Understanding the Molecular Actions of Spike Glycoprotein in SARS-CoV-2 and Issues of a Novel Therapeutic Strategy for the COVID-19 Vaccine
Abstract
1. Introduction
- (1)
- Maintaining research and development incentives: one challenge has been to sustain strong research and development incentives to drive vaccine innovation [30].
- (2)
- Financial investment and demand: while unprecedented financial investments and massive demand have accelerated vaccine development, they also pose challenges in terms of sustainability and equitable distribution [31].
- (3)
- Scientific innovations and regulatory reviews: previous scientific innovations, accelerated clinical development, and regulatory reviews have been crucial but also come with challenges in ensuring safety and efficacy standards are met.
- (4)
- Limitations in vaccination promotion: challenges exist in promoting vaccination due to factors like limitations in the target age population and breakthrough infections [32].
- (I)
- Rapid development: COVID-19 vaccines have been developed at an accelerated pace compared to traditional vaccines, with 232 vaccine candidates, 172 in preclinical development, and 60 in clinical development. This rapid development has aimed to significantly reduce the typical 10- to 15-year timeline to 12 to 24 months [33,34].
- (II)
- Vaccine types: Various types of COVID-19 vaccines have been developed, including mRNA vaccines like BNT162 by Pfizer/BioNTech and mRNA-1273 by Moderna, adenovirus vector vaccines like AstraZeneca and Jenssen, and inactivated killed vaccines like Sinopharm. These vaccines have shown high efficacy in preventing severe illness and death [35].
- (III)
- Challenges: The accelerated process of COVID-19 vaccine development and Emergency Use Authorization (EUA) have raised unanswered questions. Additionally, the emergence of new strains of SARS-CoV-2 has posed challenges for vaccine developers and governments worldwide [33].
