Assessing Curcumin Uptake and Clearance and Their Influence on Superoxide Dismutase Activity in Drosophila melanogaster
Abstract
:1. Introduction
2. Materials and Methods
2.1. Drosophila Husbandry and Curcuminoid Treatments
2.2. Larval Preparation, Extraction, and HPLC Analysis
2.3. Larval Preparation and SOD Assay
2.4. Analyses
3. Results
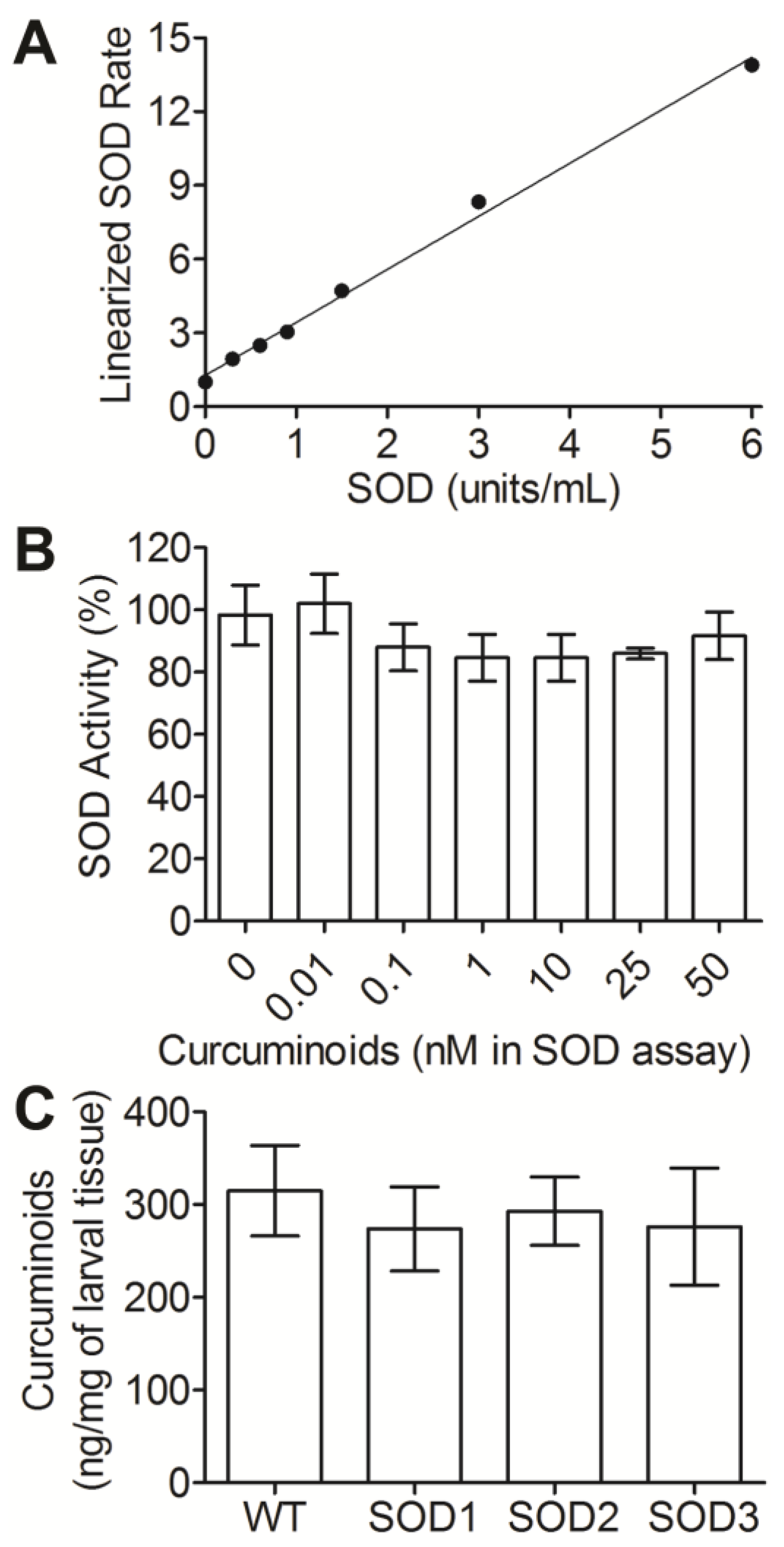
3.1. Assessing Uptake and Clearance Parameters of Curcuminoids in Drosophila Larvae
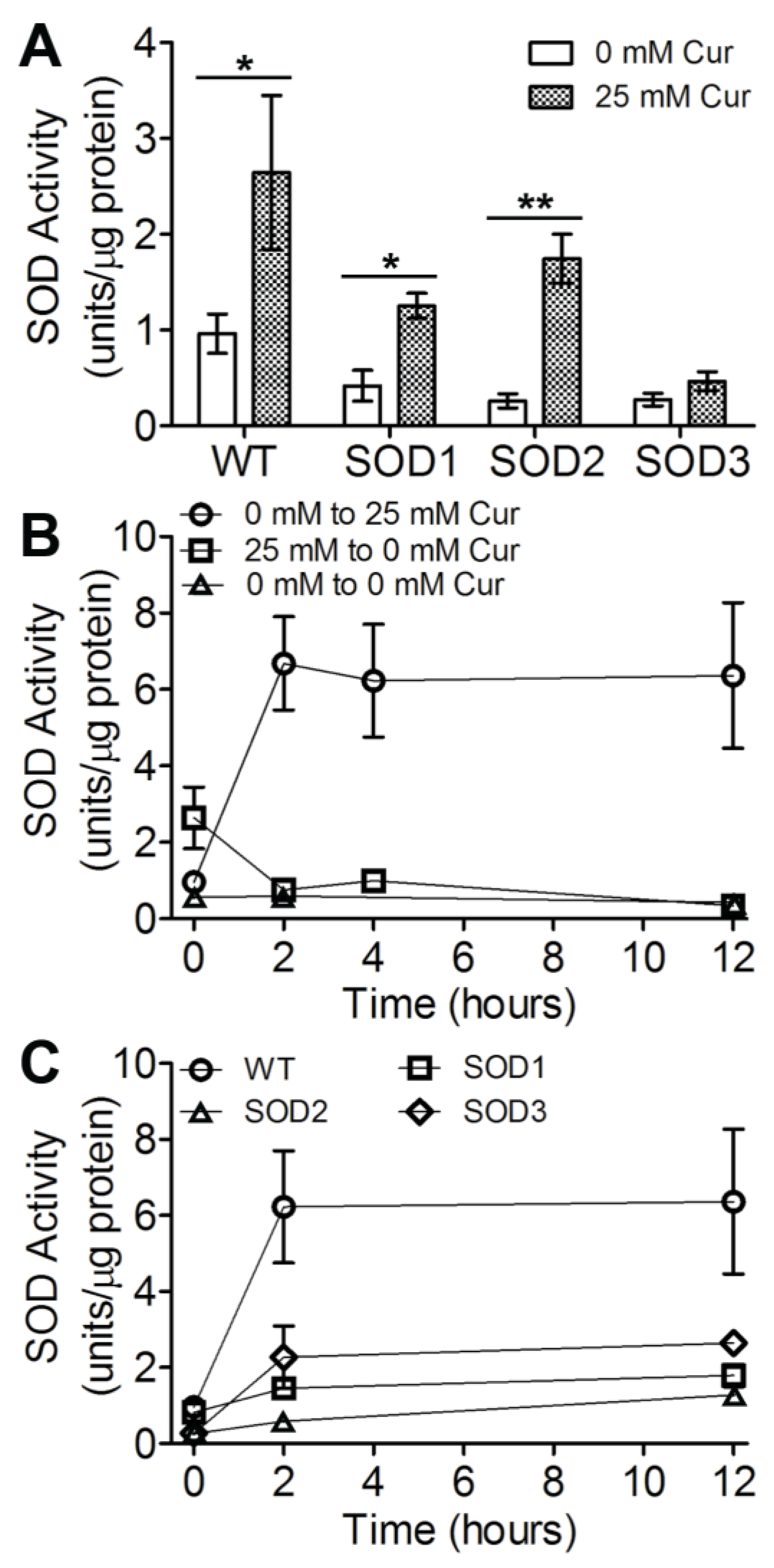
3.2. SOD Levels Are Modulated in Response to Curcuminoid Treatment
3.3. Three SOD Genes Differentially Contribute to Curcuminoid-Induced Activity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Forman, H.J.; Zhang, H. Targeting oxidative stress in disease: Promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021, 20, 689–709. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; St Clair, D.K. Regulation of superoxide dismutase genes: Implications in disease. Free Radic. Biol. Med. 2009, 47, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Vuu, M.D.; Huynh, M.A.; Yamaguchi, M.; Tran, L.T.; Dang, T.P.T. Curcumin effectively rescued Parkinson’s disease-like phenotypes in a novel Drosophila melanogaster model with dUCH knockdown. Oxid. Med. Cell. Longev. 2018, 2018, 2038267. [Google Scholar] [CrossRef] [PubMed]
- Jaroonwitchawan, T.; Chaicharoenaudomrung, N.; Namkaew, J.; Noisa, P. Curcumin attenuates paraquat-induced cell death in human neuroblastoma cells through modulating oxidative stress and autophagy. Neurosci. Lett. 2017, 636, 40–47. [Google Scholar] [CrossRef]
- Han, J.; Pan, X.-Y.; Xu, Y.; Xiao, Y.; An, Y.; Tie, L.; Pan, Y.; Li, X.-J. Curcumin induces autophagy to protect vascular endothelial cell survival from oxidative stress damage. Autophagy 2012, 8, 812–825. [Google Scholar] [CrossRef]
- Xu, X.-Y.; Meng, X.; Li, S.; Gan, R.-Y.; Li, Y.; Li, H.-B. Bioactivity, health benefits, and related molecular mechanisms of curcumin: Current progress, challenges, and perspectives. Nutrients 2018, 10, 1553. [Google Scholar] [CrossRef]
- Lin, X.; Bai, D.; Wei, Z.; Zhang, Y.; Huang, Y.; Deng, H.; Huang, X. Curcumin attenuates oxidative stress in RAW264.7 cells by increasing the activity of antioxidant enzymes and activating the Nrf2-Keap1 pathway. PLoS ONE 2019, 14, e0216711. [Google Scholar] [CrossRef]
