Identifying Potent Fat Mass and Obesity-Associated Protein Inhibitors Using Deep Learning-Based Hybrid Procedures
Abstract
1. Introduction
2. Materials and Methods
2.1. Target and Ligand Information
2.2. Virtual Screening Procedure
2.3. Molecular Dynamics Simulations
2.4. Binding Free Energy Calculation
3. Results
3.1. Virtual Screening Results
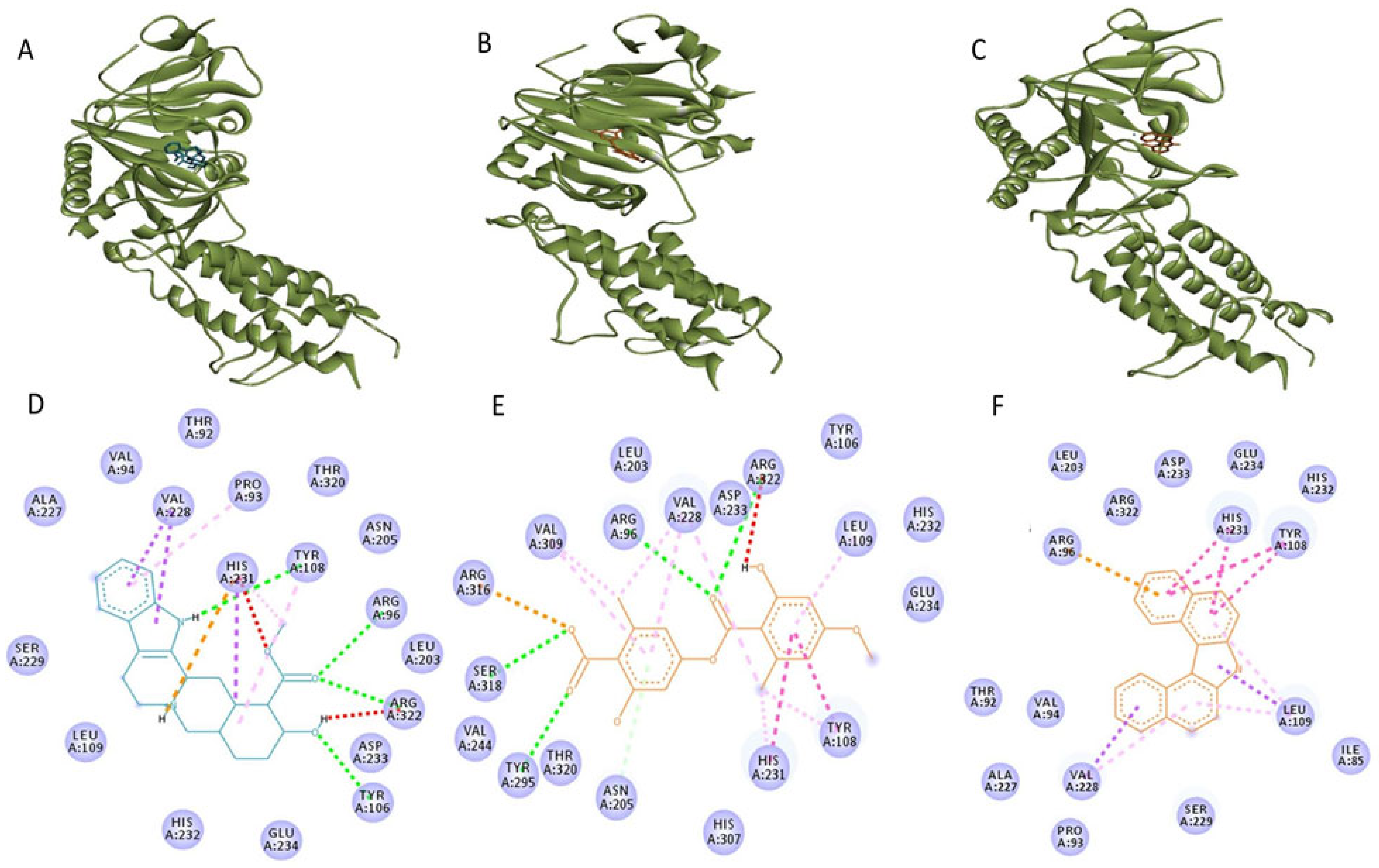
3.2. Molecular Dynamics Simulations
3.3. Binding Free Energy
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ramachandran, A.; Snehalatha, C.; Satyavani, K.; Sivasankari, S.; Vijay, V. Type 2 Diabetes in Asian-Indian Urban Children. Diabetes Care 2003, 26, 1022–1025. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, B.; Konje, J.C. The Epidemiology of Obesity in Reproduction. Best Pract. Res. Clin. Obstet. Gynaecol. 2023, 89, 102342. [Google Scholar] [CrossRef] [PubMed]
- Gross, D.C.; Cheever, C.R.; Batsis, J.A. Understanding the Development of Sarcopenic Obesity. Expert Rev. Endocrinol. Metab. 2023, 18, 469–488. [Google Scholar] [CrossRef] [PubMed]
- De Pergola, G.; Silvestris, F. Obesity as a Major Risk Factor for Cancer. J. Obes. 2013, 2013, 291546. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, K.; Nishiyama, H.; Kuriki, D.; Kawada, N.; Ochiya, T. Connecting the Dots in the Associations between Diet, Obesity, Cancer, and MicroRNAs. Semin. Cancer Biol. 2023, 93, 52–69. [Google Scholar] [CrossRef] [PubMed]
- Bupesh, G.; Saravanan, K.; Panneerselvam, G.; Meenakshi Sundaram, K.; Visvanathan, P. Role of Glucose Transporting Phytosterols in Diabetic Management. Diabetes Obes. Int. J. 2022, 7, 000261. [Google Scholar] [CrossRef]
- Relier, S.; Rivals, E.; David, A. The Multifaceted Functions of the Fat Mass and Obesity-Associated Protein (FTO) in Normal and Cancer Cells. RNA Biol. 2022, 19, 132–142. [Google Scholar] [CrossRef]
- Wei, H.; Li, Z.; Liu, F.; Wang, Y.; Ding, S.; Chen, Y.; Liu, J. The Role of FTO in Tumors and Its Research Progress. Curr. Med. Chem. 2022, 29, 924–933. [Google Scholar] [CrossRef]
- Zuidhof, H.R.; Calkhoven, C.F. Oncogenic and Tumor-Suppressive Functions of the RNA Demethylase FTO. Cancer Res. 2022, 82, 2201–2212. [Google Scholar] [CrossRef]
- Akbari, M.E.; Gholamalizadeh, M.; Doaei, S.; Mirsafa, F. FTO Gene Affects Obesity and Breast Cancer Through Similar Mechanisms: A New Insight into the Molecular Therapeutic Targets. Nutr. Cancer 2018, 70, 30–36. [Google Scholar] [CrossRef]
