Abstract
This article delves into the intersection of generative AI and digital twins within drug discovery, exploring their synergistic potential to revolutionize pharmaceutical research and development. Through various instances and examples, we illuminate how generative AI algorithms, capable of simulating vast chemical spaces and predicting molecular properties, are increasingly integrated with digital twins of biological systems to expedite drug discovery. By harnessing the power of computational models and machine learning, researchers can design novel compounds tailored to specific targets, optimize drug candidates, and simulate their behavior within virtual biological environments. This paradigm shift offers unprecedented opportunities for accelerating drug development, reducing costs, and, ultimately, improving patient outcomes. As we navigate this rapidly evolving landscape, collaboration between interdisciplinary teams and continued innovation will be paramount in realizing the promise of generative AI and digital twins in advancing drug discovery.
1. Introduction
Decades back, Alan Turing, often hailed as the visionary behind modern computer science, stirred the imagination of generations with his bold inquiry into the essence of machine cognition. In his seminal work “Computing Machinery and Intelligence”, he challenged the boundaries of human understanding by introducing the enigmatic Turing Test, sparking a timeless debate on the capacity of machines to think and emulate human intelligence honestly [1]. His question, “Can machines think?”, eventually led to the birth of artificial intelligence (AI). The term AI was coined in 1956 by John McCarthy during The Dartmouth Summer Research Project on Artificial Intelligence [2]. Progressively, in the late 1950s, two key milestones were set: Arthur Samuel created the first self-learning program for checkers, marking the introduction of machine learning (ML); and Frank Rosenblatt developed the first perceptron, representing the earliest form of a neural network [3]. One of the earliest instances of functional generative AI was the ELIZA chatbot, developed by Joseph Weizenbaum in 1961 [4]. These milestones laid the foundation for the evolution of AI and its applications in various fields, including medicine and drug discovery.
In recent years, a subset of AI, generative AI, has undergone significant advancements, transforming various domains by generating realistic content. Generative AI refers to a category of models designed to generate new content similar to, but not the same as, the input data it was trained on [5,6,7]. Unlike traditional AI systems that are often task-specific and deterministic, generative AI systems can produce novel outputs by learning the underlying patterns and structures of the training data. Initially utilizing models like hidden Markov models (HMMs) and Gaussian mixture models (GMMs), the field witnessed a breakthrough with the introduction of generative adversarial networks (GANs) by Ian Goodfellow in 2014. GANs have been implemented in cardiology and used for detecting Pneumonia, COVID-19, etc., as discussed later in this review [8,9,10,11,12,13]. Other models, like recurrent neural networks (RNNs) and transformers, have also contributed to the progress of generative AI, enabling tasks in natural language processing (NLP), computer vision (CV), and digital twins (DT). These models are being increasingly applied to improve healthcare and medicine (Table 1) [14,15,16,17,18,19,20,21,22,23,24]. Such advancements in generative AI have enhanced content generation capabilities and found applications specifically in drug discovery, where models are employed to design novel molecules with desired properties to accelerate drug development and clinical trials, which will be discussed throughout this review.

Table 1.
Concise definitions of key terms in advanced machine learning and AI.
Identifying and prioritizing chemical compounds for drug development can pose significant challenges, as determining which compounds are most promising for treating specific diseases requires extensive laboratory screening and testing. Generative AI streamlines this process by leveraging advanced chemistry models to analyze millions of known chemical compounds based on their structure and functionality. By overlaying this data with existing results from tested molecules, generative AI accelerates the screening process and aids in identifying compounds with the highest potential for successful treatment [25,26]. Research from the Tufts Centre for the Study of Drug Development indicates that bringing a single drug to market typically requires ten years and USD 1.4 billion, with about 80% of expenses attributed to clinical development [27]. This phase involves rigorous testing of a medication’s safety and efficacy in human subjects, characterized by lengthy timelines and strict regulatory requirements. Generative AI addresses these challenges by increasing efficiency across clinical development. It achieves up to 50% cost reductions by streamlining trial processes and automating document drafting, shortens trial timelines by over 12 months, and enhances net present value by at least 20% through improved health authority interactions, quality control, and signal management. The McKinsey Global Institute estimates that generative AI could yield USD 60 billion to USD 110 billion annually for the pharmaceutical and medical product sectors [28]. This potential economic value stems from its ability to enhance productivity by expediting compound identification, accelerating drug development and approval, and refining marketing strategies.
2. Drug Discovery and Generative AI
2.1. Generative Adversarial Networks (GANs)
GANs consist of two neural networks, a generator and a discriminator, which are trained simultaneously in a minimax. The generator creates synthetic data samples, while the discriminator distinguishes between natural and synthetic samples. GANs are increasingly recognized as powerful tools in drug discovery [5,29,30,31]. They offer innovative approaches to exploring chemical space, refining known compounds, and crafting new molecules. These networks find utility across various drug design and discovery stages, from creating molecules from scratch to reducing complexity and even designing peptides and proteins from scratch. GANs have proven pivotal in developing new molecules with specific attributes, aiding in developing practical drugs and expediting the drug discovery timeline [32,33,34,35,36,37].
Recent research emphasizes the benefits of GANs in drug discovery, showcasing their ability to uncover novel molecules and navigate challenges like mode collapse by encouraging exploration beyond existing data. Specialized GAN architectures like MedGAN leverage graph convolutional networks to efficiently design fresh molecules, addressing the growing need for new medications and enhancing the overall drug discovery process [31,38,39]. GANs are also applied in de novo peptide and protein design, contributing significantly to exploring new bioactive compounds [40]. Furthermore, a study demonstrated quantum advantages in small drug discovery when each component of the GAN was replaced with a variational quantum circuit (VQC). Consequently, the physicochemical properties and performance improvements were seen, with a few learnable parameters in the GAN’s generator compared to the classical approach [41,42,43].
2.2. Variational Autoencoders (VAEs)
VAEs are a probabilistic generative model that learns a latent representation of data by encoding input samples into a lower-dimensional (encoder) and decoding them back into the original space (decoder). The encoder and decoder are the only components of this neural network structure, and they are trained in conjugation with each other, employing the reparameterization technique. The function of the variational autoencoder (VAE) is defined in Equation (1), wherein autoencoder settings, Q(z|X) and P(X|z), are estimated by an encoder and a decoder, respectively.
E[logP(X∣z)] − DKL [Q(z∣X)‖P(z)]
Equation (1): variational autoencoder basic function [44].
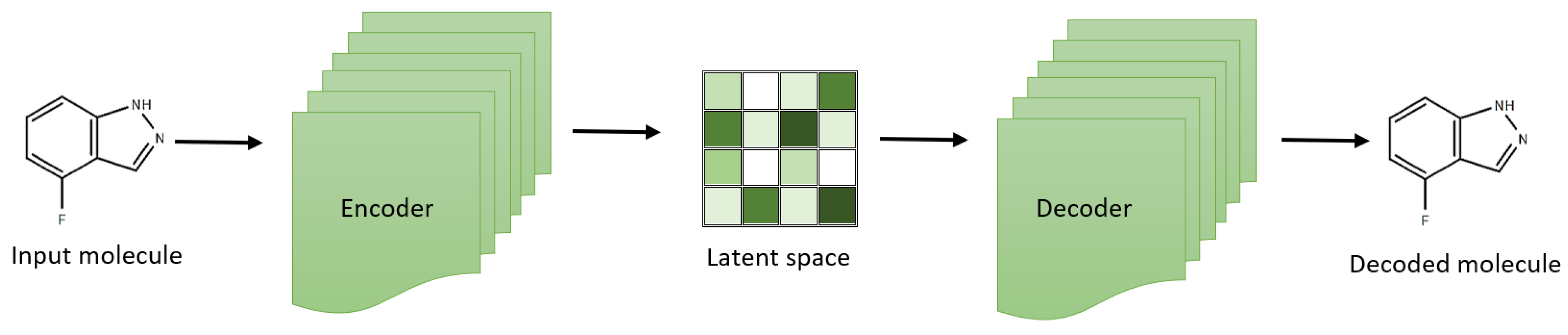
VAEs have created a chemical latent space within drug discovery, reflecting compound libraries’ structural diversity [45,46,47]. These aid in exploring a broader chemical space and facilitate the generation of novel compounds (Figure 1). For example, the development of the natural product compound variational autoencoder (NP-VAE) has enabled the handling of intricate datasets and large molecular structures, showcasing consistent performance as a generative model across multiple metrics. Employing reconstruction loss and latent loss, these models optimize reconstruction quality concurrently while exploring the latent space effectively [46,48,49]. Additionally, variants of VAEs have been used to simulate morphology and gene expression readouts induced by specific compounds accurately, allowing for the prediction of cell states affected by compounds with known polypharmacology. This inference of cell state based on drug mechanisms could assist researchers in the future by facilitating the development and identification of targeted therapeutics and the classification of off-target effects [50].