- Di Tu, Q.; Jin, J.; Hu, X.; Ren, Y.; Zhao, L.; He, Q. Curcumin improves the renal autophagy in rat experimental membranous nephropathy via regulating the PI3K/AKT/mTOR and Nrf2/HO-1 signaling pathways. Biomed. Res. Int. 2020, 2020, 7069052. [Google Scholar] [CrossRef]
- Shen, L.-R.; Xiao, F.; Yuan, P.; Chen, Y.; Gao, Q.-K.; Parnell, L.D.; Meydani, M.; Ordovas, J.M.; Li, D.; Lai, C.-Q. Curcumin-supplemented diets increase superoxide dismutase activity and mean lifespan in Drosophila. AGE 2013, 35, 1133–1142. [Google Scholar] [CrossRef]
- Huang, L.; Chen, C.; Zhang, X.; Li, X.; Chen, Z.; Yang, C.; Liang, X.; Zhu, G.; Xu, Z. Neuroprotective effect of curcumin against cerebral ischemia-reperfusion via mediating autophagy and inflammation. J. Mol. Neurosci. 2018, 64, 129–139. [Google Scholar] [CrossRef]
- Berdyński, M.; Miszta, P.; Safranow, K.; Andersen, P.M.; Morita, M.; Filipek, S.; Żekanowski, C.; Kuźma-Kozakiewicz, M. SOD1 mutations associated with amyotrophic lateral sclerosis analysis of variant severity. Sci. Rep. 2022, 12, 103. [Google Scholar] [CrossRef] [PubMed]
- Zelko, I.N.; Mariani, T.J.; Folz, R.J. Superoxide dismutase multigene family: A comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 2002, 33, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Fukai, T.; Ushio-Fukai, M. Superoxide dismutases: Role in redox signaling, vascular function, and diseases. Antioxid. Redox Signal. 2011, 15, 1583–1606. [Google Scholar] [CrossRef] [PubMed]
- Zarei, S.; Carr, K.; Reiley, L.; Diaz, K.; Guerra, O.; Altamirano, P.F.; Pagani, W.; Lodin, D.; Orozco, G.; Chinea, A. A comprehensive review of amyotrophic lateral sclerosis. Surg. Neurol. Int. 2015, 6, 171. [Google Scholar] [CrossRef]
- Valentine, J.S.; Hart, P.J. Misfolded CuZnSOD and amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 2003, 100, 3617–3622. [Google Scholar] [CrossRef]
- Dhar, S.K.; Tangpong, J.; Chaiswing, L.; Oberley, T.D.; St Clair, D.K. Manganese superoxide dismutase is a p53-regulated gene that switches cancers between early and advanced stages. Cancer Res. 2011, 71, 6684–6695. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, H.; Xu, Y.; Ding, Y.; Chen, X.; Mei, Z.; Ding, H.; Jie, Z. Association among genetic polymorphisms of GSTP1, HO-1, and SOD-3 and chronic obstructive pulmonary disease susceptibility. Int. J. Chron. Obs. Pulmon. Dis. 2019, 14, 2081–2088. [Google Scholar] [CrossRef]