- Chen, J.; Du, B. Novel Positioning from Obesity to Cancer: FTO, an M6A RNA Demethylase, Regulates Tumour Progression. J. Cancer Res. Clin. Oncol. 2019, 145, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Arvanitakis, K.; Papadakos, S.P.; Lekakis, V.; Koufakis, T.; Lempesis, I.G.; Papantoniou, E.; Kalopitas, G.; Georgakopoulou, V.E.; Stergiou, I.E.; Theocharis, S.; et al. Meeting at the Crossroad between Obesity and Hepatic Carcinogenesis: Unique Pathophysiological Pathways Raise Expectations for Innovative Therapeutic Approaches. Int. J. Mol. Sci. 2023, 24, 14704. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yu, G.; Zhu, X.; Peng, T.; Lv, Y. Critical Roles of FTO-Mediated MRNA M6A Demethylation in Regulating Adipogenesis and Lipid Metabolism: Implications in Lipid Metabolic Disorders. Genes Dis. 2022, 9, 51–61. [Google Scholar] [CrossRef]
- Zhao, L.; Kong, X.; Zhong, W.; Wang, Y.; Li, P. FTO Accelerates Ovarian Cancer Cell Growth by Promoting Proliferation, Inhibiting Apoptosis, and Activating Autophagy. Pathol.-Res. Pract. 2020, 216, 153042. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Chen, W.; Wang, X. Studies on the Fat Mass and Obesity-Associated (FTO) Gene and Its Impact on Obesity-Associated Diseases. Genes Dis. 2023, 10, 2351–2365. [Google Scholar] [CrossRef]
- Peters, T.; Ausmeier, K.; Rüther, U. Cloning of Fatso (Fto), a Novel Gene Deleted by the Fused Toes (Ft) Mouse Mutation. Mamm. Genome 1999, 10, 983–986. [Google Scholar] [CrossRef]
- Deng, X.; Su, R.; Stanford, S.; Chen, J. Critical Enzymatic Functions of FTO in Obesity and Cancer. Front. Endocrinol. 2018, 9, 396. [Google Scholar] [CrossRef]
- Scuteri, A.; Sanna, S.; Chen, W.-M.; Uda, M.; Albai, G.; Strait, J.; Najjar, S.; Nagaraja, R.; Orrú, M.; Usala, G.; et al. Genome-Wide Association Scan Shows Genetic Variants in the FTO Gene Are Associated with Obesity-Related Traits. PLOS Genet. 2007, 3, e115. [Google Scholar] [CrossRef]
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.B.; Elliott, K.S.; Lango, H.; Rayner, N.W.; et al. A Common Variant in the FTO Gene Is Associated with Body Mass Index and Predisposes to Childhood and Adult Obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef]
- Qiao, Y.; Zhou, B.; Zhang, M.; Liu, W.; Han, Z.; Song, C.; Yu, W.; Yang, Q.; Wang, R.; Wang, S.; et al. A Novel Inhibitor of the Obesity-Related Protein FTO. Biochemistry 2016, 55, 1516–1522. [Google Scholar] [CrossRef]
- Ho, T.L.; Lee, J.; Ahn, S.Y.; Lee, D.-H.; Song, W.-J.; Kang, I.; Ko, E.-J. Immunostimulatory Effects of Marine Algae Extracts on in Vitro Antigen-presenting Cell Activation and in Vivo Immune Cell Recruitment. Food Sci. Nutr. 2023, 11, 6560–6570. [Google Scholar] [CrossRef]