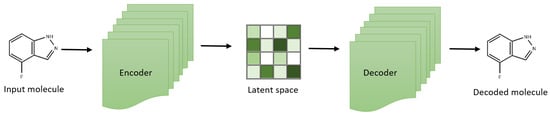
Figure 1.
Variational autoencoder architecture for effective exploration of small molecular compounds.
2.3. Transformer-Based Models
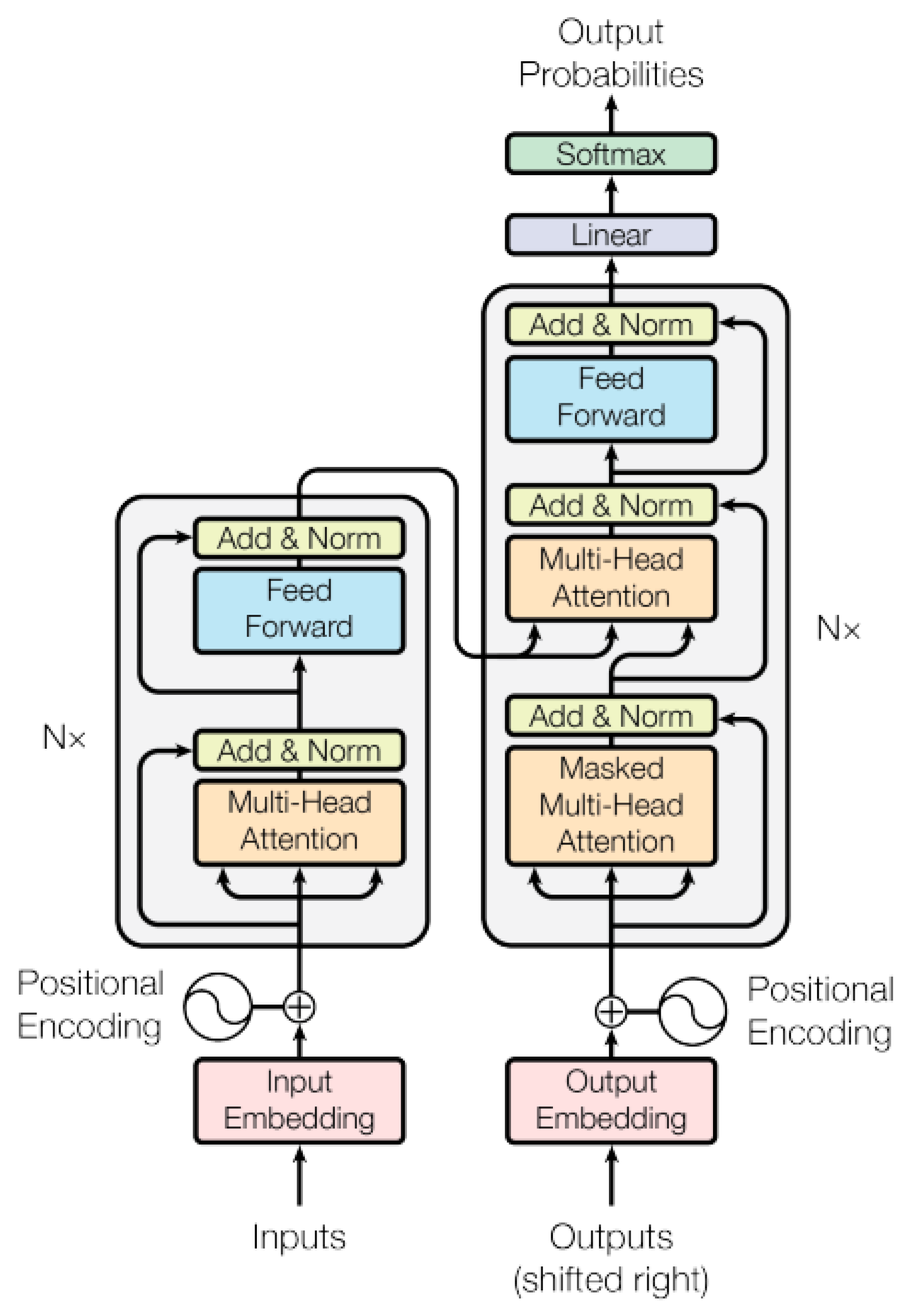
These AI models are adept at using natural language processing, or NLP, to comprehend the structure and context of language (Figure 2). They are trained with extensive datasets to grasp connections between sequential data such as words and sentences. Three recently discovered, innovative approaches to drug discovery are presented here. The first, drugAI, integrates the encoder–decoder transformer architecture with reinforcement learning via a Monte Carlo tree search to streamline the drug discovery process [51]. This method ensures the generation of valid small molecules with drug-like characteristics and robust binding affinities toward their targets. In the second approach, the authors focused more on target-specific de novo drug design, treating it as a translational challenge between the amino acid “language” and simplified molecular input line entry system representation [52]. Employing the transformer neural network architecture, known for its proficiency in sequence transduction tasks, this method captures long-range dependencies within sequences. It generates structurally diverse compounds with realistic properties within the plausible drug-like range. Finally, TransDTI is introduced by its authors as a multiclass classification and regression workflow utilizing transformer-based language models to categorize interactions between drug–target pairs as active, inactive, or intermediate. Trained on large-scale drug–target interaction datasets, these models exhibit superior performance compared to baseline methods, effectively predicting novel drug–target interactions from sequence data and outperforming existing approaches [53]. Another such model, DTSyn, utilizes its capability to extract interactions between chemicals and cell lines, depicting potential drug action mechanisms. Through the integration of attention mechanisms and pre-trained gene embeddings, DTSyn demonstrates enhanced interpretability. Consequently, this model is invaluable for prioritizing synergistic drug combinations based on chemical and cell line gene expression profiles. Similar transformer-based models are instrumental in accelerating drug discovery processes by automating tasks like retrosynthesis, generating novel molecules with desired properties, and facilitating the exploration of chemical space to develop new drugs [54,55,56,57,58] (Table 2).
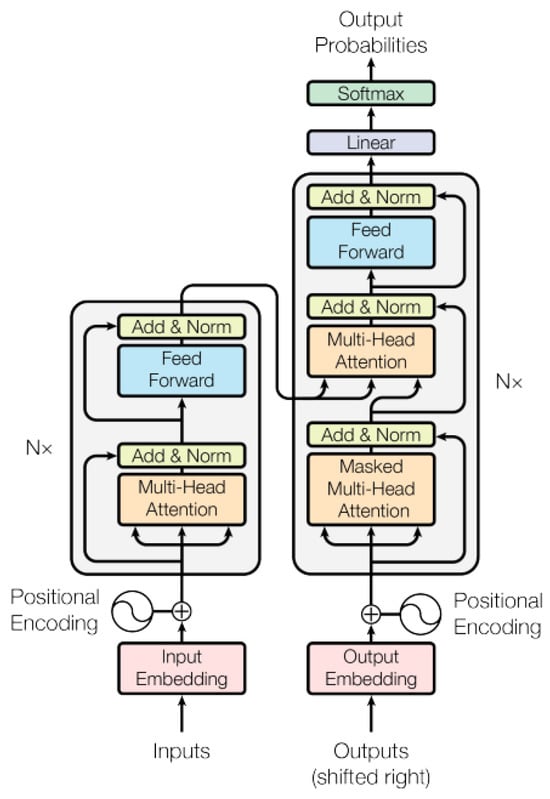
Figure 2.
Transformer model architecture [59].

Table 2.
Summary of generative AI models, their examples, salient features, metrics of scoring, and applications.
