Deep Learning Model for COVID-19-Infected Pneumonia Diagnosis Using Chest Radiography Images
Abstract
:1. Introduction
2. Related Literature
3. Proposed Methodology
3.1. Data Preprocessing
3.2. Transfer Learning
3.3. Parallel Computing
4. Experimental Environment
4.1. Performance Evaluation Metrics
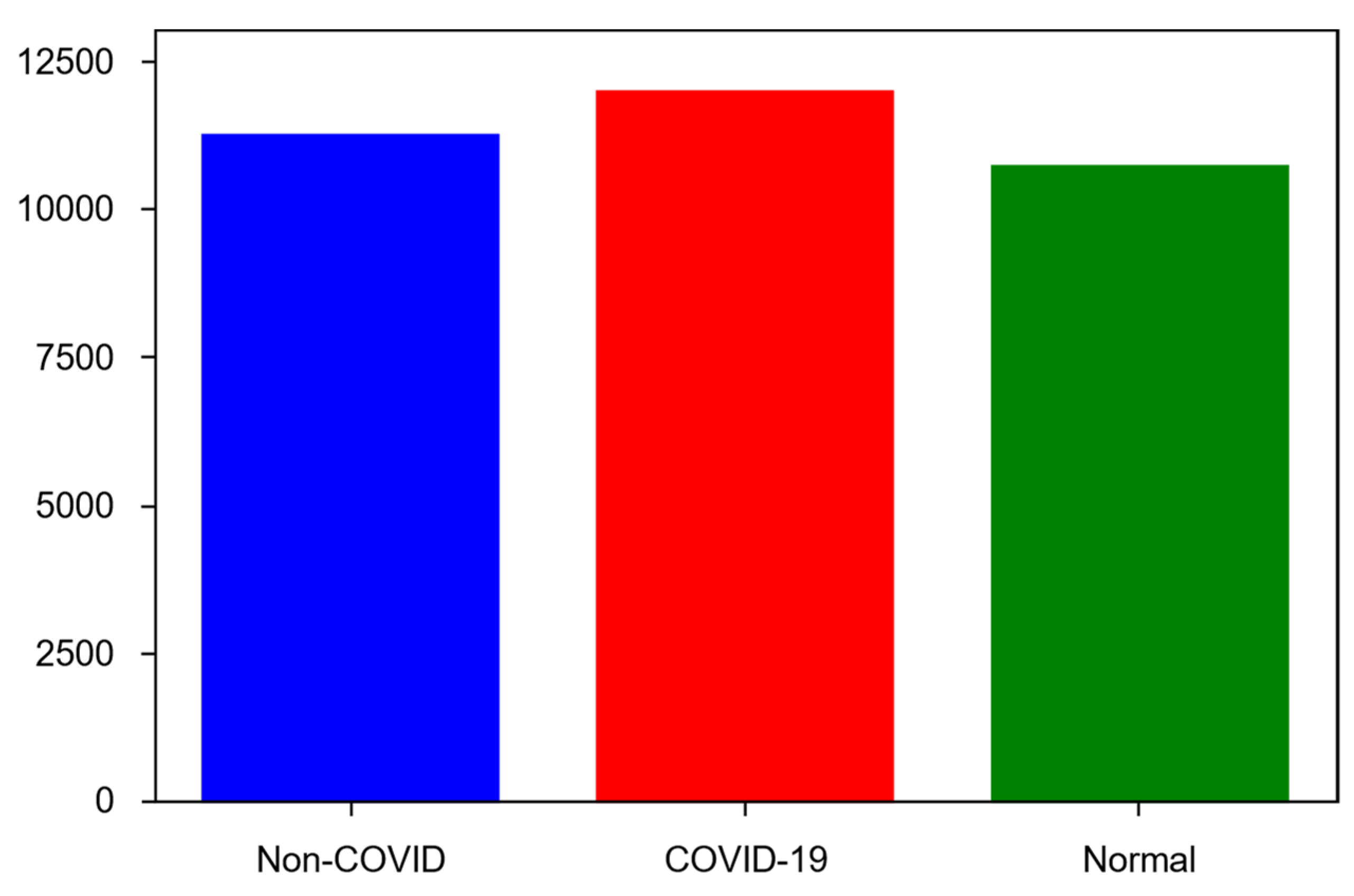
4.2. Dataset
- COVID-19—11,956 samples;
- Non-COVID infections (viral or bacterial pneumonia)—11,263 samples;
- Normal (uninfected)—10,701 samples.