- Ruud, J.; Alber, J.; Tokarska, A.; Engström Ruud, L.; Nolte, H.; Biglari, N.; Lippert, R.; Lautenschlager, Ä.; Cieślak, P.E.; Szumiec, Ł.; et al. The Fat Mass and Obesity-Associated Protein (FTO) Regulates Locomotor Responses to Novelty via D2R Medium Spiny Neurons. Cell Rep. 2019, 27, 3182–3198.e9. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhou, G.; Yu, X.; Xu, Q.; Wang, K.; Xie, D.; Yang, Q.; Wang, L. LC-MS-MS Quantitative Analysis Reveals the Association between FTO and DNA Methylation. PLoS ONE 2017, 12, e0175849. [Google Scholar] [CrossRef]
- Aik, W.; Scotti, J.S.; Choi, H.; Gong, L.; Demetriades, M.; Schofield, C.J.; McDonough, M.A. Structure of Human RNA N6-Methyladenine Demethylase ALKBH5 Provides Insights into Its Mechanisms of Nucleic Acid Recognition and Demethylation. Nucleic Acids Res. 2014, 42, 4741–4754. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Li, Z.; Li, J.; Liang, R.; Wang, Z.; Bai, Y.; Yang, Y.; Tang, Q.; Fu, Y.; Zhang, X.; et al. M 6 A MRNA Methylation: Biological Features, Mechanisms, and Therapeutic Potentials in Type 2 Diabetes Mellitus. Obes. Rev. 2023, 24, e13639. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Yan, H.-J.; Gao, C.-X.; Sun, W.-W.; Long, Y.-S. Inhibition of Hypothalamic FTO Activates STAT3 Signal through ERK1/2 Associated with Reductions in Food Intake and Body Weight. Neuroendocrinology 2023, 113, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Sebert, S.; Salonurmi, T.; Keinänen-Kiukaanniemi, S.; Savolainen, M.; Herzig, K.-H.; Symonds, M.E.; Järvelin, M.-R. Programming Effects of FTO in the Development of Obesity. Acta Physiol. 2014, 210, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Farooq, S.; Rana, S.; Siddiqui, A.J.; Iqbal, A.; Bhatti, A.A.; Musharraf, S.G. Association of Lipid Metabolism-Related Metabolites with Overweight/Obesity Based on the FTO Rs1421085. Mol. Omi. 2023, 19, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Wu, X.-N.; Ling, Y.; Rui, Y.; Wu, D.; Zhou, J.; Li, J.; Lin, S.; Peng, Q.; Li, Z.; et al. A Novel Inhibitor of N6-Methyladenosine Demethylase FTO Induces MRNA Methylation and Shows Anti-Cancer Activities. Acta Pharm. Sin. B 2022, 12, 853–866. [Google Scholar] [CrossRef]
- Zheng, Q.-K.; Ma, C.; ullah, I.; Hu, K.; Ma, R.-J.; Zhang, N.; Sun, Z.-G. Roles of N6-Methyladenosine Demethylase FTO in Malignant Tumors Progression. Onco. Targets. Ther. 2021, 14, 4837–4846. [Google Scholar] [CrossRef] [PubMed]
- Azzam, S.K.; Alsafar, H.; Sajini, A.A. FTO M6A Demethylase in Obesity and Cancer: Implications and Underlying Molecular Mechanisms. Int. J. Mol. Sci. 2022, 23, 3800. [Google Scholar] [CrossRef]