3. Restricted Boltzmann Machines (RBMs)
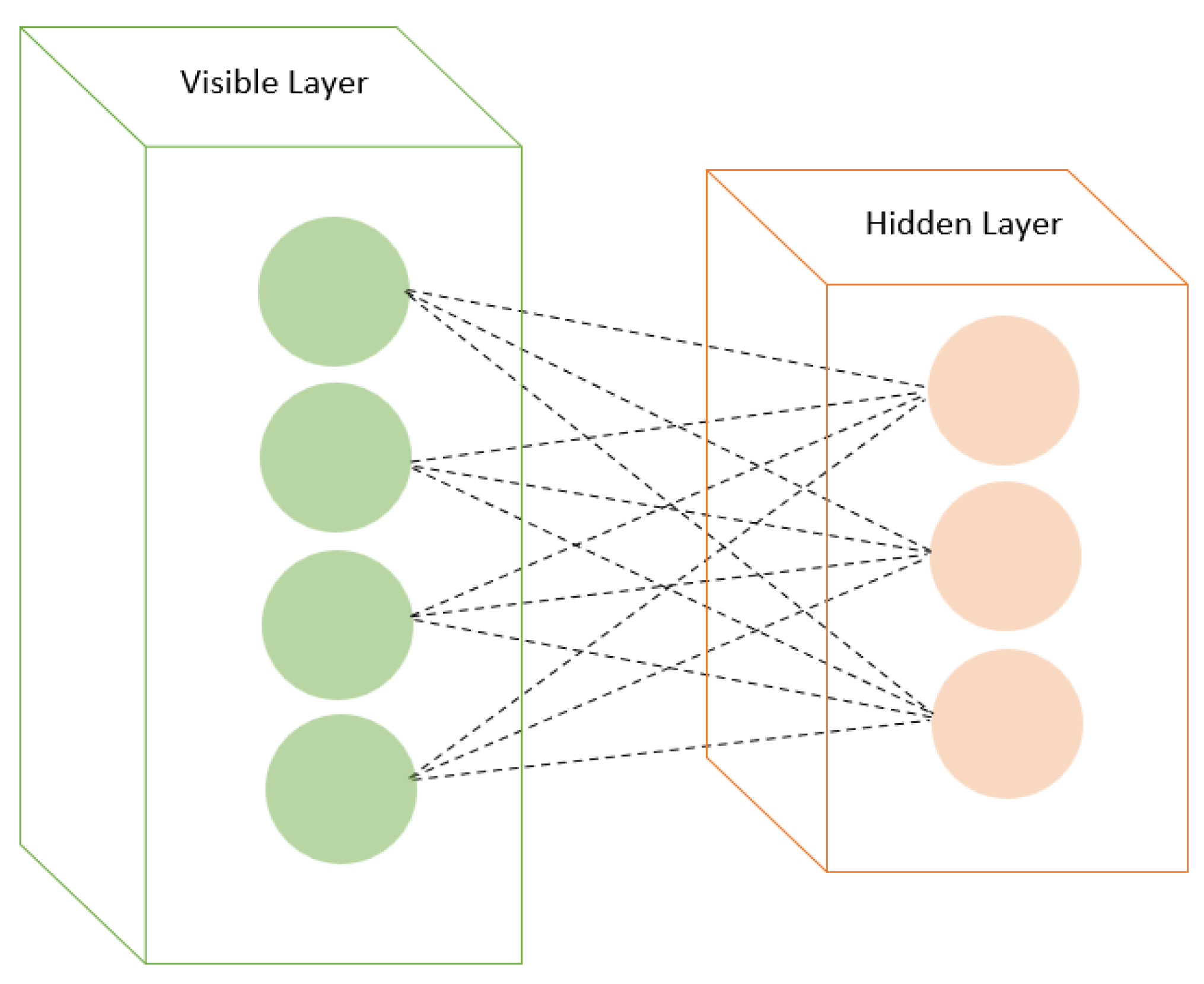
RBMs are a type of generative model based on energy-based models. They have visible and hidden layers with symmetric connections (Figure 3). RBMs are trained using contrastive divergence or other learning algorithms. RBMs have emerged as promising tools in drug discovery, specifically in forecasting drug–disease associations and drug–target interactions. In predicting drug repositioning tasks within drug–disease association networks, RBMs have displayed enhanced prediction performance compared to alternative methods. Moreover, augmenting the RBM model with momentum during weight updates has further bolstered prediction performance, positioning it as a potent tool for future drug repositioning endeavors [60,61,62].
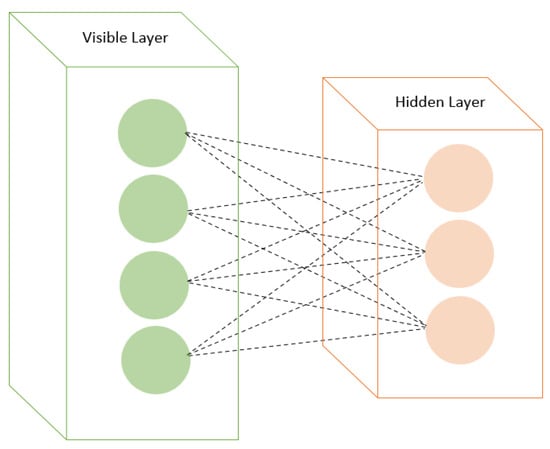
Figure 3.
Restricted Boltzmann machines with visible and hidden layers.
In the domain of drug–target interactions, RBMs have been deployed to amalgamate multiple interaction types and forecast unknown drug–target relationships or modes of action. By formulating the prediction problem into a two-layer graphical model using RBMs, researchers have adeptly captured latent features within drug–target interaction networks, resulting in high precision–recall curve values. This methodology, surpassing other prediction techniques by incorporating various interaction types, holds practical significance in predicting drug–target interactions and advancing drug repositioning efforts [63,64,65]. RBMs are demonstrating their worth as invaluable assets in drug discovery by offering innovative solutions for forecasting drug–disease associations and drug–target interactions and facilitating computational drug repositioning. However, research leveraging machine learning approaches still needs to be conducted to analyze intricate datasets, predict new relationships within biological systems, and shape the landscape of drug discovery.
4. Generative Graph Neural Networks (GNNs)
GNNs are a class of neural networks designed to operate on graph-structured data. Generative GNNs can generate new graph structures or nodes and edges within a graph. GNNs are revolutionizing drug discovery by facilitating the creation of novel molecules with specific properties and streamlining the drug design process. These networks employ graph neural network modules to construct sequential molecular graph generators like MG2N2 [66,67]. Such generators incrementally add nodes and connections to graphs, simplifying training procedures and improving interpretability. By maximizing information input at each generative step, these models effectively generalize molecular patterns learned during training without succumbing to overfitting, demonstrating competitive performance in generating molecular structures [39]. Applications of GNNs in drug discovery are rapidly expanding, particularly in conditional de novo drug design. GNNs excel at processing graph-structured data and have been pivotal in efficiently predicting drug–target interactions and designing new candidate molecules [68,69]. The fusion of GNNs with deep learning techniques is revolutionizing graph generation for molecular structures, offering promising applications in drug discovery by optimizing resource utilization and improving the efficiency of generating new bioactive molecules. One example is MM-GANN-DDI, which accurately presents a multimodal graph-agnostic neural network to forecast drug–drug interaction events [70,71,72,73]. GNNs drive innovation in drug discovery by enabling the systematic generation of novel molecules, predicting drug–target interactions, and advancing computational methods for de novo drug design. Their effectiveness in processing graph data and generating diverse chemical structures tailored for therapeutic purposes underscores their significant contribution to advancing drug discovery endeavors.
5. Language Models (LMs)
LMs are a class of generative models that learn the probability distribution of sequences of words or tokens in a language. LMs play a crucial role in drug discovery, providing innovative solutions to expedite the molecule discovery cycle, enhance de novo drug design, predict properties, and optimize chemical reactions [74]. Particularly, transformer-based architectures have showcased remarkable capabilities in comprehending and generating human-like text, extending their application into scientific domains such as protein folding, target identification, and small molecule design [75]. Within molecular discovery, Chemical LMs significantly contribute to accelerating the identification of new compounds for drug development, predicting properties, and optimizing chemical reactions [76]. These models operate on small molecules, proteins, or polymers, demonstrating promising results in early-stage drug discovery by effectively utilizing machine learning techniques to comprehend and generate scientific text [77].
Furthermore, AI-powered LMs have revolutionized natural language processing (NLP) in drug discovery and development [78]. For instance, an automatic biomedical named entity recognition (BioNER) method finds the hidden relationship among chemicals, genes, targets, and diseases from text-based documents [79]. Henceforth, it can safely be said that LM models hold the potential to transform treatment development by assisting in target identification, clinical design, regulatory decision-making, pharmacovigilance, and even aiding in the development of new treatments for diseases like COVID-19 through drug repurposing initiatives.
LM models can streamline patient recruitment processes in clinical trials by automating tasks through advanced information retrieval and prioritization mechanisms. These models learn medical terms and their synonyms to extract valuable information from clinical documents, aiding in patient stratification based on disease subtypes. They also synthesize eligibility criteria into standardized contextual queries, improving clinical trial-matching processes. By leveraging cross-model learning infrastructure, these LMs encode enrolment criteria and patient electronic health records (EHRs) for enhanced matching inference, outperforming rule-based strategies. Furthermore, they seamlessly integrate with emerging technologies like genomics and imaging data to advance precision medicine. Additionally, AI-powered LMs facilitate higher patient enrolment rates and improved site identification, considering factors such as prior site experience, connections with health non-profits, patient retention data, and cost-effectiveness to support balanced clinical decision-making, which is the key to the success of the designed drugs [80,81,82,83].