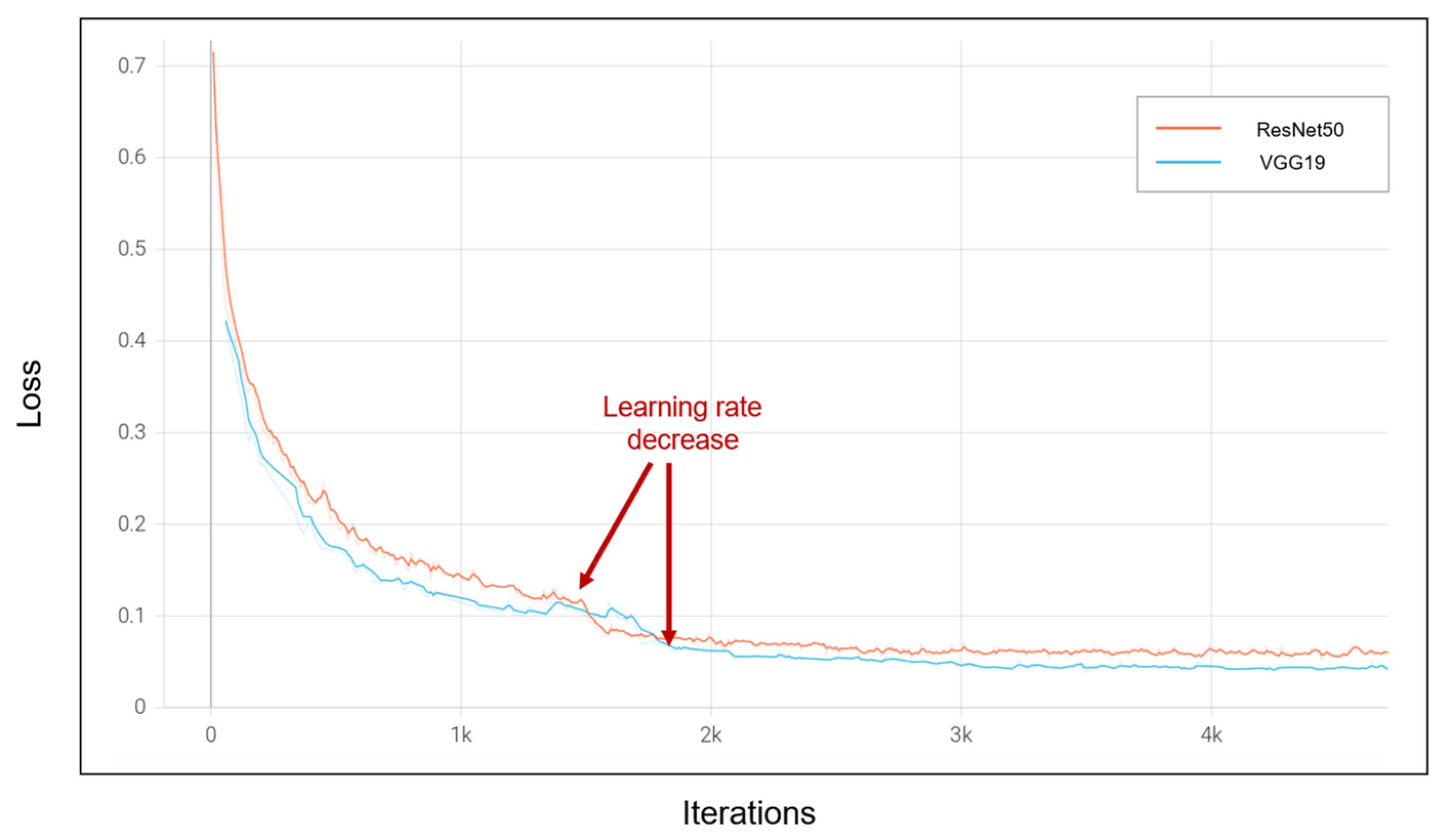
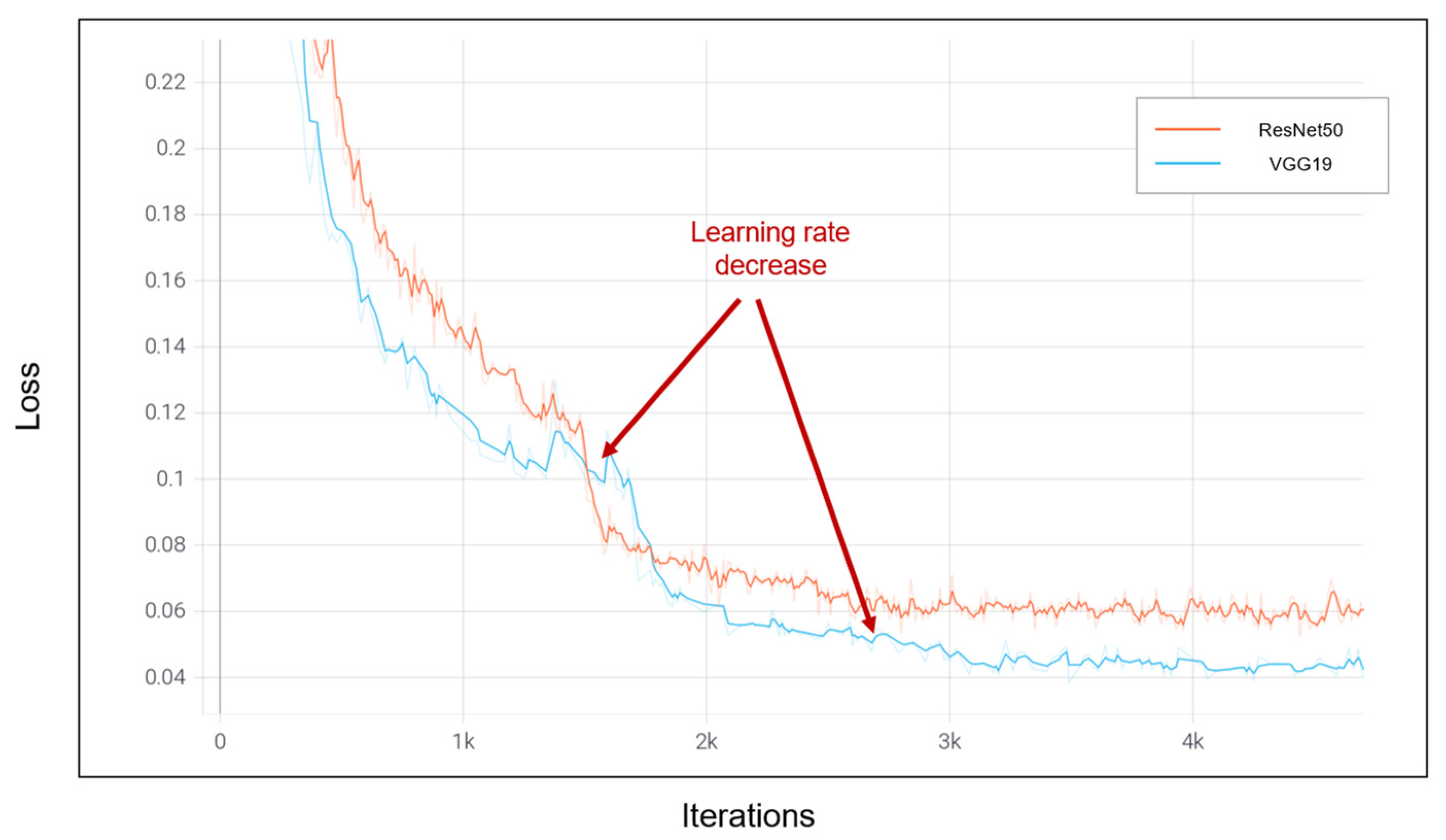
4.3. Numerical Results
5. Comparison and Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- COVID-19 Worldwide Statistics. Available online: https://www.worldometers.info/coronavirus/ (accessed on 14 November 2022).
- Coronavirus and Pneumonia. Available online: https://www.webmd.com/lung/covid-and-pneumonia#1 (accessed on 14 November 2022).