- (IV)
- Safety concerns: Studies have shown that COVID-19 vaccines are generally safe, with lower rates of death among vaccinated individuals compared to those who are not vaccinated. Adverse events post-vaccination are generally mild to moderate, with severe reactions being rare. Adverse events associated with COVID-19 vaccines include rare effects like anaphylaxis, blood clots, myocarditis, pericarditis, hearing changes, and tinnitus. However, the overall risk of severe adverse effects is low, and healthcare professionals monitor and manage any reactions carefully [36].
- (V)
- Vaccine efficacy: Studies have shown that COVID-19 vaccines significantly reduce the risk of death from COVID-19 and its complications. Vaccinated individuals are not at a greater risk of death from non-COVID causes compared to unvaccinated individuals.
- (VI)
- Long-term effects: While concerns about long-term side effects exist, current data suggest that severe effects following vaccination are rare.
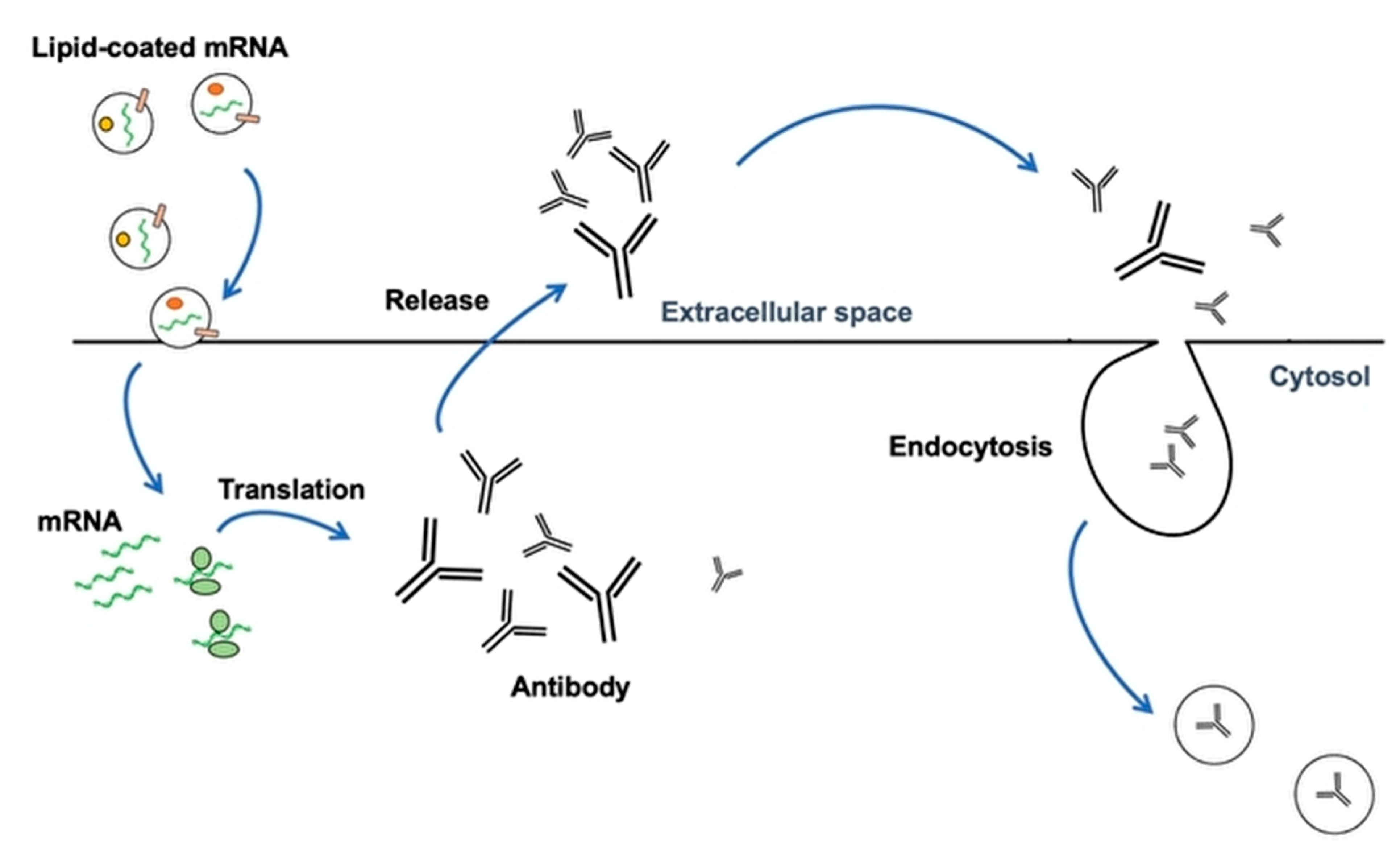
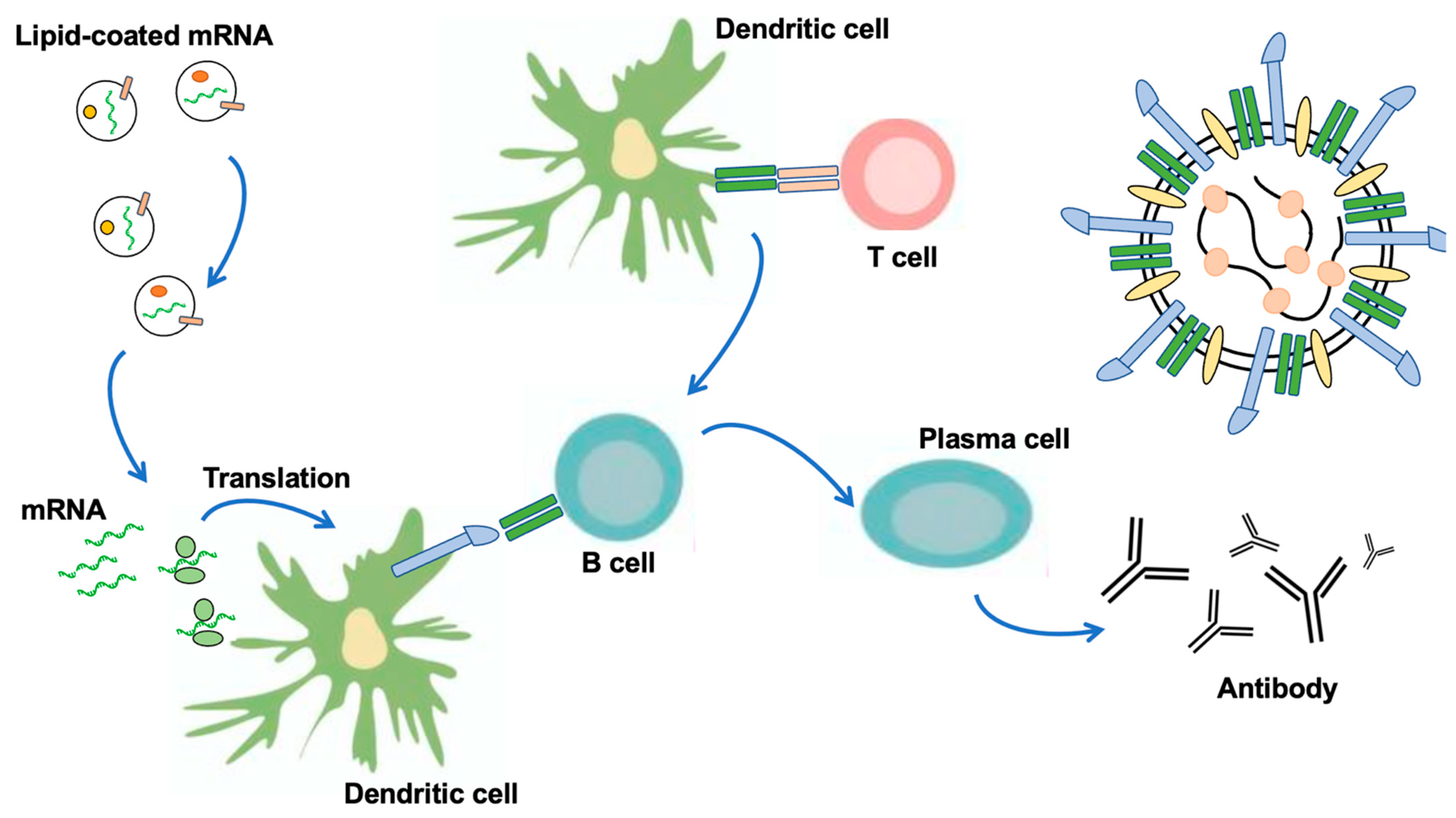
2. Vaccination with Nanoparticles (LNP) and Extracellular Vesicles as New DDS
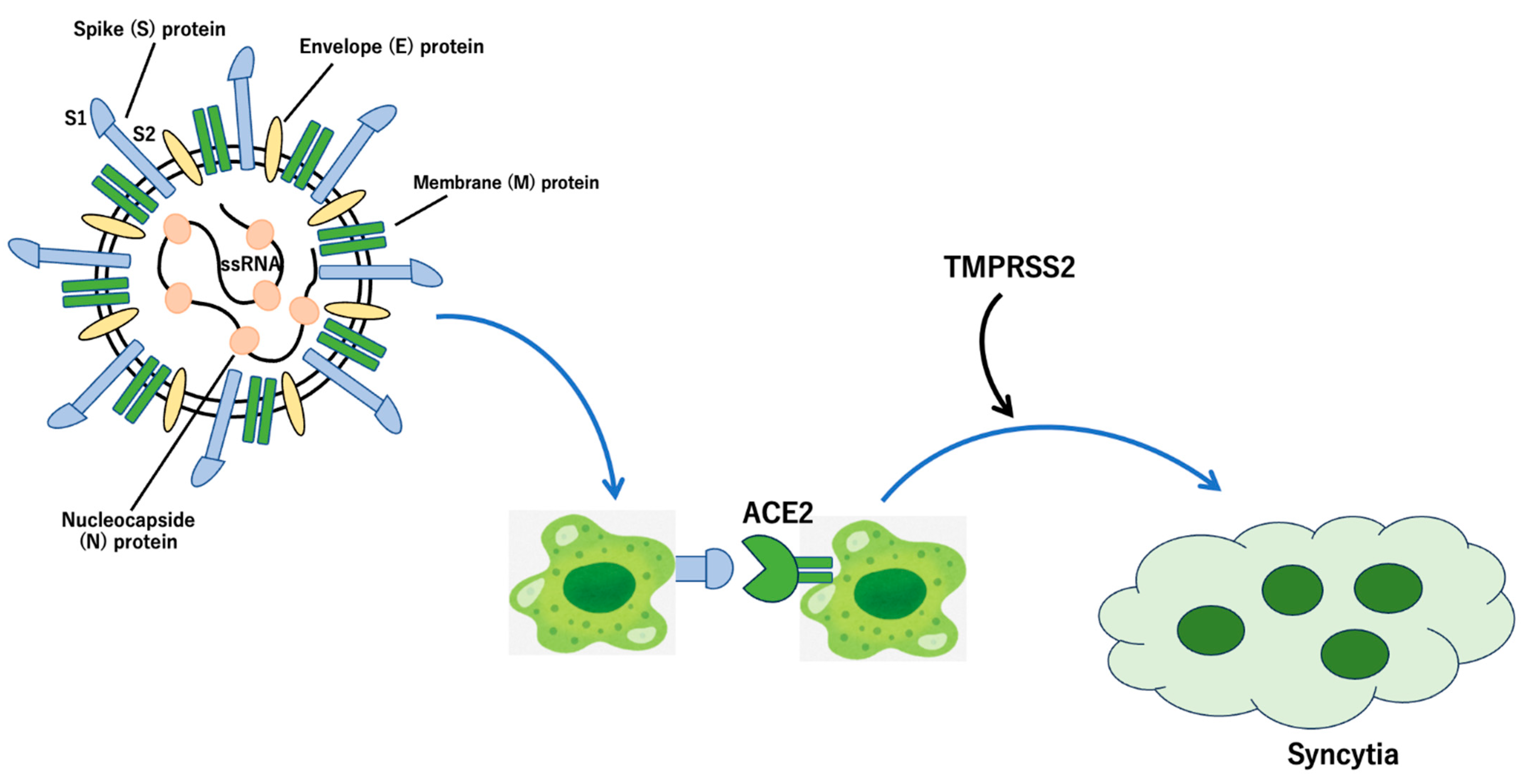
3. Spike Proteins and the Receptors of SARS-CoV-2
4. Fusion of the Viral Membrane with the Host Cell Membrane
5. RNA Genome and Translation in SARS-CoV-2
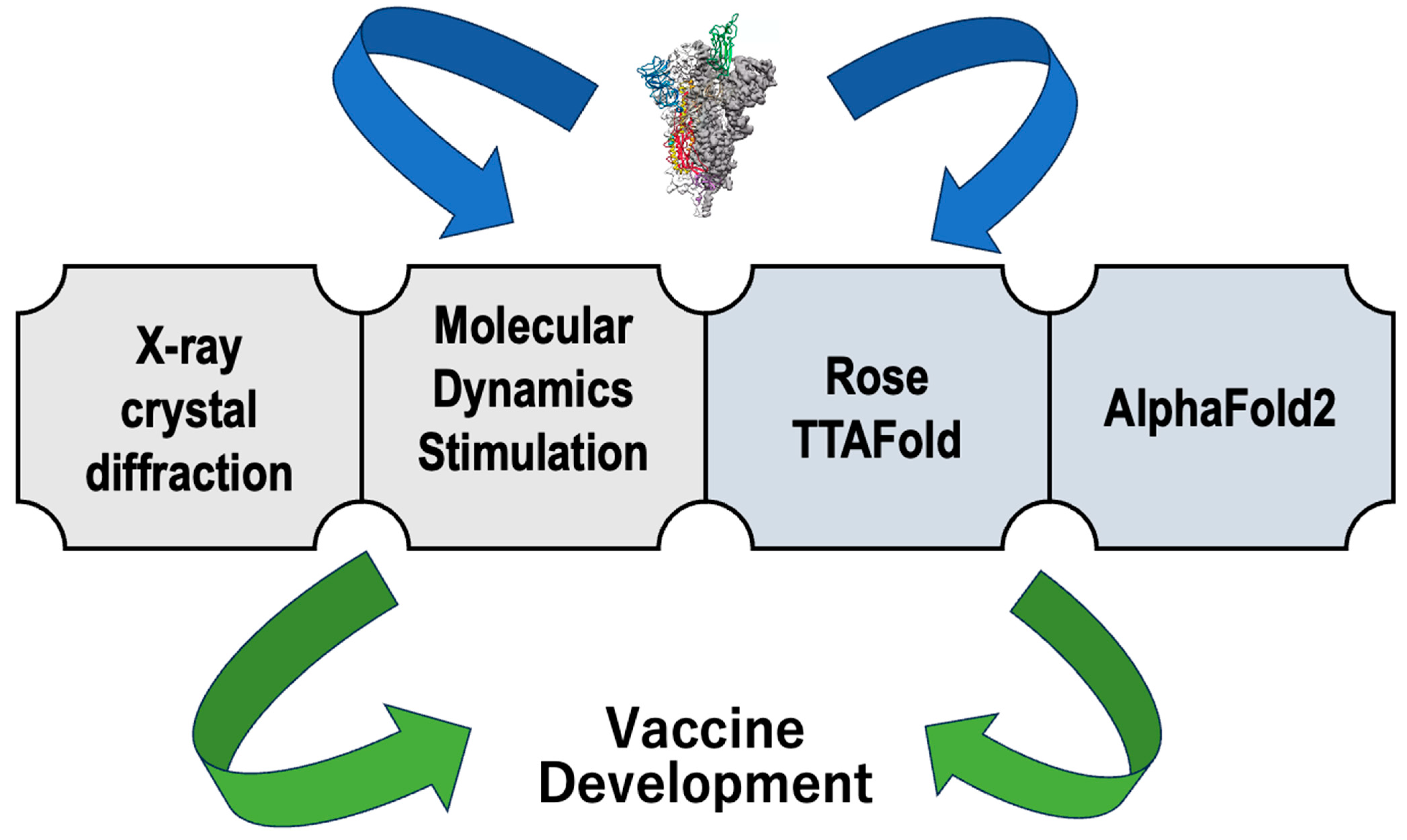
6. Importance of Structural Biology in SARS-CoV-2
7. Intracellular Changes with Virus Infections
- (1)
- (2)
- As for variants and mutations, the emergence of highly transmissible variants poses a significant challenge to vaccine efficacy. Mutations in the virus, especially in the S protein, can impact the effectiveness of existing vaccines against new variants [81].