6. Multimodal Models
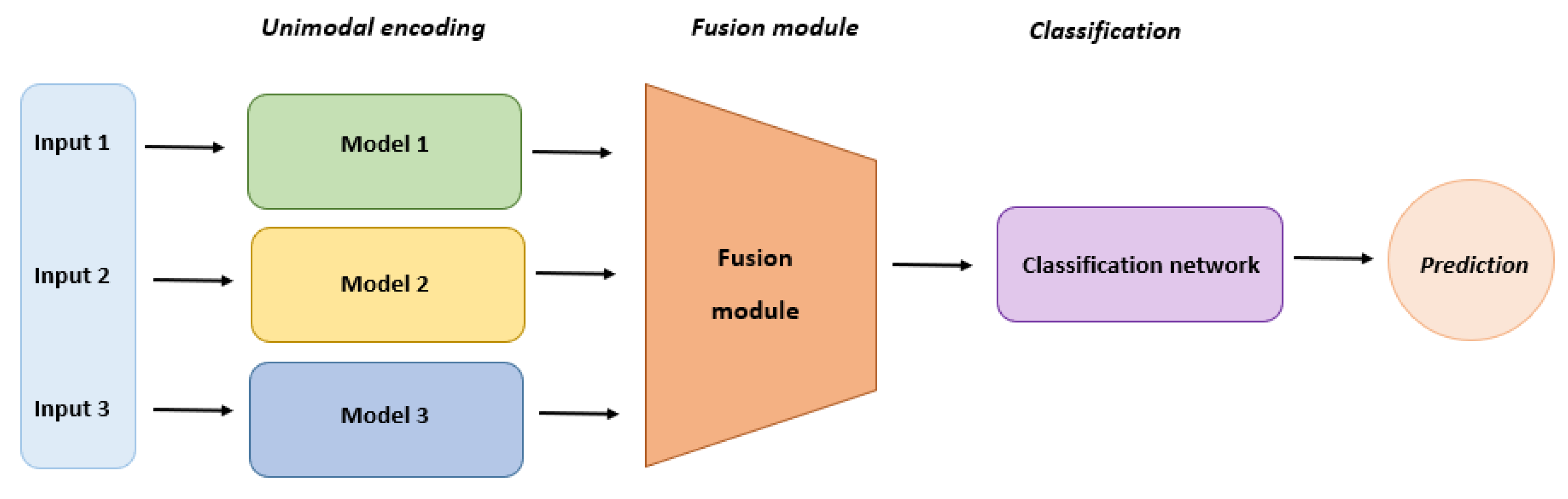
Multimodal (MM) generative AI models can simultaneously process various types of data and are essential in drug discovery and therapeutic design, utilizing a combination of deep learning techniques (Figure 4). Deep generative models using multimodal data exhibit advantages over unimodal counterparts due to the complementary insights offered by multimodal data [84,85,86]. Successful drug discovery hinges on leveraging diverse data modalities that offer complementary perspectives, aiding in the triangulation of evidence for discovery. While current studies primarily focus on molecular structural data, they underutilize other data modalities, such as drug–target interactions, drug–disease knowledge, and relevant gene expression post-drug treatment. Addressing this challenge involves exploring solutions like “modality alignment”, connecting all modalities through an intermediary modality, typically molecular structures, and “modality fusion”, where all modalities are directly mapped to a common latent space [87,88,89,90]. This hybrid data model captures diverse information during drug design, including chemical properties, drug–target interactions, drug–disease knowledge, and disease-relevant gene expression [91].
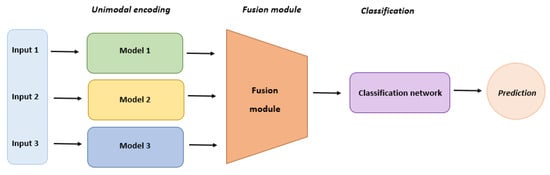
Figure 4.
Multimodal model architecture and layers.
Additionally, a multimodal generative model considers various components of the drug discovery pipeline to enhance the likelihood of success in clinical trials. By integrating structured and unstructured knowledge, frameworks like KEDD achieve a deeper understanding of biomolecules, outperforming state-of-the-art models in various predictions related to drug–target interactions, drug properties, drug–drug interactions, and protein–protein interactions [92]. Qualitative analysis reveals the promising potential of such frameworks in real-world applications, accelerating drug discovery by incorporating biomolecular expertise from multimodal knowledge.
MM models are also used to overcome the impractical size of chemical space; generative adversarial networks are proposed to generate diverse three-dimensional ligand shapes complementary to the pocket. These shapes can be decoded into a sequence of SMILES, enabling structure-based de novo drug design [93]. Evaluation shows enrichment compared to random sampling from the initial chemical space of ZINC drug-like compounds, validating the method’s effectiveness in virtual screening. Moreover, integrated with several imaging techniques, multimodal imaging provides vast anatomical, functional, and molecular information, accelerating drug discovery and development. These imaging technologies aid in understanding disease mechanisms, identifying new pharmacological targets, and assessing potential drug candidates and treatment responses. Implementing radiomics MM via targeted and untargeted methods further enhances the utility of imaging technologies in drug discovery and development, emphasizing their strengths, innovations, and future potential. Targeted approaches involve imaging specific drug molecules or targets, while untargeted approaches analyze a wide range of molecules to discover drug metabolites, effects on endogenous molecules, and disease-related changes. These imaging techniques also unveil anatomical, structural, metabolomic, lipidomic, and proteomic alterations in response to drug treatments at tissue and organ levels, advancing drug design and delivery [94,95,96].
However, the significant limitations of multimodal models are modality alignment and fusion. One of the primary challenges is the heterogeneous nature of data sources, including molecular structures, biological assays, textual literature, patient records, and medical imaging. Each modality provides valuable insights into different aspects of drug discovery and healthcare, but integrating these diverse data types into a coherent framework poses significant hurdles. Semantic misalignment between molecular structures, biological assays, and clinical data further complicates the integration process, as these modalities often represent information at different levels of abstraction. To overcome these challenges, multimodal models leverage techniques such as representation learning and feature extraction to transform each modality into a common embedding space, allowing for meaningful correlations to be captured. For example, molecular structures can be encoded into graph representations, while patient records can be represented as structured data or textual embeddings. Fusion techniques, including attention mechanisms and graph neural networks, enable information aggregation across modalities while preserving their characteristics. By effectively aligning and fusing multimodal data, these models can accelerate drug discovery processes, identify novel therapeutic targets, predict drug response, and personalize treatment strategies for better patient outcomes in healthcare settings.
7. Drug Discovery and Digital Twins
Digital twins (DT) are virtual replicas or digital representations of physical objects, processes, systems, or entities. They are created using data collected from sensors, IoT devices, and other sources, and they mimic the behavior and characteristics of their real-world counterparts in a virtual environment. DTs are increasingly utilized in drug discovery to simulate drug behavior, predict efficacy, and streamline drug development processes. These digital counterparts empower researchers with a deeper understanding of how drugs interact with the body, enabling them to anticipate potential side effects and tailor dosages more effectively. Leveraging generative AI, digital twins can model systems ranging from individual cells to entire human bodies, thereby enhancing comprehension of diseases, facilitating biomarker discovery, and expediting drug development [19,21,22]. DT’s various applications in the pharmaceutical industry include modeling cells to expedite drug discovery, forecasting patient responses to obviate placebo control arms in clinical trials, and facilitating personalized medicine by simulating organs, genomes, and patients. Furthermore, digital twins can augment drug delivery by fine-tuning drug release rates, dosages, and nanoparticle delivery efficiency [18,20,97]. With their ability to offer personalized treatment options, optimize drug delivery mechanisms, and accurately predict drug toxicity, digital twins have significant promise in revolutionizing drug discovery processes and improving clinical trial efficiency, effectiveness, and safety while reducing costs and time-to-market (Table 3).

Table 3.
Generative AI and digital twin use cases in drug discovery.
When the SARS-CoV-2 pandemic emerged in 2019, researchers quickly adapted epidemiological computer models for decision support in public health responses. However, existing tools could not predict individual COVID-19 patient outcomes. Patient-specific digital twins, akin to software replicas of engineered products, could integrate physiology, immunology, and real-time clinical data for predictive simulations. These digital twins, powered by AI, offer a promising tool against future pandemics, blending mechanistic knowledge with observational data [22,98].