- Gray, D.; Willemse, L.; Visagie, A.; Czövek, D.; Nduru, P.; Vanker, A.; Zar, H.J. Determinants of early-life lung function in African infants. Thorax 2017, 72, 445–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, J.Y.; Stern, D.A.; Guerra, S.; Wright, A.L.; Morgan, W.J.; Martinez, F.D. Pneumonia in childhood and impaired lung function in adults: A longitudinal study. Pediatrics 2015, 135, 607–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cecilie, S.; Sunyer, J.; Plana, E.; Dharmage, S.; Heinrich, J.; Jarvis, D.; de Marco, R. Early life origins of chronic obstructive pulmonary disease. Thorax 2010, 65, 14–20. [Google Scholar]
- Zar, H.J.; Andronikou, S.; Nicol, M.P. Advances in the diagnosis of pneumonia in children. BMJ 2017, 358, j2739. [Google Scholar] [CrossRef]
- Iuri, D.; De Candia, A.; Bazzocchi, M. Evaluation of the lung in children with suspected pneumonia: Usefulness of ultrasonography. La Radiol. Med. 2009, 114, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Tomà, P.; Owens, C.M. Chest ultrasound in children: Critical appraisal. Pediatr. Radiol. 2013, 43, 1427–1434. [Google Scholar] [CrossRef]
- Shah, V.P.; Tunik, M.G.; Tsung, J.W. Prospective evaluation of point-of-care ultrasonography for the diagnosis of pneumonia in children and young adults. JAMA Pediatr. 2013, 167, 119–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorycki, T.; Lasek, I.; Kamiński, K.; Studniarek, M. Evaluation of radiation doses delivered in different chest CT protocols. Pol. J. Radiol. 2014, 79, 1. [Google Scholar]
- Sodhi, K.S.; Khandelwal, N.; Saxena, A.K.; Singh, M.; Agarwal, R.; Bhatia, A.; Lee, E.Y. Rapid lung MRI in children with pulmonary infections: Time to change our diagnostic algorithms. J. Magn. Reson. Imaging 2016, 43, 1196–1206. [Google Scholar] [CrossRef]
- Biederer, J.; Mirsadraee, S.; Beer, M.; Molinari, F.; Hintze, C.; Bauman, G.; Puderbach, M. MRI of the lung (3/3)—Current applications and future perspectives. Insights Imaging 2012, 3, 373–386. [Google Scholar] [CrossRef] [Green Version]
- Hirsch, W.; Sorge, I.; Krohmer, S.; Weber, D.; Meier, K.; Till, H. MRI of the lungs in children. Eur. J. Radiol. 2008, 68, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Boiselle, P.M.; Biederer, J.; Gefter, W.B.; Lee, E.Y. Expert opinion: Why is MRI still an under-utilized modality for evaluating thoracic disorders? J. Thorac. Imaging 2013, 28, 137. [Google Scholar] [CrossRef] [PubMed]
- Aboutalib, S.S.; Mohamed, A.A.; Berg, W.A.; Zuley, M.L.; Sumkin, J.H.; Wu, S. Deep learning to distinguish recalled but benign mammography images in breast cancer screening. Clin. Cancer Res. 2018, 24, 5902–5909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.K.; Kim, H.E.; Han, K.; Kang, B.J.; Sohn, Y.M.; Woo, O.H.; Lee, C.W. Applying data-driven imaging biomarker in mammography for breast cancer screening: Preliminary study. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Shariaty, F.; Mousavi, M. Application of CAD systems for the automatic detection of lung nodules. Inform. Med. Unlocked 2019, 15, 100173. [Google Scholar] [CrossRef]
- Gu, Y.; Chi, J.; Liu, J.; Yang, L.; Zhang, B.; Yu, D.; Lu, X. A survey of computer-aided diagnosis of lung nodules from CT scans using deep learning. Comput. Biol. Med. 2021, 137, 104806. [Google Scholar] [CrossRef]
- Balyen, L.; Peto, T. Promising artificial intelligence-machine learning-deep learning algorithms in ophthalmology. Asia-Pac. J. Ophthalmol. 2019, 8, 264–272. [Google Scholar]
- Kumar, A.; Fulham, M.; Feng, D.; Kim, J. Co-learning feature fusion maps from PET-CT images of lung cancer. IEEE Trans. Med. Imaging 2019, 39, 204–217. [Google Scholar] [CrossRef] [Green Version]
- Podnar, S.; Kukar, M.; Gunčar, G.; Notar, M.; Gošnjak, N.; Notar, M. Diagnosing brain tumours by routine blood tests using machine learning. Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Ibrokhimov, B.; Kang, J.Y. Two-Stage Deep Learning Method for Breast Cancer Detection Using High-Resolution Mammogram Images. Appl. Sci. 2022, 12, 4616. [Google Scholar] [CrossRef]