- (3)
- As for glycosylation patterns, variations in the glycosylation patterns of the S protein can affect vaccine efficacy. Different degrees of glycosylation can influence binding reactivity to antibodies and the induction of immune responses, potentially impacting vaccine effectiveness [9].
- (4)
- As for cellular immune response, while neutralizing antibodies are a primary target for vaccines, the importance of cellular immune responses, particularly T-cell immunity, in controlling SARS-CoV-2 infection is significant. Understanding and harnessing these cellular responses are crucial for comprehensive vaccine development [81].
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sharma, A.; Virmani, T.; Pathak, V.; Sharma, A.; Pathak, K.; Kumar, G.; Pathak, D. Artificial Intelligence-Based Data-Driven Strategy to Accelerate Research, Development, and Clinical Trials of COVID Vaccine. BioMed Res. Int. 2022, 2022, 7205241. [Google Scholar] [CrossRef] [PubMed]
- Pollard, A.J.; Bijker, E.M. A guide to vaccinology: From basic principles to new developments. Nat. Rev. Immunol. 2021, 21, 83–100. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Flores, D.; Zepeda-Cervantes, J.; Cruz-Reséndiz, A.; Aguirre-Sampieri, S.; Sampieri, A.; Vaca, L. SARS-CoV-2 Vaccines Based on the Spike Glycoprotein and Implications of New Viral Variants. Front. Immunol. 2021, 12, 701501. [Google Scholar] [CrossRef] [PubMed]
- Mittal, A.; Khattri, A.; Verma, V. Structural and antigenic variations in the spike protein of emerging SARS-CoV-2 variants. PLoS Pathog. 2022, 18, e1010260. [Google Scholar] [CrossRef] [PubMed]
- Malik, Y.S.; Kumar, P.; Ansari, M.I.; Hemida, M.G.; El Zowalaty, M.E.; Abdel-Moneim, A.S.; Ganesh, B.; Salajegheh, S.; Natesan, S.; Sircar, S.; et al. SARS-CoV-2 Spike Protein Extrapolation for COVID Diagnosis and Vaccine Development. Front. Mol. Biosci. 2021, 8, 607886. [Google Scholar] [CrossRef] [PubMed]
- Heidary, M.; Kaviar, V.H.; Shirani, M.; Ghanavati, R.; Motahar, M.; Sholeh, M.; Ghahramanpour, H.; Khoshnood, S. A Comprehensive Review of the Protein Subunit Vaccines Against COVID-19. Front. Microbiol. 2022, 13, 927306. [Google Scholar] [CrossRef] [PubMed]
- Delgado, J.F.; Vidal-Pla, M.; Moya, M.C.; Espasa, M.; Casabella, A.; Seda, M.; Calvet, J.; Gratacós, J.; Serrano, R.M.; Peña, P. SARS-CoV-2 Spike Protein Vaccine-Induced Immune Imprinting Reduces Nucleocapsid Protein Antibody Response in SARS-CoV-2 Infection. J. Immunol. Res. 2022, 2022, 8287087. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Smatti, M.K.; Ouhtit, A.; Cyprian, F.S.; Almaslamani, M.A.; Thani, A.A.; Yassine, H.M. Antibody-Dependent Enhancement (ADE) and the role of complement system in disease pathogenesis. Mol. Immunol. 2022, 152, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Trougakos, I.P.; Terpos, E.; Alexopoulos, H.; Politou, M.; Paraskevis, D.; Scorilas, A.; Kastritis, E.; Andreakos, E.; Dimopoulos, M.A. Adverse effects of COVID-19 mRNA vaccines: The spike hypothesis. Trends Mol. Med. 2022, 28, 542–554. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zai, X.; Zhang, Z.; Xu, J.; Chen, W. Challenges and developments in universal vaccine design against SARS-CoV-2 variants. NPJ Vaccines 2022, 7, 167. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. How AlphaFold and other AI tools could help us prepare for the next pandemic. Nature 2023, 622, 440–441. [Google Scholar] [CrossRef] [PubMed]
- Teijaro, J.R.; Farber, D.L. COVID-19 vaccines: Modes of immune activation and future challenges. Nat. Rev. Immunol. 2021, 21, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Hafiz, I.; Illian, D.N.; Meila, O.; Utomo, A.R.H.; Susilowati, A.; Susetya, I.E.; Desrita, D.; Siregar, G.A.; Basyuni, M. Effectiveness and Efficacy of Vaccine on Mutated SARS-CoV-2 Virus and Post Vaccination Surveillance: A Narrative Review. Vaccines 2022, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, W.G. Impact of virus genetic variability and host immunity for the success of COVID-19 vaccines. Biomed. Pharmacother. 2021, 136, 111272. [Google Scholar] [CrossRef] [PubMed]
- Ragonnet-Cronin, M.; Nutalai, R.; Huo, J.; Dijokaite-Guraliuc, A.; Das, R.; Tuekprakhon, A.; Supasa, P.; Liu, C.; Selvaraj, M.; Groves, N.; et al. Generation of SARS-CoV-2 escape mutations by monoclonal antibody therapy. Nat. Commun. 2023, 14, 3334. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Sharma, A.R.; Bhattacharya, M.; Lee, S.S. A Detailed Overview of Immune Escape, Antibody Escape, Partial Vaccine Escape of SARS-CoV-2 and Their Emerging Variants with Escape Mutations. Front. Immunol. 2022, 13, 801522. [Google Scholar] [CrossRef] [PubMed]
- Starr, T.N.; Greaney, A.J.; Addetia, A.; Hannon, W.W.; Choudhary, M.C.; Dingens, A.S.; Li, J.Z.; Bloom, J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 2021, 371, 850–854. [Google Scholar] [CrossRef] [PubMed]
- Baloch, Z.; Ikram, A.; Hakim, M.S.; Awan, F.M. The Impact of Mutations on the Pathogenic and Antigenic Activity of SARS-CoV-2 during the First Wave of the COVID-19 Pandemic: A Comprehensive Immunoinformatics Analysis. Vaccines 2021, 9, 1410. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A. COVID-19 Genomics UK (COG-UK) Consortium; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Xu, S.; Sun, W.; Li, Q.; Wang, S.; Shi, H.; Liu, X. HA gene amino acid mutations contribute to antigenic variation and immune escape of H9N2 influenza virus. Vet. Res. 2022, 53, 43. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.S.; Reich, A.N.; Gaglione, S.; Smith, B.E.; Kim, E.J.; Dong, J.; Ronsard, L.; Okonkwo, V.; Lingwood, D.; Dougan, M.; et al. Antigen identification and high-throughput interaction mapping by reprogramming viral entry. Nat. Methods. 2022, 19, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Chen, N.; Xu, X.; Sahil, A.; Zhou, J.; Li, Z.; Zhong, H.; Gao, E.; Zhang, R.; Wang, Y.; et al. Predicting the antigenic evolution of SARS-COV-2 with deep learning. Nat. Commun. 2023, 14, 3478. [Google Scholar] [CrossRef] [PubMed]
- Umitaibatin, R.; Harisna, A.H.; Jauhar, M.M.; Syaifie, P.H.; Arda, A.G.; Nugroho, D.W.; Ramadhan, D.; Mardliyati, E.; Shalannanda, W.; Anshori, I. Immunoinformatics Study: Multi-Epitope Based Vaccine Design from SARS-CoV-2 Spike Glycoprotein. Vaccines 2023, 11, 399. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Bogdan, P.; Nazarian, S. An in silico deep learning approach to multi-epitope vaccine design: A SARS-CoV-2 case study. Sci. Rep. 2021, 11, 3238. [Google Scholar] [CrossRef] [PubMed]
- Rafi, M.O.; Al-Khafaji, K.; Sarker, M.T.; Taskin-Tok, T.; Rana, A.S.; Rahman, M.S. Design of a multi-epitope vaccine against SARS-CoV-2: Immunoinformatic and computational methods. RSC Adv. 2022, 12, 4288–4310. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Jiang, H.; Qiu, M.; Liu, L.; Zou, S.; Li, Y.; Guo, Q.; Han, N.; Sun, Y.; Wang, K.; et al. Multi-Epitope Vaccine Design Using an Immunoinformatic Approach for SARS-CoV-2. Pathogens 2021, 10, 737. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, S.; Rafi, M.O.; Paul, G.K.; Promi, M.M.; Shimu, M.S.S.; Biswas, S.; Emran, T.B.; Dhama, K.; Alyami, S.A.; Moni, M.A.; et al. Designing a multi-epitope vaccine candidate to combat MERS-CoV by employing an immunoinformatics approach. Sci. Rep. 2021, 11, 15431. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.C.; Olsen, K.S.; Gentry, K.M.; Sambade, M.; Beck, W.; Garness, J.; Entwistle, S.; Willis, C.; Vensko, S.; Woods, A.; et al. Landscape and selection of vaccine epitopes in SARS-CoV-2. Genome Med. 2021, 13, 101. [Google Scholar] [CrossRef] [PubMed]