DTs have been proposed to be used as avatars where individual simulations that match clinical criteria within a predefined margin of accuracy can be compared to real subjects. Avatars are particularly useful when an adequate population model is not feasible. Research focuses on generating avatars using pharmacometric models and exploring their properties to assess their impact on drug development stages. These avatars offer nuanced insights into a model’s ability to simulate data similar to observations at both population and individual levels [99,100,101]. Additionally, they can serve as diagnostic tools, alternatives to simulations with insurance, and measures of model fit. In another instance, DTs are utilized in single-cell RNA sequencing (scRNA-seq) to analyze time-series data in inflammatory diseases, revealing complex multi-directional gene expression networks. This complexity complicates the prioritization of upstream regulators (URs) crucial for understanding disease mechanisms and identifying potential drug targets. To address this, a quantitative approach prioritizing URs based on their predicted effects on downstream target cells has been developed, proving effective in various inflammatory diseases [23,102]. DTs are employed in high-throughput drug discovery (HDT) to enhance efficiency and reduce costs. HDT technology virtually represents organs, organ systems, and whole patients, informing target selection, drug delivery, and clinical trial design. DTs enable granular modeling of biological processes, facilitating target discovery and allowing for exploration of multiple targets for specific disease states.
Additionally, DTs replicate in vivo conditions in drug delivery to optimize solid-dosage drug parameters, decreasing costs and increasing manufacturing speed. Moreover, DTs partially virtualize control arms in clinical trials, reducing the number of physical patients needed and accelerating trial timelines, thus saving costs and expediting drug development [103,104,105].
DTs are also increasingly recognized for their potential to revolutionize various aspects of healthcare, particularly in clinical settings and drug development. These virtual replicas enable the generation of entire and realistic clinical patient trajectories, addressing the pressing need to expedite drug development processes. With only one out of ten compounds entering clinical trials achieving regulatory approval, the efficiency of phase 1 clinical trials becomes paramount. These trials aim to ascertain the efficacy and safety of compounds based on patient data, yet around 80% of them face delays due to patient enrolment issues. DTs offer a solution by augmenting clinical trials with patient replicas, significantly accelerating timelines and enhancing quality. Leveraging DT-generated data can minimize patient recruitment processes, particularly in rare conditions or oncology trials where DTs simulate comparator arms, enabling earlier efficacy assessments. Ultimately, DTs increase statistical power through simulated data, expediting clinical decision-making processes [20]. Expanding beyond their traditional application in manufacturing, DTs hold promise as integrative systems that incorporate information from diverse scientific and clinical sources to represent complex biological networks.
A notable example is the development of a digital twin of the liver, integrating knowledge gleaned from studying various liver functions, diseases, and drug effects. Based on a mathematical framework of ordinary differential equations, this twin effectively reproduces normal liver function, disease progression, and treatment impacts. Moreover, coupling the twin with experimental measurements provides valuable insights into drug-induced liver injury. This approach, applicable to other organs and biological systems, offers a generalizable strategy to enhance drug development efficiency and safety across diverse therapeutic areas [106].
8. Challenges and Considerations of Generative AI and Digital Twins in Drug Development
Generative AI and digital twins offer promising avenues for revolutionizing drug development, yet their adoption is not without significant limitations and ethical considerations. One primary challenge lies in data privacy concerns, as these technologies heavily rely on vast amounts of sensitive patient data, raising ethical questions regarding consent, ownership, and protection. Additionally, the computational resources required for training and running generative AI models can be immense, posing financial and infrastructural barriers, particularly for smaller research institutions or resource-limited settings. Furthermore, the validation and regulatory approval process for drugs generated through these technologies can be arduous and time-consuming as regulatory bodies grapple with assessing the safety and efficacy of novel compounds produced by AI algorithms. Various instances have shown successful identification of potential drug candidates using generative AI, juxtaposed with challenges in replicating and validating these findings in clinical trials. Ethically, using generative AI and digital twins raises concerns about patient data privacy, algorithmic bias, and the equitable distribution of benefits. Without robust safeguards and transparent consent mechanisms, there’s a risk of unauthorized access, data breaches, and the exploitation of patient data for commercial gain.
The potential for algorithmic bias to perpetuate disparities in healthcare outcomes must be carefully addressed through rigorous validation and ongoing monitoring. Moreover, ensuring equitable access to the benefits of these technologies requires navigating complex ethical and regulatory landscapes while safeguarding against exploitation and discrimination.
Additionally, suppose a healthcare provider integrates a digital twin system into their decision-making process for personalized medicine. However, if the underlying algorithms exhibit biases due to skewed training data or flawed assumptions, there is a risk of perpetuating disparities in healthcare outcomes. For example, if the digital twin system recommends treatments based on historical data that disproportionately favor certain demographic groups, it could exacerbate healthcare disparities rather than mitigate them. Ethical considerations dictate that AI-driven processes in drug development should be transparent and subject to independent review to ensure accountability and mitigate risks of bias or errors. Regulatory frameworks must evolve to accommodate the unique challenges of generative AI and digital twins in drug development, balancing innovation with patient safety and privacy. Democratizing access to these technologies within healthcare systems necessitates addressing infrastructure, expertise, and funding disparities while promoting transparency and collaboration across stakeholders.
Despite their potential, the limitations and potential drawbacks of generative AI and digital twins in drug discovery cannot be overlooked. Inadequate cybersecurity measures expose sensitive patient data to unauthorized access or cyberattacks. This raises ethical concerns about data security and patient privacy, emphasizing the importance of robust cybersecurity protocols and risk mitigation strategies in AI-driven healthcare systems. Strategies to mitigate these challenges include prioritizing data privacy and security, investing in computational infrastructure, fostering interdisciplinary collaboration, and promoting ethical best practices throughout the drug development pipeline. Ultimately, navigating these technologies’ ethical and regulatory complexities is essential to harnessing their full potential for advancing healthcare and improving patient outcomes.
9. Conclusions
The advancement of generative AI has significantly transformed the landscape of drug discovery, small molecule design, and clinical trials. With various model types tailored to different tasks, such as molecular generation, property optimization, and target identification, generative AI offers unprecedented efficiency and precision in drug development processes. Furthermore, integrating digital twins has revolutionized drug testing and development by providing virtual representations of patients, allowing for more accurate predictions of drug responses and potential side effects. Looking ahead, future research in this domain could explore enhanced synergies between generative AI and digital twins, potentially paving the way for personalized medicine on a scale previously unimaginable. Additionally, there is scope for deeper exploration into ethical considerations, regulatory frameworks, and the democratization of these technologies to ensure equitable access and responsible implementation in healthcare systems worldwide.
Beyond the remarkable strides already achieved, the future landscape of this domain holds even greater promise and complexity, necessitating a deeper exploration of emerging trends and interdisciplinary collaborations. One emerging trend poised to revolutionize generative AI in drug discovery is the integration of quantum computing. Quantum computers offer unparalleled computational power, capable of tackling complex optimization problems and simulating molecular interactions with unprecedented accuracy. In the realm of generative AI, quantum algorithms hold the potential to expedite molecular design processes, enabling the generation of novel drug candidates with enhanced precision and efficiency. Collaborations between experts in quantum computing, machine learning, and pharmaceutical sciences could unlock synergies that propel drug discovery into uncharted territories, accelerating the development of next-generation therapeutics.
Moreover, the integration of digital twins with real-world evidence (RWE) presents a compelling avenue for advancing personalized medicine. By leveraging vast patient data repositories, including electronic health records, genomic information, and wearables data, digital twins can be enriched with real-world insights that capture the complexities of individual patient profiles. Interdisciplinary partnerships between data scientists, clinicians, and healthcare providers can drive the seamless integration of digital twins with RWE, enabling clinicians to make data-driven decisions tailored to each patient’s unique characteristics and treatment responses. As we continue to harness the power of generative AI and digital twins, the possibilities for innovation in pharmaceutical research and development are boundless, promising a future of improved patient outcomes and transformative medical discoveries.
Author Contributions
S.K.N.: concept, structure; Z.M.: research, writing; M.M.: Research, review, and writing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
M.M. was employed by the company DEI Biopharma. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Turing, A.M. Computing Machinery and Intelligence. In The Essential Turing; Oxford Academic: Oxford, UK, 1950; Volume 59, pp. 433–460. [Google Scholar]
- Artificial Intelligence Coined at Dartmouth. Available online: https://home.dartmouth.edu/about/artificial-intelligence-ai-coined-dartmouth (accessed on 6 February 2024).