- Roy, S.; Meena, T.; Lim, S.J. Demystifying Supervised Learning in Healthcare 4.0: A New Reality of Transforming Diagnostic Medicine. Diagnostics 2022, 12, 2549. [Google Scholar] [CrossRef] [PubMed]
- Meena, T.; Roy, S. Bone fracture detection using deep supervised learning from radiological images: A paradigm shift. Diagnostics 2022, 12, 2420. [Google Scholar] [CrossRef]
- Pal, D.; Reddy, P.B.; Roy, S. Attention UW-Net: A fully connected model for automatic segmentation and annotation of chest X-ray. Comput. Biol. Med. 2022, 150, 106083. [Google Scholar] [CrossRef]
- Gunjan, V.K.; Singh, N.; Shaik, F.; Roy, S. Detection of lung cancer in CT scans using grey wolf optimization algorithm and recurrent neural network. Health Technol. 2022, 12, 1197–1210. [Google Scholar] [CrossRef]
- Gangopadhyay, T.; Halder, S.; Dasgupta, P.; Chatterjee, K.; Ganguly, D.; Sarkar, S.; Roy, S. MTSE U-Net: An architecture for segmentation, and prediction of fetal brain and gestational age from MRI of brain. Netw. Model. Anal. Health Inform. Bioinform. 2022, 11, 1–14. [Google Scholar] [CrossRef]
- Tomaszewski, M.R.; Gillies, R.J. The biological meaning of radiomic features. Radiology 2021, 298, 505–516. [Google Scholar] [CrossRef]
- Mediouni, M.; Madiouni, R.; Gardner, M.; Vaughan, N. Translational medicine: Challenges and new orthopaedic vision (Mediouni-Model). Curr. Orthop. Pract. 2020, 31, 196–200. [Google Scholar] [CrossRef]
- Mediouni, M.; RSchlatterer, D.; Madry, H.; Cucchiarini, M.; Rai, B. A review of translational medicine. The future paradigm: How can we connect the orthopedic dots better? Curr. Med. Res. Opin. 2018, 34, 1217–1229. [Google Scholar] [CrossRef]
- Rodriguez-Ruiz, A.; Lång, K.; Gubern-Merida, A.; Broeders, M.; Gennaro, G.; Clauser, P.; Sechopoulos, I. Stand-alone artificial intelligence for breast cancer detection in mammography: Comparison with 101 radiologists. JNCI J. Natl. Cancer Inst. 2019, 111, 916–922. [Google Scholar] [CrossRef]
- Shen, L.; Margolies, L.R.; Rothstein, J.H.; Fluder, E.; McBride, R.; Sieh, W. Deep learning to improve breast cancer detection on screening mammography. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Latif, S.; Usman, M.; Manzoor, S.; Iqbal, W.; Qadir, J.; Tyson, G.; Crowcroft, J. Leveraging data science to combat COVID-19: A comprehensive review. IEEE Trans. Artif. Intell. 2020, 1, 85–103. [Google Scholar] [CrossRef]
- Swapnarekha, H.; Behera, H.S.; Nayak, J.; Naik, B. Role of intelligent computing in COVID-19 prognosis: A state-of-the-art review. Chaos Solitons Fractals 2020, 138, 109947. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.U.; Aslam, N. A deep-learning-based framework for automated diagnosis of COVID-19 using X-ray images. Information 2020, 11, 419. [Google Scholar] [CrossRef]
- Brima, Y.; Atemkeng, M.; Tankio Djiokap, S.; Ebiele, J.; Tchakounté, F. Transfer Learning for the Detection and Diagnosis of Types of Pneumonia including Pneumonia Induced by COVID-19 from Chest X-ray Images. Diagnostics 2021, 11, 1480. [Google Scholar] [CrossRef]
- Narin, A.; Kaya, C.; Pamuk, Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. Pattern Anal. Appl. 2021, 24, 1207–1220. [Google Scholar] [CrossRef]
- Minaee, S.; Kafieh, R.; Sonka, M.; Yazdani, S.; Soufi, G.J. Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Med. Image Anal. 2020, 65, 101794. [Google Scholar] [CrossRef]
- Hemdan EE, D.; Shouman, M.A.; Karar, M.E. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv 2020, arXiv:2003.11055. [Google Scholar]
- Sethy, P.K.; Behera, S.K. Detection of Coronavirus Disease (COVID-19) Based on Deep Features; Preprints 2020, 2020030300. Available online: https://www.preprints.org/manuscript/202003.0300/v1 (accessed on 13 November 2022).