- Ferenc, K.; Pilžys, T.; Garbicz, D.; Marcinkowski, M.; Skorobogatov, O.; Dylewska, M.; Gajewski, Z.; Grzesiuk, E.; Zabielski, R. Intracellular and Tissue Specific Expression of FTO Protein in Pig: Changes with Age, Energy Intake and Metabolic Status. Sci. Rep. 2020, 10, 13029. [Google Scholar] [CrossRef]
- Lai, G.-Q.; Zhou, L.-L.; Yang, C.-G. RNA Methylation M6A: A New Code and Drug Target? Chin. J. Chem. 2020, 38, 420–421. [Google Scholar] [CrossRef]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell 2017, 31, 127–141. [Google Scholar] [CrossRef]
- Huang, Y.; Su, R.; Sheng, Y.; Dong, L.; Dong, Z.; Xu, H.; Ni, T.; Zhang, Z.S.; Zhang, T.; Li, C.; et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell 2019, 35, 677–691. [Google Scholar] [CrossRef]
- He, W.; Zhou, B.; Liu, W.; Zhang, M.; Shen, Z.; Han, Z.; Jiang, Q.; Yang, Q.; Song, C.; Wang, R.; et al. Identification of A Novel Small-Molecule Binding Site of the Fat Mass and Obesity Associated Protein (FTO). J. Med. Chem. 2015, 58, 7341–7348. [Google Scholar] [CrossRef]
- Gao, S.; Li, X.; Zhang, M.; Zhang, N.; Wang, R.; Chang, J. Structural Characteristics of Small-Molecule Inhibitors Targeting FTO Demethylase. Future Med. Chem. 2021, 13, 1475–1489. [Google Scholar] [CrossRef] [PubMed]
- Shishodia, S.; Demetriades, M.; Zhang, D.; Tam, N.Y.; Maheswaran, P.; Clunie-O’Connor, C.; Tumber, A.; Leung, I.K.H.; Ng, Y.M.; Leissing, T.M.; et al. Structure-Based Design of Selective Fat Mass and Obesity Associated Protein (FTO) Inhibitors. J. Med. Chem. 2021, 64, 16609–16625. [Google Scholar] [CrossRef] [PubMed]
- Huff, S.; Kummetha, I.R.; Zhang, L.; Wang, L.; Bray, W.; Yin, J.; Kelley, V.; Wang, Y.; Rana, T.M. Rational Design and Optimization of M6A-RNA Demethylase FTO Inhibitors as Anticancer Agents. J. Med. Chem. 2022, 65, 10920–10937. [Google Scholar] [CrossRef] [PubMed]
- Askr, H.; Elgeldawi, E.; Aboul Ella, H.; Elshaier, Y.A.M.M.; Gomaa, M.M.; Hassanien, A.E. Deep Learning in Drug Discovery: An Integrative Review and Future Challenges. Artif. Intell. Rev. 2023, 56, 5975–6037. [Google Scholar] [CrossRef]
- Zhang, H.; Saravanan, K.M.; Yang, Y.; Hossain, M.T.; Li, J.; Ren, X.; Pan, Y.; Wei, Y. Deep Learning Based Drug Screening for Novel Coronavirus 2019-NCov. Interdiscip. Sci. Comput. Life Sci. 2020, 12, 368–376. [Google Scholar] [CrossRef]