- Kitada, M.; Xu, J.; Ogura, Y.; Monno, I.; Koya, D. Manganese superoxide dismutase dysfunction and the pathogenesis of kidney disease. Front. Physiol. 2020, 11, 755. [Google Scholar] [CrossRef]
- Sundararajan, V.; Venkatasubbu, G.D.; Sheik Mohideen, S. Investigation of therapeutic potential of cerium oxide nanoparticles in Alzheimer’s disease using transgenic Drosophila. 3 Biotech. 2021, 11, 159. [Google Scholar] [CrossRef]
- Layalle, S.; They, L.; Ourghani, S.; Raoul, C.; Soustelle, L. Amyotrophic Lateral Sclerosis Genes in Drosophila melanogaster. Int. J. Mol. Sci. 2021, 22, 904. [Google Scholar] [CrossRef]
- Surguchov, A. Invertebrate models untangle the mechanism of neurodegeneration in Parkinson’s disease. Cells 2021, 10, 407. [Google Scholar] [CrossRef] [PubMed]
- Akinyemi, A.J.; Oboh, G.; Ogunsuyi, O.; Abolaji, A.O.; Udofia, A. Curcumin-supplemented diets improve antioxidant enzymes and alter acetylcholinesterase genes expression level in Drosophila melanogaster model. Metab. Brain Dis. 2018, 33, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, X.; Jiang, C.; Liu, L.; Ordovas, J.M.; Lai, C.-Q.; Shen, L. Curcumin supplementation increases survival and lifespan in Drosophila under heat stress conditions. BioFactors 2018, 44, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Suckow, B.K.; Suckow, M.A. Lifespan extension by the antioxidant curcumin in Drosophila melanogaster. Int. J. Biomed. Sci. 2006, 2, 402–405. [Google Scholar]
- Seong, K.M.; Yu, M.; Lee, K.S.; Park, S.; Jin, Y.W.; Min, K.J. Curcumin mitigates accelerated aging after irradiation in Drosophila by reducing oxidative stress. Biomed. Res. Int. 2015, 2015, 425380. [Google Scholar] [CrossRef]
- Daverey, A.; Agrawal, S.K. Curcumin alleviates oxidative stress and mitochondrial dysfunction in astrocytes. Neuroscience 2016, 333, 92–103. [Google Scholar] [CrossRef]
- Abolaji, A.O.; Fasae, K.D.; Iwezor, C.E.; Aschner, M.; Farombi, E.O. Curcumin attenuates copper-induced oxidative stress and neurotoxicity in Drosophila melanogaster. Toxicol. Rep. 2020, 7, 261–268. [Google Scholar] [CrossRef]
- Esquivel, A.R.; Douglas, J.C.; Loughran, R.M.; Rezendes, T.E.; Reed, K.R.; Cains, T.H.L.; Emsley, S.A.; Paddock, W.A.; Videau, P.; Koyack, M.J.; et al. Assessing the influence of curcumin in sex-specific oxidative stress, survival and behavior in Drosophila melanogaster. J. Exp. Biol. 2020, 223, jeb223867. [Google Scholar] [CrossRef]
