.; Das, R.C.; Das, M.C. Computational Approaches for Structure-Based Molecular Characterization and Functional Annotation of the Fusion Protein of Nipah henipavirus. Chem. Proc. 2022, 12, 32. [Google Scholar] [CrossRef]
- Dubey, S.P.N.; Kini, N.G.; Balaji, S.; Kumar, M.S. A Review of Protein Structure Prediction Using Lattice Model. Crit. Rev. Biomed. Eng. 2018, 46, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Saikat, A.S.M. An In Silico Approach for Potential Natural Compounds as Inhibitors of Protein CDK1/Cks2. Chem. Proc. 2022, 8, 5. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).