- Palma, M. Epitopes and Mimotopes Identification Using Phage Display for Vaccine Development against Infectious Pathogens. Vaccines 2023, 11, 1176. [Google Scholar] [CrossRef] [PubMed]
- Forman, R.; Shah, S.; Jeurissen, P.; Jit, M.; Mossialos, E. COVID-19 vaccine challenges: What have we learned so far and what remains to be done? Health Policy 2021, 125, 553–567. [Google Scholar] [CrossRef] [PubMed]
- Excler, J.L.; Saville, M.; Privor-Dumm, L.; Gilbert, S.; Hotez, P.J.; Thompson, D.; Abdool-Karim, S.; Kim, J.H. Factors, enablers and challenges for COVID-19 vaccine development. BMJ Glob. Health 2023, 8, e011879. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.C.; Rai, C.I.; Chen, Y.C. Challenges and Recent Advancements in COVID-19 Vaccines. Microorganisms 2023, 11, 787. [Google Scholar] [CrossRef] [PubMed]
- Kashte, S.; Gulbake, A.; El-Amin Iii, S.F.; Gupta, A. COVID-19 vaccines: Rapid development, implications, challenges and future prospects. Hum. Cell 2021, 34, 711–733. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Pang, Y.; Lyu, Z.; Wang, R.; Wu, X.; You, C.; Zhao, H.; Manickam, S.; Lester, E.; Wu, T.; et al. The COVID-19 Vaccines: Recent Development, Challenges and Prospects. Vaccines 2021, 9, 349. [Google Scholar] [CrossRef] [PubMed]
- Amer, S.A.; Al-Zahrani, A.; Imam, E.A.; Ishteiwy, E.M.; Djelleb, I.F.; Abdullh, L.R.; Ballaj, D.; Amer, Y.A.; El-Sokkary, R.H.; Elshabrawy, A.M.; et al. Exploring the reported adverse effects of COVID-19 vaccines among vaccinated Arab populations: A multi-national survey study. Sci. Rep. 2024, 14, 4785. [Google Scholar] [CrossRef] [PubMed]
- Kaura, A.; Trickey, A.; Shah, A.S.V.; Benedetto, U.; Glampson, B.; Mulla, A.; Mercuri, L.; Gautama, S.; Costelloe, C.E.; Goodman, I.; et al. Comparing the longer-term effectiveness of a single dose of the Pfizer-BioNTech and Oxford-AstraZeneca COVID-19 vaccines across the age spectrum. EClinicalMedicine. 2022, 46, 101344. [Google Scholar] [CrossRef] [PubMed]
- Sa, S.; Lee, C.W.; Shim, S.R.; Yoo, H.; Choi, J.; Kim, J.H.; Lee, K.; Hong, M.; Han, H.W. The Safety of mRNA-1273, BNT162b2 and JNJ-78436735 COVID-19 Vaccines: Safety Monitoring for Adverse Events Using Real-World Data. Vaccines 2022, 10, 320. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Khan, S.; Imran, I.; Al Mughairbi, F.; Sheikh, F.S.; Hussain, J.; Khan, A.; Al-Harrasi, A. Vaccine Development against COVID-19: Study from Pre-Clinical Phases to Clinical Trials and Global Use. Vaccines 2021, 9, 836. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Minn, D.; Chang, S.H.; Suh, J.S. Comparing SARS-CoV-2 Antibody Responses after Various COVID-19 Vaccinations in Healthcare Workers. Vaccines 2022, 10, 193. [Google Scholar] [CrossRef] [PubMed]
- Firouzabadi, N.; Ghasemiyeh, P.; Moradishooli, F.; Mohammadi-Samani, S. Update on the effectiveness of COVID-19 vaccines on different variants of SARS-CoV-2. Int. Immunopharmacol. 2023, 117, 109968. [Google Scholar] [CrossRef] [PubMed]
- Kyriakidis, N.C.; López-Cortés, A.; González, E.V.; Grimaldos, A.B.; Prado, E.O. SARS-CoV-2 vaccines strategies: A comprehensive review of phase 3 candidates. NPJ Vaccines 2021, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Beatty, A.L.; Peyser, N.D.; Butcher, X.E.; Cocohoba, J.M.; Lin, F.; Olgin, J.E.; Pletcher, M.J.; Marcus, G.M. Analysis of COVID-19 Vaccine Type and Adverse Effects Following Vaccination. JAMA Netw. Open. 2021, 4, e2140364. [Google Scholar] [CrossRef] [PubMed]
- Yau, K.; Chan, C.T.; Abe, K.T.; Jiang, Y.; Atiquzzaman, M.; Mullin, S.I.; Shadowitz, E.; Liu, L.; Kostadinovic, E.; Sukovic, T.; et al. Differences in mRNA-1273 (Moderna) and BNT162b2 (Pfizer-BioNTech) SARS-CoV-2 vaccine immunogenicity among patients undergoing dialysis. CMAJ 2022, 194, E297–E305. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.; Hasan, M.R.; Chemaitelly, H.; Yassine, H.M.; Benslimane, F.M.; Al Khatib, H.A.; AlMukdad, S.; Coyle, P.; Ayoub, H.H.; Al Kanaani, Z.; et al. BNT162b2 and mRNA-1273 COVID-19 vaccine effectiveness against the SARS-CoV-2 Delta variant in Qatar. Nat. Med. 2021, 27, 2136–2143. [Google Scholar] [CrossRef] [PubMed]
- Dickerman, B.A.; Gerlovin, H.; Madenci, A.L.; Kurgansky, K.E.; Ferolito, B.R.; Figueroa Muñiz, M.J.; Gagnon, D.R.; Gaziano, J.M.; Cho, K.; Casas, J.P.; et al. Comparative Effectiveness of BNT162b2 and mRNA-1273 Vaccines in U.S. Veterans. N. Engl. J. Med. 2022, 386, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Yaamika, H.; Muralidas, D.; Elumalai, K. Review of adverse events associated with COVID-19 vaccines, highlighting their frequencies and reported cases. J. Taibah Univ. Med. Sci. 2023, 18, 1646–1661. [Google Scholar] [CrossRef] [PubMed]
- Byrne, P.O.; McLellan, J.S. Principles and practical applications of structure-based vaccine design. Curr. Opin. Immunol. 2022, 77, 102209. [Google Scholar] [CrossRef] [PubMed]
- Bryant, P.; Pozzati, G.; Elofsson, A. Improved prediction of protein-protein interactions using AlphaFold2. Nat. Commun. 2022, 13, 1265. [Google Scholar] [CrossRef] [PubMed]
- Hartley, S.M.; Tiernan, K.A.; Ahmetaj, G.; Cretu, A.; Zhuang, Y.; Zimmer, M. AlphaFold2 and RoseTTAFold predict posttranslational modifications. Chromophore formation in GFP-like proteins. PLoS ONE 2022, 17, e0267560. [Google Scholar] [CrossRef] [PubMed]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.; Geetha, K.M. Lipid nanoparticles in the development of mRNA vaccines for COVID-19. J. Drug Deliv. Sci. Technol. 2022, 74, 103553. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, S.J.; Han, X.; Mukalel, A.J.; El-Mayta, R.; Thatte, A.S.; Wu, J.; Padilla, M.S.; Alameh, M.G.; Srikumar, N.; Lee, D.; et al. Throughput-scalable manufacturing of SARS-CoV-2 mRNA lipid nanoparticle vaccines. Proc. Natl. Acad. Sci. USA 2023, 120, e2303567120. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Hosn, R.R.; Remba, T.; Yun, D.; Li, N.; Abraham, W.; Melo, M.B.; Cortes, M.; Li, B.; Zhang, Y.; et al. Optimization of storage conditions for lipid nanoparticle-formulated self-replicating RNA vaccines. J. Control Release 2023, 353, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Young, R.E.; Hofbauer, S.I.; Riley, R.S. Overcoming the challenge of long-term storage of mRNA-lipid nanoparticle vaccines. Mol. Ther. 2022, 30, 1792–1793. [Google Scholar] [CrossRef] [PubMed]
- Brader, M.L.; Williams, S.J.; Banks, J.M.; Hui, W.H.; Zhou, Z.H.; Jin, L. Encapsulation state of messenger RNA inside lipid nanoparticles. Biophys. J. 2021, 120, 2766–2770. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, M.J.; Alishetty, S.; Alameh, M.G.; Said, H.; Wright, L.; Paige, M.; Soliman, O.; Weissman, D.; Cleveland, T.E., 4th; Grishaev, A.; et al. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun. Biol. 2021, 4, 956. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Tian, Y.; Zheng, A.; Cui, C. Design Strategies for and Stability of mRNA-Lipid Nanoparticle COVID-19 Vaccines. Polymers 2022, 14, 4195. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef] [PubMed]