- Blackney, M.J.; Cox, R.; Shepherd, D.; Parker, J.D. Cloning and expression analysis of Drosophila extracellular Cu Zn superoxide dismutase. Biosci. Rep. 2014, 34, e00164. [Google Scholar] [CrossRef]
- Phillips, J.P.; Campbell, S.D.; Michaud, D.; Charbonneau, M.; Hilliker, A.J. Null mutation of copper/zinc superoxide dismutase in Drosophila confers hypersensitivity to paraquat and reduced longevity. Proc. Natl. Acad. Sci. USA 1989, 86, 2761–2765. [Google Scholar] [CrossRef]
- Celotto, A.M.; Liu, Z.; Vandemark, A.P.; Palladino, M.J. A novel Drosophila SOD2 mutant demonstrates a role for mitochondrial ROS in neurodevelopment and disease. Brain Behav. 2012, 2, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.-R.; Parnell, L.D.; Ordovas, J.M.; Lai, C.-Q. Curcumin and aging. BioFactors 2013, 39, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-S.; Lee, B.-S.; Semnani, S.; Avanesian, A.; Um, C.-Y.; Jeon, H.-J.; Seong, K.-M.; Yu, K.; Min, K.-J.; Jafari, M. Curcumin extends life span, improves health span, and modulates the expression of age-associated aging genes in Drosophila melanogaster. Rejuvenation Res. 2010, 13, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Parks, A.L.; Cook, K.R.; Belvin, M.; Dompe, N.A.; Fawcett, R.; Huppert, K.; Tan, L.R.; Winter, C.G.; Bogart, K.P.; Deal, J.E.; et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 2004, 36, 288–292. [Google Scholar] [CrossRef]
- Shishodia, S. Molecular mechanisms of curcumin action: Gene expression. BioFactors 2013, 39, 37–55. [Google Scholar] [CrossRef]
- Ming, J.; Ye, J.; Zhang, Y.; Xu, Q.; Yang, X.; Shao, X.; Qiang, J.; Xu, P. Optimal dietary curcumin improved growth performance, and modulated innate immunity, antioxidant capacity and related genes expression of NF-κB and Nrf2 signaling pathways in grass carp (Ctenopharyngodon idella) after infection with Aeromonas hydrophila. Fish Shellfish Immunol. 2020, 97, 540–553. [Google Scholar] [CrossRef]
- Shahcheraghi, S.H.; Salemi, F.; Peirovi, N.; Ayatollahi, J.; Alam, W.; Khan, H.; Saso, L. Nrf2 regulation by curcumin: Molecular aspects for therapeutic prospects. Molecules 2021, 27, 167. [Google Scholar] [CrossRef]
