Abstract
The Epiretinal Membrane (ERM) is an ocular disease that appears as a fibro-cellular layer of tissue over the retina, specifically, over the Inner Limiting Membrane (ILM). It causes vision blurring and distortion, and its presence can be indicative of other ocular pathologies, such as diabetic macular edema. The ERM diagnosis is usually performed by visually inspecting Optical Coherence Tomography (OCT) images, a manual process which is tiresome and prone to subjectivity. In this work, we present a methodology for the automatic segmentation and visualisation of the ERM in OCT volumes using deep learning. By employing a Densely Connected Convolutional Network, every pixel in the ILM can be classified into either healthy or pathological. Thus, a segmentation of the region susceptible to ERM appearance can be produced. This methodology also produces an intuitive colour map representation of the ERM presence over a visualisation of the eye fundus created from the OCT volume. In a series of representative experiments conducted to evaluate this methodology, it achieved a Dice score of and a Jaccard index of . The results that were obtained demonstrate the competitive performance of the proposed methodology when compared to other works in the state of the art.
1. Introduction
The Epiretinal Membrane (ERM) is an ocular disease that consists of scar tissue that is formed over the boundary between the retina and the vitreous body of the eye, an area known as the Inner Limiting Membrane (ILM). As the ERM appears over the retina, it may start to contract, exerting a traction and producing puckers or wrinkles over the underlying tissue. This may cause vision blurring, distortion, and metamorphopsia.
Several authors have approached the automatic detection of the ERM using Optical Coherence Tomography (OCT) images. In References [1,2], the authors identify the ILM layer using classical machine learning algorithms and local luminosity patterns; Lo et al. [3] proposes the use of a Residual Neural Network for the screening of ERM in cross-sectional OCT images, while, in Sonobe et al. [4], the authors compare the use of classical machine learning algorithms and deep learning for the detection of ERM, with deep learning outperforming the classical methods. These works, however, deal only with the screening of ERM, a simpler problem than its precise segmentation. In this regard, only Baamonde et al. [5,6] has approached the problem of segmenting the ERM in OCT images, using classical machine learning methods in this case.
In this work, we present an automatic methodology for the segmentation of the ERM in OCT volumes by using deep learning. This methodology consists of three phases, corresponding to the detection of the region of interest, the segmentation of the ERM via the classification of window samples of the ILM, and the visualisation and post-processing of the segmentation map [7]. This process produces a representation of the ERM presence and absence over the eye fundus in the form of a 2D colour map. This map can be used to aid clinicians in the detection and posterior removal of the ERM via pars plana vitrectomy.