- Gatto, M.S.; Najahi-Missaoui, W. Lyophilization of Nanoparticles, Does It Really Work? Overview of the Current Status and Challenges. Int. J. Mol. Sci. 2023, 24, 14041. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, H.; Lam, K.; Bajusz, C.; Laczkó, D.; Karikó, K.; Schreiner, P.; Martin, A.; Lutwyche, P.; Heyes, J.; Pardi, N. Lyophilization provides long-term stability for a lipid nanoparticle-formulated, nucleoside-modified mRNA vaccine. Mol. Ther. 2022, 30, 1941–1951. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.; Imai, S.; Yamano, T.; Hanayama, R. Preventing SARS-CoV-2 Infection Using Anti-spike Nanobody-IFN-β Conjugated Exosomes. Pharm. Res. 2023, 40, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Scott, T.A.; Supramaniam, A.; Idris, A.; Cardoso, A.A.; Shrivastava, S.; Kelly, G.; Grepo, N.A.; Soemardy, C.; Ray, R.M.; McMillan, N.A.J.; et al. Engineered extracellular vesicles directed to the spike protein inhibit SARS-CoV-2. Mol. Ther. Methods Clin. Dev. 2022, 24, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Raghav, A.; Giri, R.; Agarwal, S.; Kala, S.; Jeong, G.B. Protective role of engineered extracellular vesicles loaded quercetin nanoparticles as anti-viral therapy against SARS-CoV-2 infection: A prospective review. Front. Immunol. 2022, 13, 1040027. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Díazcouder, A.; Díaz-Godínez, C.; Carrero, J.C. Extracellular vesicles in COVID-19 prognosis, treatment, and vaccination: An update. Appl. Microbiol. Biotechnol. 2023, 107, 2131–2141. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Qiu, Y.; Jiang, W.; Shen, J.; Yao, X.; He, X.; Li, L.; Fu, B.; Liu, X. Biological Features of Extracellular Vesicles and Challenges. Front. Cell Dev. Biol. 2022, 10, 816698. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Brocchini, S.; Williams, G.R. Extracellular vesicle-embedded materials. J. Control Release 2023, 361, 280–296. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, I.K.; Wood, M.J.A.; Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 2021, 16, 748–759. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Wang, Y.; Fa, S.; Yuan, C.; Yang, L. Extracellular Vesicles as Natural Delivery Carriers Regulate Oxidative Stress Under Pathological Conditions. Front. Bioeng. Biotechnol. 2021, 9, 752019. [Google Scholar] [CrossRef] [PubMed]
- Syromiatnikova, V.; Prokopeva, A.; Gomzikova, M. Methods of the Large-Scale Production of Extracellular Vesicles. Int. J. Mol. Sci. 2022, 23, 10522. [Google Scholar] [CrossRef] [PubMed]
- Chhoy, P.; Brown, C.W.; Amante, J.J.; Mercurio, A.M. Protocol for the separation of extracellular vesicles by ultracentrifugation from in vitro cell culture models. STAR Protoc. 2021, 2, 100303. [Google Scholar] [CrossRef] [PubMed]
- Hahm, J.; Kim, J.; Park, J. Strategies to Enhance Extracellular Vesicle Production. Tissue Eng. Regen. Med. 2021, 18, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Esmaeili, A.; Alini, M.; Baghaban Eslaminejad, M.; Hosseini, S. Engineering strategies for customizing extracellular vesicle uptake in a therapeutic context. Stem Cell Res. Ther. 2022, 13, 129. [Google Scholar] [CrossRef] [PubMed]
- Hassanpour, M.; Rezaie, J.; Nouri, M.; Panahi, Y. The role of extracellular vesicles in COVID-19 virus infection. Infect. Genet. Evol. 2020, 85, 104422. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Pan, X.; Luo, R.H.; Shen, X.; Li, S.; Wang, Y.; Zuo, X.; Wu, Y.; Guo, Y.; Xiao, G.; et al. Extracellular vesicles mediate antibody-resistant transmission of SARS-CoV-2. Cell Discov. 2023, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Bernal, C.; How-Volkman, C.; Spencer, M.; El-Shamy, A.; Mohieldin, A.M. The Role of Extracellular Vesicles in SARS-CoV-2-Induced Acute Kidney Injury: An Overview. Life 2024, 14, 163. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Nova-Lamperti, E.; Labarca, G.; Kulasinghe, A.; Short, K.R.; Carrión, F.; Salomon, C. Genomic communication via circulating extracellular vesicles and long-term health consequences of COVID-19. J. Transl. Med. 2023, 21, 709. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, Y.; Yoshida, T.; Takagi, Y.; Tsukamoto, H.; Takashima, K.; Kouwaki, T.; Makino, K.; Fukushima, S.; Nakamura, K.; Oshiumi, H. Circulating extracellular vesicle microRNAs associated with adverse reactions, proinflammatory cytokine, and antibody production after COVID-19 vaccination. NPJ Vaccines 2022, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Yonker, L.M.; Swank, Z.; Bartsch, Y.C.; Burns, M.D.; Kane, A.; Boribong, B.P.; Davis, J.P.; Loiselle, M.; Novak, T.; Senussi, Y.; et al. Circulating Spike Protein Detected in Post-COVID-19 mRNA Vaccine Myocarditis. Circulation 2023, 147, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Casner, R.G.; Guo, Y.; Wang, Q.; Iketani, S.; Chan, J.F.; Yu, J.; Dadonaite, B.; Nair, M.S.; Mohri, H.; et al. Antibodies targeting a quaternary site on SARS-CoV-2 spike glycoprotein prevent viral receptor engagement by conformational locking. Immunity 2023, 56, 2442–2455.e8. [Google Scholar] [CrossRef] [PubMed]
- Dhuli, K.; Medori, M.C.; Micheletti, C.; Donato, K.; Fioretti, F.; Calzoni, A.; Praderio, A.; De Angelis, M.G.; Arabia, G.; Cristoni, S.; et al. Presence of viral spike protein and vaccinal spike protein in the blood serum of patients with long-COVID syndrome. Eur. Rev. Med. Pharmacol. Sci. 2023, 27 (Suppl. 6), 13–19. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.H.; Patel, N.; Haupt, R.; Zhou, H.; Weston, S.; Hammond, H.; Logue, J.; Portnoff, A.D.; Norton, J.; Guebre-Xabier, M.; et al. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat. Commun. 2021, 12, 372. [Google Scholar] [CrossRef] [PubMed]
- Troyer, Z.; Alhusaini, N.; Tabler, C.O.; Sweet, T.; de Carvalho, K.I.L.; Schlatzer, D.M.; Carias, L.; King, C.L.; Matreyek, K.; Tilton, J.C. Extracellular vesicles carry SARS-CoV-2 spike protein and serve as decoys for neutralizing antibodies. J. Extracell. Vesicles 2021, 10, e12112. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Mendonça, L.; Allen, E.R.; Howe, A.; Lee, M.; Allen, J.D.; Chawla, H.; Pulido, D.; Donnellan, F.; Davies, H.; et al. Native-like SARS-CoV-2 Spike Glycoprotein Expressed by ChAdOx1 nCoV-19/AZD1222 Vaccine. ACS Cent. Sci. 2021, 7, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Sauer, M.M.; Tortorici, M.A.; Park, Y.J.; Walls, A.C.; Homad, L.; Acton, O.J.; Bowen, J.E.; Wang, C.; Xiong, X.; de van der Schueren, W.; et al. Structural basis for broad coronavirus neutralization. Nat. Struct. Mol. Biol. 2021, 28, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, F.; Crivelli, V.; Abernathy, M.E.; Guerra, C.; Palus, M.; Muri, J.; Marcotte, H.; Piralla, A.; Pedotti, M.; De Gasparo, R.; et al. Human neutralizing antibodies to cold linear epitopes and subdomain 1 of the SARS-CoV-2 spike glycoprotein. Sci. Immunol. 2023, 8, eade0958. [Google Scholar] [CrossRef] [PubMed]