- Zhang, H.; Li, J.; Saravanan, K.M.; Wu, H.; Wang, Z.; Wu, D.; Wei, Y.; Lu, Z.; Chen, Y.H.; Wan, X.; et al. An Integrated Deep Learning and Molecular Dynamics Simulation-Based Screening Pipeline Identifies Inhibitors of a New Cancer Drug Target TIPE2. Front. Pharmacol. 2021, 12, 772296. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, T.; Saravanan, K.M.; Liao, L.; Wu, H.; Zhang, H.; Zhang, H.; Pan, Y.; Wu, X.; Wei, Y. DeepBindBC: A Practical Deep Learning Method for Identifying Native-like Protein-Ligand Complexes in Virtual Screening. Methods 2022, 205, 247–262. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Saravanan, K.M.; Lin, J.; Liao, L.; Ng, J.T.-Y.; Zhou, J.; Wei, Y. DeepBindPoc: A Deep Learning Method to Rank Ligand Binding Pockets Using Molecular Vector Representation. PeerJ 2020, 8, e8864. [Google Scholar] [CrossRef] [PubMed]
- Bhatti, U.A.; Tang, H.; Wu, G.; Marjan, S.; Hussain, A. Deep Learning with Graph Convolutional Networks: An Overview and Latest Applications in Computational Intelligence. Int. J. Intell. Syst. 2023, 2023, 8342104. [Google Scholar] [CrossRef]
- Zhou, J.; Cui, G.; Hu, S.; Zhang, Z.; Yang, C.; Liu, Z.; Wang, L.; Li, C.; Sun, M. Graph Neural Networks: A Review of Methods and Applications. AI Open 2020, 1, 57–81. [Google Scholar] [CrossRef]
- Puentes, P.R.; Rueda-Gensini, L.; Valderrama, N.; Hernández, I.; González, C.; Daza, L.; Muñoz-Camargo, C.; Cruz, J.C.; Arbeláez, P. Predicting Target–Ligand Interactions with Graph Convolutional Networks for Interpretable Pharmaceutical Discovery. Sci. Rep. 2022, 12, 8434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Saravanan, K.M.; Zhang, J.Z.H. DeepBindGCN: Integrating Molecular Vector Representation with Graph Convolutional Neural Networks for Protein–Ligand Interaction Prediction. Molecules 2023, 28, 4691. [Google Scholar] [CrossRef]
- Dudek, G.; Pełka, P.; Smyl, S. A Hybrid Residual Dilated LSTM and Exponential Smoothing Model for Midterm Electric Load Forecasting. IEEE Trans. Neural Netw. Learn. Syst. 2022, 33, 2879–2891. [Google Scholar] [CrossRef]
- Jalali, S.M.J.; Osório, G.J.; Ahmadian, S.; Lotfi, M.; Campos, V.M.A.; Shafie-khah, M.; Khosravi, A.; Catalão, J.P.S. New Hybrid Deep Neural Architectural Search-Based Ensemble Reinforcement Learning Strategy for Wind Power Forecasting. IEEE Trans. Ind. Appl. 2022, 58, 15–27. [Google Scholar] [CrossRef]
- Feng, Y.; Cheng, X.; Wu, S.; Saravanan, K.M.; Liu, W. Hybrid Drug-Screening Strategy Identifies Potential SARS-CoV-2 Cell-Entry Inhibitors Targeting Human Transmembrane Serine Protease. Struct. Chem. 2022, 33, 1503–1515. [Google Scholar] [CrossRef]
- Dai, G.; Tian, Z.; Fan, J.; Sunil, C.K.; Dewi, C. DFN-PSAN: Multi-Level Deep Information Feature Fusion Extraction Network for Interpretable Plant Disease Classification. Comput. Electron. Agric. 2024, 216, 108481. [Google Scholar] [CrossRef]