2. Methodology
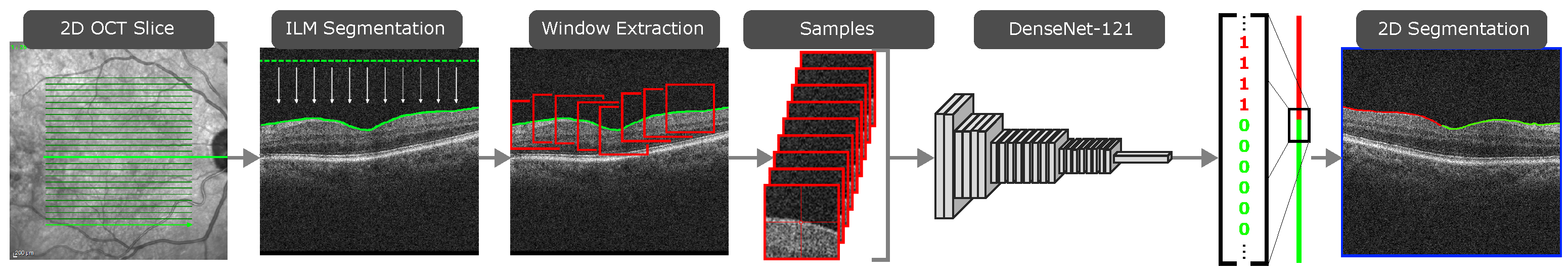
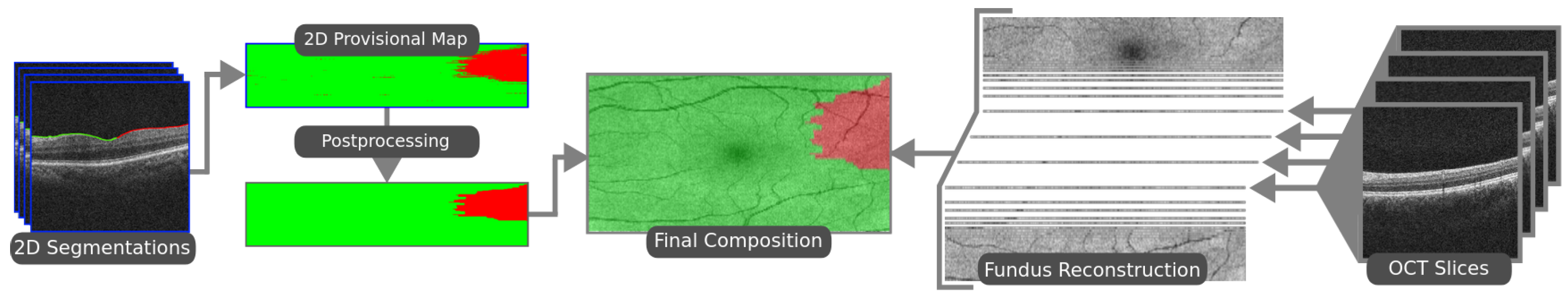
The ERM segmentation methodology consists of three steps. In the first one, the ILM is segmented using active contours [8]. The position of the ILM is modeled as a height value for every image column, since the retina appears as an irregular horizontal line in OCT images. The active contour models are allowed to contract downwards until they converge over the ILM. With the region of interest segmented, the ILM can be sampled via sliding window. The next step consists of sample classification. A Densely Connected Convolutional Neural Network [9] is then used to classify each of the pixel sliding window samples into either healthy or pathological. These classes are then assigned to the central pixel around which each sliding window was extracted, effectively producing a segmentation of each OCT scan. This process is illustrated in Figure 1. In the final step, all the slice segmentations are combined to produce a 2D preliminary segmentation map of the whole eye fundus. A post-processing step is then applied to this map in order to soften the boundary of the ERM and eliminate some misclassifications caused by image artifacts. This post-processed map is then overlaid on a reconstruction of the eye fundus to produce an intuitive visualisation of the ERM presence over the eye tissue, as illustrated in Figure 2.
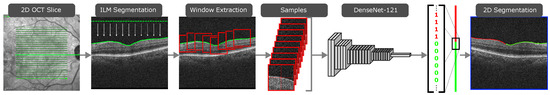
Figure 1.
The first two steps of the proposed methodology, producing a 2D segmentation of the ERM in a single OCT slice.
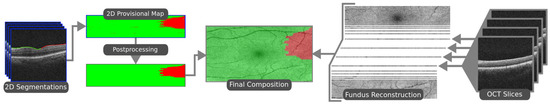
Figure 2.
The final steps of the proposed methodology. Slice segmentations are stacked to form a map. This map is post-processed and overlaid on a reconstruction of the eye fundus.
3. Results and Conclusions
The proposed methodology was evaluated in terms of its ability to correctly segment the ERM, with preliminary maps achieving a Dice score of and a Jaccard index of . When applying the post-processing stage, these results were improved up to and . A comparison between the proposed methodology and the state-of-the-art proposal [6], which only takes into account ERM-positive eyes, can be found in Table 1. These results show that the proposed system is able to accurately segment the ERM, even surpassing the current state of the art before the application of the post-processing stage.

Table 1.
Comparison of Dice and Jaccard indexes of the previous work and the deep learning method proposed in this work for the segmentation and post-processing stages.