- Zhu, H.; Itoh, K.; Yamamoto, M.; Zweier, J.L.; Li, Y. Role of Nrf2 signaling in regulation of antioxidants and phase 2 enzymes in cardiac fibroblasts: Protection against reactive oxygen and nitrogen species-induced cell injury. FEBS Lett. 2005, 579, 3029–3036. [Google Scholar] [CrossRef]
- Ma, Q.; He, X. Molecular basis of electrophilic and oxidative defense: Promises and perils of Nrf2. Pharmacol. Rev. 2012, 64, 1055–1081. [Google Scholar] [CrossRef]
- Signor, S.; Nuzhdin, S. Dynamic changes in gene expression and alternative splicing mediate the response to acute alcohol exposure in Drosophila melanogaster. Heredity 2018, 121, 342–360. [Google Scholar] [CrossRef]
- McGraw, L.A.; Clark, A.G.; Wolfner, M.F. Post-mating gene expression profiles of female Drosophila melanogaster in response to time and to four male accessory gland proteins. Genetics 2008, 179, 1395–1408. [Google Scholar] [CrossRef] [PubMed]
- Telonis-Scott, M.; van Heerwaarden, B.; Johnson, T.K.; Hoffmann, A.A.; Sgrò, C.M. New levels of transcriptome complexity at upper thermal limits in wild Drosophila revealed by exon expression analysis. Genetics 2013, 195, 809–830. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoffman, T.R.; Emsley, S.A.; Douglas, J.C.; Reed, K.R.; Esquivel, A.R.; Koyack, M.J.; Paddock, B.E.; Videau, P. Assessing Curcumin Uptake and Clearance and Their Influence on Superoxide Dismutase Activity in Drosophila melanogaster. BioTech 2023, 12, 58. https://doi.org/10.3390/biotech12030058
Hoffman TR, Emsley SA, Douglas JC, Reed KR, Esquivel AR, Koyack MJ, Paddock BE, Videau P. Assessing Curcumin Uptake and Clearance and Their Influence on Superoxide Dismutase Activity in Drosophila melanogaster. BioTech. 2023; 12(3):58. https://doi.org/10.3390/biotech12030058
Chicago/Turabian StyleHoffman, Tammy R., Sarah A. Emsley, Jenna C. Douglas, Kaela R. Reed, Abigail R. Esquivel, Marc J. Koyack, Brie E. Paddock, and Patrick Videau. 2023. "Assessing Curcumin Uptake and Clearance and Their Influence on Superoxide Dismutase Activity in Drosophila melanogaster" BioTech 12, no. 3: 58. https://doi.org/10.3390/biotech12030058
APA StyleHoffman, T. R., Emsley, S. A., Douglas, J. C., Reed, K. R., Esquivel, A. R., Koyack, M. J., Paddock, B. E., & Videau, P. (2023). Assessing Curcumin Uptake and Clearance and Their Influence on Superoxide Dismutase Activity in Drosophila melanogaster. BioTech, 12(3), 58. https://doi.org/10.3390/biotech12030058