- Cantera, J.L.; Cate, D.M.; Golden, A.; Peck, R.B.; Lillis, L.L.; Domingo, G.J.; Murphy, E.; Barnhart, B.C.; Anderson, C.A.; Alonzo, L.F.; et al. Screening Antibodies Raised against the Spike Glycoprotein of SARS-CoV-2 to Support the Development of Rapid Antigen Assays. ACS Omega 2021, 6, 20139–20148. [Google Scholar] [CrossRef] [PubMed]
- Glauninger, H.; Wong Hickernell, C.J.; Bard, J.A.M.; Drummond, D.A. Stressful steps: Progress and challenges in understanding stress-induced mRNA condensation and accumulation in stress granules. Mol. Cell 2022, 82, 2544–2556. [Google Scholar] [CrossRef] [PubMed]
- Seneff, S.; Nigh, G.; Kyriakopoulos, A.M.; McCullough, P.A. Innate immune suppression by SARS-CoV-2 mRNA vaccinations: The role of G-quadruplexes, exosomes, and MicroRNAs. Food Chem. Toxicol. 2022, 164, 113008. [Google Scholar] [CrossRef] [PubMed]
- Gillen, S.L.; Giacomelli, C.; Hodge, K.; Zanivan, S.; Bushell, M.; Wilczynska, A. Differential regulation of mRNA fate by the human Ccr4-Not complex is driven by coding sequence composition and mRNA localization. Genome Biol. 2021, 22, 284. [Google Scholar] [CrossRef] [PubMed]
- Batista-Roche, J.L.; Gómez-Gil, B.; Lund, G.; Berlanga-Robles, C.A.; García-Gasca, A. Global m6A RNA Methylation in SARS-CoV-2 Positive Nasopharyngeal Samples in a Mexican Population: A First Approximation Study. Epigenomes 2022, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Pantazi, P.; Clements, T.; Venø, M.; Abrahams, V.M.; Holder, B. Distinct non-coding RNA cargo of extracellular vesicles from M1 and M2 human primary macrophages. J. Extracell. Vesicles 2022, 11, e12293. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Bhattacharya, M.; Dhama, K. SARS-CoV-2 Vaccines, Vaccine Development Technologies, and Significant Efforts in Vaccine Development during the Pandemic: The Lessons Learned Might Help to Fight against the Next Pandemic. Vaccines 2023, 11, 682. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Xie, C.; Bu, G.L.; Zhong, L.Y.; Zeng, M.S. Molecular characteristics, immune evasion, and impact of SARS-CoV-2 variants. Signal Transduct. Target. Ther. 2022, 7, 202. [Google Scholar] [CrossRef] [PubMed]
- Bellavite, P.; Ferraresi, A.; Isidoro, C. Immune Response and Molecular Mechanisms of Cardiovascular Adverse Effects of Spike Proteins from SARS-CoV-2 and mRNA Vaccines. Biomedicines 2023, 11, 451. [Google Scholar] [CrossRef] [PubMed]
- Roessler, J.; Pich, D.; Albanese, M.; Wratil, P.R.; Krähling, V.; Hellmuth, J.C.; Scherer, C.; von Bergwelt-Baildon, M.; Becker, S.; Keppler, O.T.; et al. Quantitation of SARS-CoV-2 neutralizing antibodies with a virus-free, authentic test. PNAS Nexus 2022, 1, pgac045. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Loo, L.; Tran, A.; Lin, D.M.; Moreno, C.; Hesselson, D.; Neely, G.G.; Yang, J.Y.H. Scalable workflow for characterization of cell-cell communication in COVID-19 patients. PLoS Comput. Biol. 2022, 18, e1010495. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Yao, X.; Li, W.; Wang, C.; Xu, W.; Gan, Z.; Yang, Y.; Zhong, A.; Wang, B.; He, Z.; et al. Novel insight into the underlying dysregulation mechanisms of immune cell-to-cell communication by analyzing multitissue single-cell atlas of two COVID-19 patients. Cell Death Dis. 2023, 14, 286. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Banerjea, A.C. SARS-CoV-2 Spike Targets USP33-IRF9 Axis via Exosomal miR-148a to Activate Human Microglia. Front. Immunol. 2021, 12, 656700. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Fang, D.; Gao, X.; Deng, X.; Chen, N.; Wu, J.; Zeng, M.; Luo, M. Circulating microRNAs as emerging regulators of COVID-19. Theranostics 2023, 13, 125–147. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.H.; Hong, Y.; Heo, J.; Kang, S.; Lillehoj, H.S.; Hong, Y.H. Chicken miR-148a-3p regulates immune responses against AIV by targeting the MAPK signalling pathway and IFN-γ. Vet. Res. 2023, 54, 110. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chen, H.; Wang, H. Glycans of SARS-CoV-2 Spike Protein in Virus Infection and Antibody Production. Front. Mol. Biosci. 2021, 8, 629873. [Google Scholar] [CrossRef] [PubMed]
- Aloor, A.; Aradhya, R.; Venugopal, P.; Gopalakrishnan Nair, B.; Suravajhala, R. Glycosylation in SARS-CoV-2 variants: A path to infection and recovery. Biochem. Pharmacol. 2022, 206, 115335. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Rodriguez, P.; Francés-Gómez, C.; Chiner-Oms, Á.; López, M.G.; Jiménez-Serrano, S.; Cancino-Muñoz, I.; Ruiz-Hueso, P.; Torres-Puente, M.; Bracho, M.A.; D’Auria, G.; et al. Evolutionary and Phenotypic Characterization of Two Spike Mutations in European Lineage 20E of SARS-CoV-2. mBio 2021, 12, e0231521. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, R.L.; Ostrov, D.A. The Elusive Coreceptors for the SARS-CoV-2 Spike Protein. Viruses 2022, 15, 67. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.K.; Chang, W.C.; Prakash, E.; Peng, Y.J.; Tu, Z.J.; Lin, C.H.; Hsu, P.H.; Chang, C.F. Carbohydrate Ligands for COVID-19 Spike Proteins. Viruses 2022, 14, 330. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Gutiérrez, S.; Buckley, J.; Battaglia, G. The Role of Host Cell Glycans on Virus Infectivity: The SARS-CoV-2 Case. Adv. Sci. 2022, 10, e2201853. [Google Scholar] [CrossRef] [PubMed]
- Mathez, G.; Cagno, V. Viruses Like Sugars: How to Assess Glycan Involvement in Viral Attachment. Microorganisms 2021, 9, 1238. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, C.J.; Gaunt, B.; Harrison, P.J.; Yang, Y.; Liu, J.; Khan, A.; Giltrap, A.M.; Le Bas, A.; Ward, P.N.; Gupta, K.; et al. Pathogen-sugar interactions revealed by universal saturation transfer analysis. Science 2022, 377, eabm3125. [Google Scholar] [CrossRef] [PubMed]
- Van Egeren, D.; Stoddard, M.; White, L.F.; Hochberg, N.S.; Rogers, M.S.; Zetter, B.; Joseph-McCarthy, D.; Chakravarty, A. Vaccines Alone Cannot Slow the Evolution of SARS-CoV-2. Vaccines 2023, 11, 853. [Google Scholar] [CrossRef] [PubMed]
- Nassar, A.; Ibrahim, I.M.; Amin, F.G.; Magdy, M.; Elgharib, A.M.; Azzam, E.B.; Nasser, F.; Yousry, K.; Shamkh, I.M.; Mahdy, S.M.; et al. A Review of Human Coronaviruses’ Receptors: The Host-Cell Targets for the Crown Bearing Viruses. Molecules 2021, 26, 6455. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiang, R.; Huo, S.; Zhou, Y.; Jiang, S.; Wang, Q.; Yu, F. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct. Target. Ther. 2021, 6, 233. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhu, J.; Zhang, D.; Liu, G. Ribosomal control in RNA virus-infected cells. Front. Microbiol. 2022, 13, 1026887. [Google Scholar] [CrossRef] [PubMed]
- Overduin, M.; Kervin, T.A.; Tran, A. Progressive membrane-binding mechanism of SARS-CoV-2 variant spike proteins. iScience 2022, 25, 104722. [Google Scholar] [CrossRef] [PubMed]