- Chen, R.-C.; Dewi, C.; Zhuang, Y.-C.; Chen, J.-K. Contrast Limited Adaptive Histogram Equalization for Recognizing Road Marking at Night Based on Yolo Models. IEEE Access 2023, 11, 92926–92942. [Google Scholar] [CrossRef]
- Fadafen, M.K.; Rezaee, K. Ensemble-Based Multi-Tissue Classification Approach of Colorectal Cancer Histology Images Using a Novel Hybrid Deep Learning Framework. Sci. Rep. 2023, 13, 8823. [Google Scholar] [CrossRef]
- Dewi, C.; Chen, R.-C. Automatic Medical Face Mask Detection Based on Cross-Stage Partial Network to Combat COVID-19. Big Data Cogn. Comput. 2022, 6, 106. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Niu, T.; Chang, J.; Lei, X.; Zhao, M.; Wang, Q.; Cheng, W.; Wang, J.; Feng, Y.; Chai, J. Crystal Structure of the FTO Protein Reveals Basis for Its Substrate Specificity. Nature 2010, 464, 1205–1209. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yan, J.; Li, Q.; Li, J.; Gong, S.; Zhou, H.; Gan, J.; Jiang, H.; Jia, G.-F.; Luo, C.; et al. Meclofenamic Acid Selectively Inhibits FTO Demethylation of M6A over ALKBH5. Nucleic Acids Res. 2015, 43, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Shoichet, B.K. ZINC—A Free Database of Commercially Available Compounds for Virtual Screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Sterling, T.; Mysinger, M.M.; Bolstad, E.S.; Coleman, R.G. ZINC: A Free Tool to Discover Chemistry for Biology. J. Chem. Inf. Model. 2012, 52, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Zhao, S.; Gilvary, C.; Elemento, O.; Zhou, J.; Wang, F. Graph Convolutional Networks for Computational Drug Development and Discovery. Brief. Bioinform. 2020, 21, 919–935. [Google Scholar] [CrossRef]
- Son, J.; Kim, D. Development of a Graph Convolutional Neural Network Model for Efficient Prediction of Protein-Ligand Binding Affinities. PLoS ONE 2021, 16, e0249404. [Google Scholar] [CrossRef]
- Zhang, H.; Liao, L.; Saravanan, K.M.; Yin, P.; Wei, Y. DeepBindRG: A Deep Learning Based Method for Estimating Effective Protein–Ligand Affinity. PeerJ 2019, 7, e7362. [Google Scholar] [CrossRef]
- Brooks, B.R., III; Mackerell, J.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; Caflisch, A.; et al. Autodock Vina. J. Comput. Chem. 2009, 30, 1545. [Google Scholar] [CrossRef]
- Goodsell, D.S.; Sanner, M.F.; Olson, A.J.; Forli, S. The AutoDock Suite at 30. Protein Sci. 2021, 30, 31–43. [Google Scholar] [CrossRef]
- Kaminski, G.; Jorgensen, W.L. Performance of the AMBER94, MMFF94, and OPLS-AA Force Fields for Modeling Organic Liquids. J. Phys. Chem. 1996, 100, 18010–18013. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Discovery Studio Visualizer, V4.0.100.13345; Accelrys Softw. Inc.: San Diego, CA, USA, 2005.