- Wiederhold, J.M.G. Arthur Samuel: Pioneer in Machine Learning. IBM J. Res. Dev. 1992, 36, 329–331. [Google Scholar] [CrossRef]
- Natale, S. The ELIZA Effect: Joseph Weizenbaum and the Emergence of Chatbots. In Deceitful Media: Artificial Intelligence and Social Life after the Turing Test; Oxford Academic: New York, NY, USA, 2021. [Google Scholar] [CrossRef]
- Bian, Y.; Wang, J.; Jun, J.J.; Xie, X.Q. Deep Convolutional Generative Adversarial Network (dcGAN) Models for Screening and Design of Small Molecules Targeting Cannabinoid Receptors. Mol. Pharm. 2019, 16, 4451–4460. [Google Scholar] [CrossRef] [PubMed]
- Parrot, M.; Tajmouati, H.; da Silva, V.B.R.; Atwood, B.R.; Fourcade, R.; Gaston-Mathe, Y.; Do Huu, N.; Perron, Q. Integrating synthetic accessibility with AI-based generative drug design. J. Cheminform. 2023, 15, 83. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.B.; Karmazyn-Raz, H. Episodes of experience and generative intelligence. Trends Cogn. Sci. 2022, 26, 1064–1065. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.H.D.; Chatterjee, S.; Rasmussen, L.K. Powering Hidden Markov Model by Neural Network based Generative Models. arXiv 2019. [Google Scholar] [CrossRef]
- Cao, S.L.Y.; Liu, Y.; Yan, Z.; Dai, Y.; Yu, P.S.; Sun, L. A Comprehensive Survey of AI-Generated Content (AIGC): A History of Generative AI from GAN to ChatGPT. arXiv 2023. [Google Scholar] [CrossRef]
- Lendasse, E.E.A. Gaussian Mixture Models for Time Series Modelling, Forecasting, and Interpolation. In Advances in Intelligent Data Analysis XII; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Jeong, J.J.; Tariq, A.; Adejumo, T.; Trivedi, H.; Gichoya, J.W.; Banerjee, I. Systematic Review of Generative Adversarial Networks (GANs) for Medical Image Classification and Segmentation. J. Digit. Imaging 2022, 35, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Skandarani, Y.; Lalande, A.; Afilalo, J.; Jodoin, P.M. Generative Adversarial Networks in Cardiology. Can. J. Cardiol. 2022, 38, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Motamed, S.; Rogalla, P.; Khalvati, F. Data augmentation using Generative Adversarial Networks (GANs) for GAN-based detection of Pneumonia and COVID-19 in chest X-ray images. Inform. Med. Unlocked 2021, 27, 100779. [Google Scholar] [CrossRef]
- Goulas, A.; Damicelli, F.; Hilgetag, C.C. Bio-instantiated recurrent neural networks: Integrating neurobiology-based network topology in artificial networks. Neural Netw. 2021, 142, 608–618. [Google Scholar] [CrossRef]
- Hossain, E.; Rana, R.; Higgins, N.; Soar, J.; Barua, P.D.; Pisani, A.R.; Turner, K. Natural Language Processing in Electronic Health Records in relation to healthcare decision-making: A systematic review. Comput. Biol. Med. 2023, 155, 106649. [Google Scholar] [CrossRef] [PubMed]
- Nath, S.; Marie, A.; Ellershaw, S.; Korot, E.; Keane, P.A. New meaning for NLP: The trials and tribulations of natural language processing with GPT-3 in ophthalmology. Br. J. Ophthalmol. 2022, 106, 889–892. [Google Scholar] [CrossRef] [PubMed]
- Jungmann, F.; Kuhn, S.; Kampgen, B. Basics and applications of Natural Language Processing (NLP) in radiology. Radiologe 2018, 58, 764–768. [Google Scholar] [CrossRef] [PubMed]
- An, G.; Cockrell, C. Drug Development Digital Twins for Drug Discovery, Testing and Repurposing: A Schema for Requirements and Development. Front. Syst. Biol. 2022, 2, 928387. [Google Scholar] [CrossRef] [PubMed]
- Bjornsson, B.; Borrebaeck, C.; Elander, N.; Gasslander, T.; Gawel, D.R.; Gustafsson, M.; Jornsten, R.; Lee, E.J.; Li, X.; Lilja, S.; et al. Digital twins to personalize medicine. Genome Med. 2019, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Bordukova, M.; Makarov, N.; Rodriguez-Esteban, R.; Schmich, F.; Menden, M.P. Generative artificial intelligence empowers digital twins in drug discovery and clinical trials. Expert Opin. Drug Discov. 2024, 19, 33–42. [Google Scholar] [CrossRef]
- Croatti, A.; Gabellini, M.; Montagna, S.; Ricci, A. On the Integration of Agents and Digital Twins in Healthcare. J. Med. Syst. 2020, 44, 161. [Google Scholar] [CrossRef] [PubMed]
- Laubenbacher, R.; Sluka, J.P.; Glazier, J.A. Using digital twins in viral infection. Science 2021, 371, 1105–1106. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lee, E.J.; Lilja, S.; Loscalzo, J.; Schafer, S.; Smelik, M.; Strobl, M.R.; Sysoev, O.; Wang, H.; Zhang, H.; et al. A dynamic single cell-based framework for digital twins to prioritize disease genes and drug targets. Genome Med. 2022, 14, 48. [Google Scholar] [CrossRef]
- Ward, T.M.; Mascagni, P.; Ban, Y.; Rosman, G.; Padoy, N.; Meireles, O.; Hashimoto, D.A. Computer vision in surgery. Surgery 2021, 169, 1253–1256. [Google Scholar] [CrossRef]
- Kather, J.N.; Ghaffari Laleh, N.; Foersch, S.; Truhn, D. Medical domain knowledge in domain-agnostic generative AI. npj Digit. Med. 2022, 5, 90. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Li, W.; Moon, H.; Roell, G.W.; Chen, Y.; Tang, Y.J. Generative Artificial Intelligence GPT-4 Accelerates Knowledge Mining and Machine Learning for Synthetic Biology. ACS Synth. Biol. 2023, 12, 2973–2982. [Google Scholar] [CrossRef]
- Di Masi, J.A.; Grabowski, H.G.; Hansen, R.W. Innovation in the pharmaceutical industry: New estimates of R&D costs. J. Health Econ. 2016, 47, 20–33. [Google Scholar] [CrossRef] [PubMed]
- MGI. Generative AI in the Pharmaceutical Industry: Moving from Hype to Reality. Available online: https://www.mckinsey.com/industries/life-sciences/our-insights/generative-ai-in-the-pharmaceutical-industry-moving-from-hype-to-reality#/ (accessed on 6 February 2024).