Author Contributions
Author Contributions: Conceptualization, J.N., J.d.M., and M.O.; methodology, M.G.; software, M.G.; validation, M.G.; formal analysis, M.G.; investigation, M.G.; resources, J.N., J.d.M., P.C., and M.O.; data curation, P.C. and M.G-; writing—original draft preparation, M.G.; writing—review and editing, J.N., J.d.M., and M.O.; visualization, M.G.; supervision, J.N., J.d.M. and M.O.; project administration, J.N., J.d.M., and M.O.; funding acquisition, J.N., J.d.M., and M.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Instituto de Salud Carlos III, Government of Spain, DTS18/00136 research project; Ministerio de Ciencia e Innovación y Universidades, Government of Spain, RTI2018-095894-B-I00 research project; Ministerio de Ciencia e Innovación, Government of Spain through the research project with reference PID2019-108435RB-I00; Consellería de Cultura, Educación e Universidade, Xunta de Galicia through the predoctoral and postdoctoral grant contracts ref. ED481A 2021/161 and ED481B 2021/059, respectively; and Grupos de Referencia Competitiva, grant ref. ED431C 2020/24; Axencia Galega de Innovación (GAIN), Xunta de Galicia, grant ref. IN845D 2020/38; CITIC, Centro de Investigación de Galicia ref. ED431G 2019/01, receives financial support from Consellería de Educación, Universidade e Formación Profesional, Xunta de Galicia, through the ERDF (80%) and Secretaría Xeral de Universidades (20%).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the local Ethics Committee of Investigation from A Coruña/Ferrol (2014/437) the 24 November 2014.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Baamonde, S.; de Moura, J.; Novo, J.; Rouco, J.; Ortega, M. Feature Definition and Selection for Epiretinal Membrane Characterization in Optical Coherence Tomography Images. In Image Analysis and Processing—ICIAP 2017; Springer International Publishing: Basel, Switzerland, 2017; pp. 456–466. [Google Scholar] [CrossRef]
- Baamonde, S.; de Moura, J.; Novo, J.; Ortega, M. Automatic Detection of Epiretinal Membrane in OCT Images by Means of Local Luminosity Patterns. In Advances in Computational Intelligence; Springer International Publishing: Basel, Switzerland, 2017; pp. 222–235. [Google Scholar] [CrossRef]
- Lo, Y.C.; Lin, K.H.; Bair, H.; Sheu, W.H.H.; Chang, C.S.; Shen, Y.C.; Hung, C.L. Epiretinal Membrane Detection at the Ophthalmologist Level using Deep Learning of Optical Coherence Tomography. Sci. Rep. 2020, 10, 8424. [Google Scholar] [CrossRef] [PubMed]
- Sonobe, T.; Tabuchi, H.; Ohsugi, H.; Masumoto, H.; Ishitobi, N.; Morita, S.; Enno, H.; Nagasato, D. Comparison between support vector machine and deep learning, machine-learning technologies for detecting epiretinal membrane using 3D-OCT. Int. Ophthalmol. 2018, 39, 1871–1877. [Google Scholar] [CrossRef] [PubMed]
- Baamonde, S.; de Moura, J.; Novo, J.; Charlón, P.; Ortega, M. Automatic identification and characterization of the epiretinal membrane in OCT images. Biomed. Opt. Express 2019, 10, 4018. [Google Scholar] [CrossRef] [PubMed]
- Baamonde, S.; de Moura, J.; Novo, J.; Charlón, P.; Ortega, M. Automatic Identification and Intuitive Map Representation of the Epiretinal Membrane Presence in 3D OCT Volumes. Sensors 2019, 19, 5269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gende, M.; De Moura, J.; Novo, J.; Charlón, P.; Ortega, M. Automatic Segmentation and Intuitive Visualisation of the Epiretinal Membrane in 3D OCT Images Using Deep Convolutional Approaches. IEEE Access 2021, 9, 75993–76004. [Google Scholar] [CrossRef]
- Gawlik, K.; Hausser, F.; Paul, F.; Brandt, A.U.; Kadas, E.M. Active contour method for ILM segmentation in ONH volume scans in retinal OCT. Biomed. Opt. Express 2018, 9, 6497–6518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, G.; Liu, Z.; Maaten, L.V.D.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).