- Lill, M.A.; Danielson, M.L. Computer-Aided Drug Design Platform Using PyMOL. J. Comput. Aided. Mol. Des. 2011, 25, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [PubMed]
- da Silva, A.W.S.; Vranken, W.F. ACPYPE—AnteChamber PYthon Parser InterfacE. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Homeyer, N.; Gohlke, H. Free Energy Calculations by the Molecular Mechanics Poisson−Boltzmann Surface Area Method. Mol. Inform. 2012, 31, 114–122. [Google Scholar] [CrossRef]
- Ferreira de Freitas, R.; Schapira, M. A Systematic Analysis of Atomic Protein–Ligand Interactions in the PDB. Medchemcomm 2017, 8, 1970–1981. [Google Scholar] [CrossRef]
- Zhao, Z.; Bourne, P.E. Harnessing Systematic Protein–Ligand Interaction Fingerprints for Drug Discovery. Drug Discov. Today 2022, 27, 103319. [Google Scholar] [CrossRef]
- Kuhlman, B.; Bradley, P. Advances in Protein Structure Prediction and Design. Nat. Rev. Mol. Cell Biol. 2019, 20, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Skolnick, J.; Zhou, H. Implications of the Essential Role of Small Molecule Ligand Binding Pockets in Protein–Protein Interactions. J. Phys. Chem. B 2022, 126, 6853–6867. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Pang, B.; Shyu, C.-R.; Korkin, D. Charged Residues at Protein Interaction Interfaces: Unexpected Conservation and Orchestrated Divergence. Protein Sci. 2011, 20, 1275–1284. [Google Scholar] [CrossRef] [PubMed]
- Hirano, A.; Kameda, T.; Arakawa, T.; Shiraki, K. Arginine-Assisted Solubilization System for Drug Substances: Solubility Experiment and Simulation. J. Phys. Chem. B 2010, 114, 13455–13462. [Google Scholar] [CrossRef]
- Shiammala, P.N.; Duraimutharasan, N.K.B.; Vaseeharan, B.; Alothaim, A.S.; Al-Malki, E.S.; Snekaa, B.; Safi, S.Z.; Singh, S.K.; Velmurugan, D.; Selvaraj, C. Exploring the Artificial Intelligence and Machine Learning Models in the Context of Drug Design Difficulties and Future Potential for the Pharmaceutical Sectors. Methods 2023, 219, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Murugesan, A.; Mani, S.K.; Thiyagarajan, R.; Palanivel, S.; Gurbanov, A.V.; Zubkov, F.I.; Kandhavelu, M. Benzenesulfonamide Analogs: Synthesis, Anti-GBM Activity and Pharmacoprofiling. Int. J. Mol. Sci. 2023, 24, 12276. [Google Scholar] [CrossRef]
- Kumar, S.U.; Rajan, B.; Kumar, D.T.; Cathryn, R.H.; Das, V.S.; Zayed, H.; Walter, C.E.J.; Ramanathan, G.; Doss, C.P.G. Comparison of Potential Inhibitors and Targeting Fat Mass and Obesity-Associated Protein Causing Diabesity through Docking and Molecular Dynamics Strategies. J. Cell. Biochem. 2021, 122, 1625–1638. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, B. Strategies of Polypharmacology BT—Polypharmacology: Principles and Methodologies; Wang, Z., Yang, B., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 43–72. ISBN 978-3-031-04998-9. [Google Scholar]