- Oh, Y.; Park, S.; Ye, J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging 2020, 39, 2688–2700. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Mpesiana, T.A. Covid-19: Automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef] [Green Version]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Peng, Y.; Xu, H.; Cui, Z.; Williams, R.O. The COVID-19 vaccine race: Challenges and opportunities in vaccine formulation. AAPS PharmSciTech 2020, 21, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, B.N.; Hardie, R.C.; Krishnaraja, V.; Karam, C.; Davuluru VS, P. Transfer-to-transfer learning approach for computer aided detection of COVID-19 in chest radiographs. AI 2020, 1, 539–557. [Google Scholar] [CrossRef]
- Tahir, A.M.; Chowdhury, M.E.; Khandakar, A.; Rahman, T.; Qiblawey, Y.; Khurshid, U.; Hamid, T. COVID-19 infection localization and severity grading from chest X-ray images. Comput. Biol. Med. 2021, 139, 105002. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Zhao, L.; Zhou, Y.; Lu, H.; Fujita, H. Parallel computing method of deep belief networks and its application to traffic flow prediction. Knowl. -Based Syst. 2019, 163, 972–987. [Google Scholar] [CrossRef]











| Dataset | Normal | Non-COVID Infections | COVID-19 | Total |
|---|---|---|---|---|
| Train | 6849 | 7208 | 7658 | 21,715 |
| Validation | 1712 | 1802 | 1903 | 5417 |
| Test | 2140 | 2253 | 2395 | 6788 |
| Study | Dataset | Method | Accuracy |
|---|---|---|---|
| Oh et al. [42] | 191 Normal 74 Pneumonia 57 Tuberculosis 180 COVID-19 | ResNet18 | 88.9% |
| Ozturk et al. [44] | 1000 Normal 500 Pneumonia 125 COVID-19 | DarkCovidNet | 87% |
| Wang et al. [45] | 8066 Normal 5538 Pneumonia 358 COVID-19 | COVIDNet, VGG19, ResNet50 | 93.3% |
| Narayanan et al. [46] | 1583 Normal 1493 Viral Pneumonia 2780 Bacterial Pneumonia | ResNet50, Inceptionv3, Xception, DenseNet201 | 98% |
| Apostolopoulos et al. [43] | 504 Normal 714 Pneumonia 224 COVID-19 | VGG19, Inception, Xception, MobileNet | 98.75% |
| Brima et al. [37] | 10,192 Normal 1345 Pneumonia 6012 Lung opacity 3616 COVID-19 | VGG19, DenseNet121, ResNet50 | 94% |
| Khan et al. [36] | 802 Normal 790 COVID-19 | VGG16, VGG19 | 99.38% |
| Our method | 10,701 Normal 11,263 Pneumonia 11,956 COVID-19 | VGG19, ResNet50 | 96.6% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibrokhimov, B.; Kang, J.-Y. Deep Learning Model for COVID-19-Infected Pneumonia Diagnosis Using Chest Radiography Images. BioMedInformatics 2022, 2, 654-670. https://doi.org/10.3390/biomedinformatics2040043
Ibrokhimov B, Kang J-Y. Deep Learning Model for COVID-19-Infected Pneumonia Diagnosis Using Chest Radiography Images. BioMedInformatics. 2022; 2(4):654-670. https://doi.org/10.3390/biomedinformatics2040043
Chicago/Turabian StyleIbrokhimov, Bunyodbek, and Justin-Youngwook Kang. 2022. "Deep Learning Model for COVID-19-Infected Pneumonia Diagnosis Using Chest Radiography Images" BioMedInformatics 2, no. 4: 654-670. https://doi.org/10.3390/biomedinformatics2040043