- Wenzel, M. Generative Adversarial Networks and Other Generative Models. In Machine Learning for Brain Disorders; Colliot, O., Ed.; Humana: New York, NY, USA, 2023; pp. 139–192. [Google Scholar]
- Li, C.; Xu, K.; Zhu, J.; Liu, J.; Zhang, B. Triple Generative Adversarial Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 9629–9640. [Google Scholar] [CrossRef]
- Zhong, G.; Gao, W.; Liu, Y.; Yang, Y.; Wang, D.H.; Huang, K. Generative adversarial networks with decoder-encoder output noises. Neural Netw. 2020, 127, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, A.E.; Stanley, C.; Bhowmik, D. Using GANs with adaptive training data to search for new molecules. J. Cheminform. 2021, 13, 14. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Li, K.; Shi, J. DGANDDI: Double Generative Adversarial Networks for Drug-Drug Interaction Prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2023, 20, 1854–1863. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Anees, A.; Das, A.; Nguyen, B.P.; Marzuki, M.; Lin, S.; Wright, G.; Singhal, A. High-content image generation for drug discovery using generative adversarial networks. Neural Netw. 2020, 132, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Bian, Y.; Xie, X.Q. Generative chemistry: Drug discovery with deep learning generative models. J. Mol. Model. 2021, 27, 71. [Google Scholar] [CrossRef]
- Tong, X.; Liu, X.; Tan, X.; Li, X.; Jiang, J.; Xiong, Z.; Xu, T.; Jiang, H.; Qiao, N.; Zheng, M. Generative Models for De Novo Drug Design. J. Med. Chem. 2021, 64, 14011–14027. [Google Scholar] [CrossRef]
- Lin, E.; Lin, C.H.; Lane, H.Y. De Novo Peptide and Protein Design Using Generative Adversarial Networks: An Update. J. Chem. Inf. Model. 2022, 62, 761–774. [Google Scholar] [CrossRef]
- Macedo, B.; Ribeiro Vaz, I.; Taveira Gomes, T. MedGAN: Optimized generative adversarial network with graph convolutional networks for novel molecule design. Sci. Rep. 2024, 14, 1212. [Google Scholar] [CrossRef]
- Zhang, Z.; Cui, P.; Zhu, W. Deep Learning on Graphs: A Survey. IEEE Pulse 2022, 34, 249–270. [Google Scholar] [CrossRef]
- Lin, E.; Lin, C.H.; Lane, H.Y. Relevant Applications of Generative Adversarial Networks in Drug Design and Discovery: Molecular De Novo Design, Dimensionality Reduction, and De Novo Peptide and Protein Design. Molecules 2020, 25, 3250. [Google Scholar] [CrossRef]
- Kao, P.Y.; Yang, Y.C.; Chiang, W.Y.; Hsiao, J.Y.; Cao, Y.; Aliper, A.; Ren, F.; Aspuru-Guzik, A.; Zhavoronkov, A.; Hsieh, M.H.; et al. Exploring the Advantages of Quantum Generative Adversarial Networks in Generative Chemistry. J. Chem. Inf. Model. 2023, 63, 3307–3318. [Google Scholar] [CrossRef]
- Niu, M.Y.; Zlokapa, A.; Broughton, M.; Boixo, S.; Mohseni, M.; Smelyanskyi, V.; Neven, H. Entangling Quantum Generative Adversarial Networks. Phys. Rev. Lett. 2022, 128, 220505. [Google Scholar] [CrossRef]
- Tian, J.; Sun, X.; Du, Y.; Zhao, S.; Liu, Q.; Zhang, K.; Yi, W.; Huang, W.; Wang, C.; Wu, X.; et al. Recent Advances for Quantum Neural Networks in Generative Learning. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 12321–12340. [Google Scholar] [CrossRef]
- Lim, J.; Ryu, S.; Kim, J.W.; Kim, W.Y. Molecular generative model based on conditional variational autoencoder for de novo molecular design. J. Cheminform. 2018, 10, 31. [Google Scholar] [CrossRef]
- Marino, J. Predictive Coding, Variational Autoencoders, and Biological Connections. Neural Comput. 2021, 34, 1–44. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Y.; Li, H.; Liu, X. Drug-protein interaction prediction via variational autoencoders and attention mechanisms. Front. Genet. 2022, 13, 1032779. [Google Scholar] [CrossRef]
- Li, T.; Zhao, X.M.; Li, L. Co-VAE: Drug-Target Binding Affinity Prediction by Co-Regularized Variational Autoencoders. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 8861–8873. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, S.; Yu, L. Predicting new drug indications based on double variational autoencoders. Comput. Biol. Med. 2023, 164, 107261. [Google Scholar] [CrossRef]
- Ochiai, T.; Inukai, T.; Akiyama, M.; Furui, K.; Ohue, M.; Matsumori, N.; Inuki, S.; Uesugi, M.; Sunazuka, T.; Kikuchi, K.; et al. Variational autoencoder-based chemical latent space for large molecular structures with 3D complexity. Commun. Chem. 2023, 6, 249. [Google Scholar] [CrossRef]
- Chow, Y.L.; Singh, S.; Carpenter, A.E.; Way, G.P. Predicting drug polypharmacology from cell morphology readouts using variational autoencoder latent space arithmetic. PLoS Comput. Biol. 2022, 18, e1009888. [Google Scholar] [CrossRef]
- Ang, D.; Rakovski, C.; Atamian, H.S. De Novo Drug Design Using Transformer-Based Machine Translation and Reinforcement Learning of an Adaptive Monte Carlo Tree Search. Pharmaceuticals 2024, 17, 161. [Google Scholar] [CrossRef]
- Grechishnikova, D. Transformer neural network for protein-specific de novo drug generation as a machine translation problem. Sci. Rep. 2021, 11, 321. [Google Scholar] [CrossRef]
- Kalakoti, Y.; Yadav, S.; Sundar, D. TransDTI: Transformer-Based Language Models for Estimating DTIs and Building a Drug Recommendation Workflow. ACS Omega 2022, 7, 2706–2717. [Google Scholar] [CrossRef]
- Shiju, A.; He, Z. Classifying Drug Ratings Using User Reviews with Transformer-Based Language Models. In Proceedings of the 2022 IEEE 10th International Conference on Healthcare Informatics (ICHI), Rochester, MN, USA, 11–14 June 2022; Volume 2022, pp. 163–169. [Google Scholar] [CrossRef]
- Zhang, S.; Fan, R.; Liu, Y.; Chen, S.; Liu, Q.; Zeng, W. Applications of transformer-based language models in bioinformatics: A survey. Bioinform. Adv. 2023, 3, vbad001. [Google Scholar] [CrossRef]
- Jiang, L.; Jiang, C.; Yu, X.; Fu, R.; Jin, S.; Liu, X. DeepTTA: A transformer-based model for predicting cancer drug response. Brief. Bioinform. 2022, 23, bbac100. [Google Scholar] [CrossRef]
- Hu, J.; Gao, J.; Fang, X.; Liu, Z.; Wang, F.; Huang, W.; Wu, H.; Zhao, G. DTSyn: A dual-transformer-based neural network to predict synergistic drug combinations. Brief. Bioinform. 2022, 23, bbac302. [Google Scholar] [CrossRef]
- Mao, J.; Wang, J.; Zeb, A.; Cho, K.H.; Jin, H.; Kim, J.; Lee, O.; Wang, Y.; No, K.T. Transformer-Based Molecular Generative Model for Antiviral Drug Design. J. Chem. Inf. Model. 2023, 64, 2733–2745. [Google Scholar] [CrossRef]
- Vaswani, N.S.A.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. arXiv 2017. [Google Scholar] [CrossRef]
- Hugo Larochelle, P. Classification Using Discriminative Restricted Boltzmann Machines. In Proceedings of the 25th International Conference on Machine Learning, Helsinki, Finland, 5–9 July 2008. [Google Scholar] [CrossRef]
- Max Welling, G.E.H. A New Learning Algorithm for Mean Field Boltzmann Machines. In Artificial Neural Networks—ICANN 2002; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar] [CrossRef]
- Salakhutdinov, R.; Mnih, A.; Hinton, G. Restricted Boltzmann Machines for Collaborative Filtering. In Proceedings of the 24th International Conference on Machine Learning, Corvalis, OR, USA, 20–24 June 2007. [Google Scholar]
- Wang, Y.; Zeng, J. Predicting drug-target interactions using restricted Boltzmann machines. Bioinformatics 2013, 29, i126–i134. [Google Scholar] [CrossRef]
- Qian, Y.; Ding, Y.; Zou, Q.; Guo, F. Identification of drug-side effect association via restricted Boltzmann machines with penalized term. Brief. Bioinform. 2022, 23, bbac458. [Google Scholar] [CrossRef]
- Cheng, X.; Qu, J.; Song, S.; Bian, Z. Neighborhood-based inference and restricted Boltzmann machine for microbe and drug associations prediction. PeerJ 2022, 10, e13848. [Google Scholar] [CrossRef]
- Bongini, P.; Messori, E.; Pancino, N.; Bianchini, M. A Deep Learning Approach to the Prediction of Drug Side-Effects on Molecular Graphs. IEEE/ACM Trans. Comput. Biol. Bioinform. 2023, 20, 3681–3690. [Google Scholar] [CrossRef]
- Wu, Z.; Pan, S.; Chen, F.; Long, G.; Zhang, C.; Yu, P.S. A Comprehensive Survey on Graph Neural Networks. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 4–24. [Google Scholar] [CrossRef]
- Abate, C.; Decherchi, S.; Cavalli, A. Graph neural networks for conditional de novo drug design. WIREs Comput. Mol. Sci. 2023, 13, e1651. [Google Scholar] [CrossRef]