- Correa, Y.; Waldie, S.; Thépaut, M.; Micciulla, S.; Moulin, M.; Fieschi, F.; Pichler, H.; Trevor Forsyth, V.; Haertlein, M.; Cárdenas, M. SARS-CoV-2 spike protein removes lipids from model membranes and interferes with the capacity of high density lipoprotein to exchange lipids. J. Colloid Interface Sci. 2021, 602, 732–739. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Nomura, N.; Muramoto, Y.; Ekimoto, T.; Uemura, T.; Liu, K.; Yui, M.; Kono, N.; Aoki, J.; Ikeguchi, M.; et al. Structure of SARS-CoV-2 membrane protein essential for virus assembly. Nat. Commun. 2022, 13, 4399. [Google Scholar] [CrossRef] [PubMed]
- Dolan, K.A.; Dutta, M.; Kern, D.M.; Kotecha, A.; Voth, G.A.; Brohawn, S.G. Structure of SARS-CoV-2 M protein in lipid nanodiscs. Elife 2022, 11, e81702. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yuan, H.; Li, X.; Wang, H. Spike protein mediated membrane fusion during SARS-CoV-2 infection. J. Med. Virol. 2023, 95, e28212. [Google Scholar] [CrossRef] [PubMed]
- Olukitibi, T.A.; Ao, Z.; Warner, B.; Unat, R.; Kobasa, D.; Yao, X. Significance of Conserved Regions in Coronavirus Spike Protein for Developing a Novel Vaccine against SARS-CoV-2 Infection. Vaccines 2023, 11, 545. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.; Sangeet, S.; Roy, S. Evolution of Sequence and Structure of SARS-CoV-2 Spike Protein: A Dynamic Perspective. ACS Omega 2023, 8, 23283–23304. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Qian, Z.; Lu, X.; Lu, J. Adaptive Evolution of the Spike Protein in Coronaviruses. Mol. Biol. Evol. 2023, 40, msad089. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, Z.; Wei, Z.; Schapiro, I.; Li, J. Binding affinity and mechanisms of SARS-CoV-2 variants. Comput. Struct. Biotechnol. J. 2021, 19, 4184–4191. [Google Scholar] [CrossRef] [PubMed]
- Verma, J.; Subbarao, N. Insilico study on the effect of SARS-CoV-2 RBD hotspot mutants’ interaction with ACE2 to understand the binding affinity and stability. Virology 2021, 561, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Jawad, B.; Adhikari, P.; Podgornik, R.; Ching, W.Y. Key Interacting Residues between RBD of SARS-CoV-2 and ACE2 Receptor: Combination of Molecular Dynamics Simulation and Density Functional Calculation. J. Chem. Inf. Model. 2021, 61, 4425–4441. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; Sun, X.; Lin, Y.; Gu, C.; Ding, L.; Lu, X.; Yang, Z.; Zhang, Y.; Ma, L.; Gu, W.; et al. Comprehensive mapping of binding hot spots of SARS-CoV-2 RBD-specific neutralizing antibodies for tracking immune escape variants. Genome Med. 2021, 13, 164. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Srivastava, Y.; Muthuramalingam, P.; Singh, S.K.; Verma, G.; Tiwari, S.; Tandel, N.; Beura, S.K.; Panigrahi, A.R.; Maji, S.; et al. Understanding Mutations in Human SARS-CoV-2 Spike Glycoprotein: A Systematic Review & Meta-Analysis. Viruses 2023, 15, 856. [Google Scholar] [CrossRef] [PubMed]
- Magazine, N.; Zhang, T.; Wu, Y.; McGee, M.C.; Veggiani, G.; Huang, W. Mutations and Evolution of the SARS-CoV-2 Spike Protein. Viruses 2022, 14, 640. [Google Scholar] [CrossRef] [PubMed]
- Borkotoky, S.; Dey, D.; Hazarika, Z. Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): A structural perspective. Mol. Biol. Rep. 2023, 50, 2713–2721. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.C.; Cheng, Y.W.; Chao, C.H.; Chang, Y.Y.; Chen, C.D.; Tsai, W.J.; Wang, S.; Lin, Y.S.; Chang, C.P.; Chuang, W.J.; et al. Antigenic Cross-Reactivity Between SARS-CoV-2 S1-RBD and Its Receptor ACE2. Front. Immunol. 2022, 13, 868724. [Google Scholar] [CrossRef] [PubMed]
- Hoter, A.; Naim, H.Y. Biochemical Characterization of SARS-CoV-2 Spike RBD Mutations and Their Impact on ACE2 Receptor Binding. Front. Mol. Biosci. 2022, 9, 893843. [Google Scholar] [CrossRef] [PubMed]
- da Costa, C.H.S.; de Freitas, C.A.B.; Alves, C.N.; Lameira, J. Assessment of mutations on RBD in the Spike protein of SARS-CoV-2 Alpha, Delta and Omicron variants. Sci. Rep. 2022, 12, 8540. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, J.; Wang, J. Surface charge changes in spike RBD mutations of SARS-CoV-2 and its variant strains alter the virus evasiveness via HSPGs: A review and mechanistic hypothesis. Front. Public Health 2022, 10, 952916. [Google Scholar] [CrossRef] [PubMed]
- Candido, K.L.; Eich, C.R.; de Fariña, L.O.; Kadowaki, M.K.; da Conceição Silva, J.L.; Maller, A.; Simão, R.C.G. Spike protein of SARS-CoV-2 variants: A brief review and practical implications. Braz. J. Microbiol. 2022, 53, 1133–1157. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, D.; Pinto, S.N.; Fernandes, F. Extracellular Vesicles and Infection: From Hijacked Machinery to Therapeutic Tools. Pharmaceutics 2023, 15, 1738. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Xiong, S. Tagged extracellular vesicles with the RBD of the viral spike protein for delivery of antiviral agents against SARS-COV-2 infection. J. Control Release 2021, 335, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Verta, R.; Grange, C.; Skovronova, R.; Tanzi, A.; Peruzzi, L.; Deregibus, M.C.; Camussi, G.; Bussolati, B. Generation of Spike-Extracellular Vesicles (S-EVs) as a Tool to Mimic SARS-CoV-2 Interaction with Host Cells. Cells 2022, 11, 146. [Google Scholar] [CrossRef] [PubMed]
- Martins, S.T.; Alves, L.R. Extracellular Vesicles in Viral Infections: Two Sides of the Same Coin? Front. Cell Infect. Microbiol. 2020, 10, 593170. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Zeng, J.; Marcink, T.C.; Porotto, M.; Moscona, A.; O’Shaughnessy, B. Host Cell Membrane Capture by the SARS-CoV-2 Spike Protein Fusion Intermediate. ACS Cent. Sci. 2023, 9, 1213–1228. [Google Scholar] [CrossRef] [PubMed]
- Lazebnik, Y. Cell fusion as a link between the SARS-CoV-2 spike protein, COVID-19 complications, and vaccine side effects. Oncotarget 2021, 12, 2476–2488. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Wu, L.A.; Wang, Q.; Qi, J.; Gao, G.F. Cell entry by SARS-CoV-2. Trends Biochem. Sci. 2021, 46, 848–860. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.M.; Yang, W.L.; Yang, F.Y.; Zhang, L.; Huang, W.J.; Hou, W.; Fan, C.F.; Jin, R.H.; Feng, Y.M.; Wang, Y.C.; et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct. Target. Ther. 2021, 6, 134. [Google Scholar] [CrossRef] [PubMed]
- Berdowska, I.; Matusiewicz, M. Cathepsin L, transmembrane peptidase/serine subfamily member 2/4, and other host proteases in COVID-19 pathogenesis—With impact on gastrointestinal tract. World J. Gastroenterol. 2021, 27, 6590–6600. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, R.; Sakamoto, A.; Deguchi, S.; Yi, R.; Sano, E.; Hotta, A.; Takahashi, K.; Yamanaka, S.; Takayama, K. Dual inhibition of TMPRSS2 and Cathepsin Bprevents SARS-CoV-2 infection in iPS cells. Mol. Ther. Nucleic Acids 2021, 26, 1107–1114. [Google Scholar] [CrossRef] [PubMed]
- Murigneux, E.; Softic, L.; Aubé, C.; Grandi, C.; Judith, D.; Bruce, J.; Le Gall, M.; Guillonneau, F.; Schmitt, A.; Parissi, V.; et al. Proteomic analysis of SARS-CoV-2 particles unveils a key role of G3BP proteins in viral assembly. Nat. Commun. 2024, 15, 640. [Google Scholar] [CrossRef] [PubMed]
- Sauter, D.; Kirchhoff, F. Evolutionary conflicts and adverse effects of antiviral factors. Elife 2021, 10, e65243. [Google Scholar] [CrossRef] [PubMed]