- Isert, C.; Atz, K.; Schneider, G. Structure-Based Drug Design with Geometric Deep Learning. Curr. Opin. Struct. Biol. 2023, 79, 102548. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Le, H.; Quinn, T.P.; Nguyen, T.; Le, T.D.; Venkatesh, S. GraphDTA: Predicting Drug Target Binding Affinity with Graph Neural Networks. Bioinformatics 2021, 37, 1140–1147. [Google Scholar] [CrossRef] [PubMed]
- Sreeraman, S.; Kannan, P.M.; Singh Kushwah, R.B.; Sundaram, V.; Veluchamy, A.; Thirunavukarasou, A.; Saravanan, M.K. Drug Design and Disease Diagnosis: The Potential of Deep Learning Models in Biology. Curr. Bioinform. 2023, 18, 208–220. [Google Scholar] [CrossRef]



| S.No | Compound | DeepBindGCN_BC | DeepBindGCN_RG | Binding Energy (kJ/mol) |
|---|---|---|---|---|
| 1 | MolPort-001-741-269_ZINC000003643476 | 1 | 9.079 | −10.0 |
| 2 | MolPort-002-508-662_ZINC000000517415 | 1 | 9.025 | −9.7 |
| 3 | MolPort-002-476-943_ZINC000001562130 | 1 | 7.666 | −9.0 |
| 4 | MolPort-002-507-418_ZINC000142857948 | 1 | 6.040 | −7.1 |
| 5 | MolPort-002-801-687_ZINC000001022034 | 1 | 9.036 | −7.1 |
| 6 | MolPort-001-739-485_ZINC000229938091 | 1 | 9.052 | −6.9 |
| 7 | MolPort-005-945-631_ZINC000013485421 | 1 | 9.002 | −6.8 |
| 8 | MolPort-001-736-557_ZINC000005447704 | 1 | 9.334 | −6.8 |
| 9 | MolPort-001-741-121_ZINC000075906045 | 1 | 9.031 | −6.8 |
| 10 | MolPort-001-832-299_ZINC000004521756 | 1 | 9.045 | −6.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mayuri, K.; Varalakshmi, D.; Tharaheswari, M.; Somala, C.S.; Priya, S.S.; Bharathkumar, N.; Senthil, R.; Kushwah, R.B.S.; Vickram, S.; Anand, T.; et al. Identifying Potent Fat Mass and Obesity-Associated Protein Inhibitors Using Deep Learning-Based Hybrid Procedures. BioMedInformatics 2024, 4, 347-359. https://doi.org/10.3390/biomedinformatics4010020
Mayuri K, Varalakshmi D, Tharaheswari M, Somala CS, Priya SS, Bharathkumar N, Senthil R, Kushwah RBS, Vickram S, Anand T, et al. Identifying Potent Fat Mass and Obesity-Associated Protein Inhibitors Using Deep Learning-Based Hybrid Procedures. BioMedInformatics. 2024; 4(1):347-359. https://doi.org/10.3390/biomedinformatics4010020
Chicago/Turabian StyleMayuri, Kannan, Durairaj Varalakshmi, Mayakrishnan Tharaheswari, Chaitanya Sree Somala, Selvaraj Sathya Priya, Nagaraj Bharathkumar, Renganathan Senthil, Raja Babu Singh Kushwah, Sundaram Vickram, Thirunavukarasou Anand, and et al. 2024. "Identifying Potent Fat Mass and Obesity-Associated Protein Inhibitors Using Deep Learning-Based Hybrid Procedures" BioMedInformatics 4, no. 1: 347-359. https://doi.org/10.3390/biomedinformatics4010020
APA StyleMayuri, K., Varalakshmi, D., Tharaheswari, M., Somala, C. S., Priya, S. S., Bharathkumar, N., Senthil, R., Kushwah, R. B. S., Vickram, S., Anand, T., & Saravanan, K. M. (2024). Identifying Potent Fat Mass and Obesity-Associated Protein Inhibitors Using Deep Learning-Based Hybrid Procedures. BioMedInformatics, 4(1), 347-359. https://doi.org/10.3390/biomedinformatics4010020