- Zhou, G.C.J.; Hu, S.; Zhang, Z.; Yang, C.; Liu, Z.; Wang, L.; Li, C.; Sun, M. Graph neural networks: A review of methods and applications. AI Open 2020, 1, 57–81. [Google Scholar] [CrossRef]
- Xiong, J.; Xiong, Z.; Chen, K.; Jiang, H.; Zheng, M. Graph neural networks for automated de novo drug design. Drug Discov. Today 2021, 26, 1382–1393. [Google Scholar] [CrossRef]
- Feng, J.; Liang, Y.; Yu, T. MM-GANN-DDI: Multimodal Graph-Agnostic Neural Networks for Predicting Drug-Drug Interaction Events. Comput. Biol. Med. 2023, 166, 107492. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, S.; Kv, P.; Balaji, S. Training recurrent neural networks as generative neural networks for molecular structures: How does it impact drug discovery? Expert Opin. Drug Discov. 2022, 17, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Hu, J.; Wang, Y.; Zhang, L.; Liu, Z. Graph-based generative models for de Novo drug design. Drug Discov. Today Technol. 2019, 32–33, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Moret, M.; Pachon Angona, I.; Cotos, L.; Yan, S.; Atz, K.; Brunner, C.; Baumgartner, M.; Grisoni, F.; Schneider, G. Leveraging molecular structure and bioactivity with chemical language models for de novo drug design. Nat. Commun. 2023, 14, 114. [Google Scholar] [CrossRef] [PubMed]
- Schenone, M.; Dancik, V.; Wagner, B.K.; Clemons, P.A. Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 2013, 9, 232–240. [Google Scholar] [CrossRef]
- Janakarajan, N.; Erdmann, T.; Swaminathan, S.; Laino, T.; Born, J. Language models in molecular discovery. arXiv 2023. [Google Scholar] [CrossRef]
- Bajorath, J. Chemical language models for molecular design. Mol. Inform. 2024, 43, e202300288. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Roberts, R.A.; Lal-Nag, M.; Chen, X.; Huang, R.; Tong, W. AI-based language models powering drug discovery and development. Drug Discov. Today 2021, 26, 2593–2607. [Google Scholar] [CrossRef]
- Giorgi, J.M.; Bader, G.D. Towards reliable named entity recognition in the biomedical domain. Bioinformatics 2020, 36, 280–286. [Google Scholar] [CrossRef]
- Blanco, A.; Perez-de-Vinaspre, O.; Perez, A.; Casillas, A. Boosting ICD multi-label classification of health records with contextual embeddings and label-granularity. Comput. Methods Programs Biomed. 2020, 188, 105264. [Google Scholar] [CrossRef]
- Dias, R.; Torkamani, A. Artificial intelligence in clinical and genomic diagnostics. Genome Med. 2019, 11, 70. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.L.; Ryan, J.J.; Bray, B.E.; Brown, C.; Lanfear, D.; Newby, L.K.; Relling, M.V.; Risch, N.J.; Roden, D.M.; Shaw, S.Y.; et al. Merging Electronic Health Record Data and Genomics for Cardiovascular Research: A Science Advisory from the American Heart Association. Circ. Cardiovasc. Genet. 2016, 9, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Harrer, S.; Shah, P.; Antony, B.; Hu, J. Artificial Intelligence for Clinical Trial Design. Trends Pharmacol. Sci. 2019, 40, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Wang, F.; Luo, Y.; Kang, S.-G.; Tang, J.; Lightstone, F.C.; Fang, E.F.; Cornell, W.; Nussinov, R.; Cheng, F. Deep generative molecular design reshapes drug discovery. Cell Rep. Med. 2022, 3, 100794. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Eran, A.; Palmer, N.; Avillach, P.; Levy-Moonshine, A.; Szolovits, P.; Kohane, I.S. A multidimensional precision medicine approach identifies an autism subtype characterized by dyslipidemia. Nat. Med. 2020, 26, 1375–1379. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, H.; Liu, W.; Lasenby, J.; Guo, H.; Tang, J. Pre-training Molecular Graph Representation with 3D Geometry. arXiv 2021. [Google Scholar] [CrossRef]
- Manica, M.; Oskooei, A.; Born, J.; Subramanian, V.; Sáez-Rodríguez, J.; Martínez, M.R. Toward explainable anticancer compound sensitivity prediction via multimodal attention-based convolutional encoders. Mol. Pharm. 2019, 16, 4797–4806. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Yang, K.; Barzilay, R.; Jaakkola, T. Learning Multimodal Graph-to-Graph Translation for Molecular Optimization. arXiv 2018. [Google Scholar] [CrossRef]
- Ma, M.; Ren, J.; Zhao, L.; Tulyakov, S.; Wu, C.; Peng, X. SMIL: Multimodal Learning with Severely Missing Modality. AAAI Tech. Track Comput. Vis. II 2021, 35, 2302–2310. [Google Scholar] [CrossRef]
- Baltrusaitis, T.; Ahuja, C.; Morency, L.P. Multimodal Machine Learning: A Survey and Taxonomy. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 41, 423–443. [Google Scholar] [CrossRef]
- Steyaert, S.; Pizurica, M.; Nagaraj, D.; Khandelwal, P.; Hernandez-Boussard, T.; Gentles, A.J.; Gevaert, O. Multimodal data fusion for cancer biomarker discovery with deep learning. Nat. Mach. Intell. 2023, 5, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Liu, X.Y.; Yang, K.; Huang, K.; Hong, M.; Zhang, J.; Wu, Y.; Nie, Z. Toward Unified AI Drug Discovery with Multimodal Knowledge. Health Data Sci. 2024, 4, 0113. [Google Scholar] [CrossRef] [PubMed]
- Skalic, M.; Sabbadin, D.; Sattarov, B.; Sciabola, S.; De Fabritiis, G. From Target to Drug: Generative Modeling for the Multimodal Structure-Based Ligand Design. Mol. Pharm. 2019, 16, 4282–4291. [Google Scholar] [CrossRef] [PubMed]
- Buchberger, A.R.; DeLaney, K.; Johnson, J.; Li, L. Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights. Anal. Chem. 2018, 90, 240–265. [Google Scholar] [CrossRef] [PubMed]
- Loscher, W. Single-Target Versus Multi-Target Drugs Versus Combinations of Drugs with Multiple Targets: Preclinical and Clinical Evidence for the Treatment or Prevention of Epilepsy. Front. Pharmacol. 2021, 12, 730257. [Google Scholar] [CrossRef] [PubMed]
- Marecek, R.; Riha, P.; Bartonova, M.; Kojan, M.; Lamos, M.; Gajdos, M.; Vojtisek, L.; Mikl, M.; Barton, M.; Dolezalova, I.; et al. Automated fusion of multimodal imaging data for identifying epileptogenic lesions in patients with inconclusive magnetic resonance imaging. Hum. Brain Mapp. 2021, 42, 2921–2930. [Google Scholar] [CrossRef] [PubMed]
- Laubenbacher, R.; Niarakis, A.; Helikar, T.; An, G.; Shapiro, B.; Malik-Sheriff, R.S.; Sego, T.J.; Knapp, A.; Macklin, P.; Glazier, J.A. Building digital twins of the human immune system: Toward a roadmap. npj Digit. Med. 2022, 5, 64. [Google Scholar] [CrossRef] [PubMed]
- Cockrell, C.; An, G. Utilizing the Heterogeneity of Clinical Data for Model Refinement and Rule Discovery Through the Application of Genetic Algorithms to Calibrate a High-Dimensional Agent-Based Model of Systemic Inflammation. Front. Physiol. 2021, 12, 662845. [Google Scholar] [CrossRef] [PubMed]
- Polasek, T.M.; Rostami-Hodjegan, A. Virtual Twins: Understanding the Data Required for Model-Informed Precision Dosing. Clin. Pharmacol. Ther. 2020, 107, 742–745. [Google Scholar] [CrossRef]
- Patel, N.; Wisniowska, B.; Jamei, M.; Polak, S. Real Patient and its Virtual Twin: Application of Quantitative Systems Toxicology Modelling in the Cardiac Safety Assessment of Citalopram. AAPS J. 2017, 20, 6. [Google Scholar] [CrossRef]
- Chasseloup, E.; Hooker, A.C.; Karlsson, M.O. Generation and application of avatars in pharmacometric modelling. J. Pharmacokinet. Pharmacodyn. 2023, 50, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Shalek, A.K.; Benson, M. Single-cell analyses to tailor treatments. Sci. Transl. Med. 2017, 9, eaan4730. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, K.P.; Brito, G.; Kamel Boulos, M.N. Health Digital Twins in Life Science and Health Care Innovation. Annu. Rev. Pharmacol. Toxicol. 2024, 64, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Fogel, D.B. Factors associated with clinical trials that fail and opportunities for improving the likelihood of success: A review. Contemp. Clin. Trials Commun. 2018, 11, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Schutt, M.; Stamatopoulos, K.; Batchelor, H.K.; Simmons, M.J.H.; Alexiadis, A. Development of a digital twin of a tablet that mimics a real solid dosage form: Differences in the dissolution profile in conventional mini-USP II and a biorelevant colon model. Eur. J. Pharm. Sci. 2022, 179, 106310. [Google Scholar] [CrossRef]
- Subramanian, K. Digital Twin for Drug Discovery and Development—The Virtual Liver. J. Indian Inst. Sci. 2020, 100, 653–662. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).