- Shapira, T.; Monreal, I.A.; Dion, S.P.; Buchholz, D.W.; Imbiakha, B.; Olmstead, A.D.; Jager, M.; Désilets, A.; Gao, G.; Martins, M.; et al. A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature 2022, 605, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Koch, J.; Uckeley, Z.M.; Doldan, P.; Stanifer, M.; Boulant, S.; Lozach, P.Y. TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells. EMBO J. 2021, 40, e107821. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.P.; Goldhill, D.H.; Zhou, J.; Baillon, L.; Frise, R.; Swann, O.C.; Kugathasan, R.; Penn, R.; Brown, J.C.; Sanchez-David, R.Y.; et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021, 6, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Essalmani, R.; Jain, J.; Susan-Resiga, D.; Andréo, U.; Evagelidis, A.; Derbali, R.M.; Huynh, D.N.; Dallaire, F.; Laporte, M.; Delpal, A.; et al. Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity. J. Virol. 2022, 96, e0012822. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. Domains and Functions of Spike Protein in Sars-Cov-2 in the Context of Vaccine Design. Viruses 2021, 13, 109. [Google Scholar] [CrossRef] [PubMed]
- Akkız, H. The Biological Functions and Clinical Significance of SARS-CoV-2 Variants of Corcern. Front. Med. 2022, 9, 849217. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.C.; Wang, Y.H.; Chen, Y.L.; Tsai, W.C.; Lee, C.H.; Chuang, K.P.; Chen, Y.A.; Yuan, C.H.; Ho, S.Y.; Yang, M.H.; et al. Chloroquine and Hydroxychloroquine: Efficacy in the Treatment of the COVID-19. Pathogens 2021, 10, 217. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jing, X.; Hua, L.; Zheng, Y.; Hu, S.; Xiao, J.; Guo, D.; Wu, W.; Ji, H.; Peng, L.; et al. Hypertension related toxicity of chloroquine explains its failure against COVID-19: Based on rat model. Front. Pharmacol. 2022, 13, 1051694. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Hofmann-Winkler, H.; Smith, J.C.; Krüger, N.; Arora, P.; Sørensen, L.K.; Søgaard, O.S.; Hasselstrøm, J.B.; Winkler, M.; Hempel, T.; et al. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine 2021, 65, 103255. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Cheng, Y.; Zhou, H.; Sun, C.; Zhang, S. The SARS-CoV-2 nucleocapsid protein: Its role in the viral life cycle, structure and functions, and use as a potential target in the development of vaccines and diagnostics. Virol. J. 2023, 20, 6. [Google Scholar] [CrossRef] [PubMed]
- Dicker, K.; Järvelin, A.I.; Garcia-Moreno, M.; Castello, A. The importance of virion-incorporated cellular RNA-Binding Proteins in viral particle assembly and infectivity. Semin. Cell Dev. Biol. 2021, 111, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Lisy, S.; Rothamel, K.; Ascano, M. RNA Binding Proteins as Pioneer Determinants of Infection: Protective, Proviral, or Both? Viruses 2021, 13, 2172. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, N.; Lareau, C.A.; Keshishian, H.; Ganskih, S.; Schneider, C.; Hennig, T.; Melanson, R.; Werner, S.; Wei, Y.; Zimmer, M.; et al. The SARS-CoV-2 RNA-protein interactome in infected human cells. Nat. Microbiol. 2021, 6, 339–353. [Google Scholar] [CrossRef] [PubMed]
- Finkel, Y.; Gluck, A.; Nachshon, A.; Winkler, R.; Fisher, T.; Rozman, B.; Mizrahi, O.; Lubelsky, Y.; Zuckerman, B.; Slobodin, B.; et al. SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature 2021, 594, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Aminpour, M.; Hameroff, S.; Tuszynski, J.A. How COVID-19 Hijacks the Cytoskeleton: Therapeutic Implications. Life 2022, 12, 814. [Google Scholar] [CrossRef] [PubMed]
- Gómez, S.A.; Rojas-Valencia, N.; Gómez, S.; Egidi, F.; Cappelli, C.; Restrepo, A. Binding of SARS-CoV-2 to Cell Receptors: A Tale of Molecular Evolution. ChemBioChem 2021, 22, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Bujanic, L.; Shevchuk, O.; von Kügelgen, N.; Kalinina, A.; Ludwik, K.; Koppstein, D.; Zerna, N.; Sickmann, A.; Chekulaeva, M. The key features of SARS-CoV-2 leader and NSP1 required for viral escape of NSP1-mediated repression. RNA 2022, 28, 766–779. [Google Scholar] [CrossRef] [PubMed]
- Tardivat, Y.; Sosnowski, P.; Tidu, A.; Westhof, E.; Eriani, G.; Martin, F. SARS-CoV-2 NSP1 induces mRNA cleavages on the ribosome. Nucleic Acids Res. 2023, 51, 8677–8690. [Google Scholar] [CrossRef] [PubMed]
- Kozai, T.; Fernandez-Martinez, J.; van Eeuwen, T.; Gallardo, P.; Kapinos, L.E.; Mazur, A.; Zhang, W.; Tempkin, J.; Panatala, R.; Delgado-Izquierdo, M.; et al. Dynamic molecular mechanism of the nuclear pore complex permeability barrier. bioRxiv 2023, 2023.03.31.535055. [Google Scholar] [CrossRef] [PubMed]
- Samuel, C.E. Interferon at the crossroads of SARS-CoV-2 infection and COVID-19 disease. J. Biol. Chem. 2023, 299, 104960. [Google Scholar] [CrossRef] [PubMed]
- Znaidia, M.; Demeret, C.; van der Werf, S.; Komarova, A.V. Characterization of SARS-CoV-2 Evasion: Interferon Pathway and Therapeutic Options. Viruses 2022, 14, 1247. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.M.; Shin, E.C. Type I and III interferon responses in SARS-CoV-2 infection. Exp. Mol. Med. 2021, 53, 750–760. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Barrett, B.S.; Morrison, J.H.; Mickens, K.L.; Vladar, E.K.; Hasenkrug, K.J.; Poeschla, E.M.; Santiago, M.L. Interferon resistance of emerging SARS-CoV-2 variants. Proc. Natl. Acad. Sci. USA 2022, 119, e2203760119. [Google Scholar] [CrossRef] [PubMed]
- Gasmi, A.; Mujawdiya, P.K.; Lysiuk, R.; Shanaida, M.; Peana, M.; Gasmi Benahmed, A.; Beley, N.; Kovalska, N.; Bjørklund, G. Quercetin in the Prevention and Treatment of Coronavirus Infections: A Focus on SARS-CoV-2. Pharmaceuticals 2022, 15, 1049. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Lokugamage, K.G.; Zhang, X.; Vu, M.N.; Muruato, A.E.; Menachery, V.D.; Shi, P.Y. Engineering SARS-CoV-2 using a reverse genetic system. Nat. Protoc. 2021, 16, 1761–1784. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, B.; Shirafkan, F.; Ripperger, K.; Rattay, K. The Role of Viral Infections in the Onset of Autoimmune Diseases. Viruses 2023, 15, 782. [Google Scholar] [CrossRef] [PubMed]
- Pasrija, R.; Naime, M. The deregulated immune reaction and cytokines release storm (CRS) in COVID-19 disease. Int. Immunopharmacol. 2021, 90, 107225. [Google Scholar] [CrossRef] [PubMed]
- Karousis, E.D.; Schubert, K.; Ban, N. Coronavirus takeover of host cell translation and intracellular antiviral response: A molecular perspective. EMBO J. 2024, 43, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Kloc, M.; Uosef, A.; Wosik, J.; Kubiak, J.Z.; Ghobrial, R.M. Virus interactions with the actin cytoskeleton-what we know and do not know about SARS-CoV-2. Arch. Virol. 2022, 167, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Rajah, M.M.; Bernier, A.; Buchrieser, J.; Schwartz, O. The Mechanism and Consequences of SARS-CoV-2 Spike-Mediated Fusion and Syncytia Formation. J. Mol. Biol. 2022, 434, 167280. [Google Scholar] [CrossRef]
- Li, Q.; Wang, Y.; Shi, Y. Syncytia formation during SARS-CoV-2 lung infection: A disastrous unity to eliminate lymphocytes. Cell Death Differ. 2021, 28, 2019–2021. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Frascaroli, G.; Zhou, X.; Knickmann, J.; Brune, W. Cell Fusion and Syncytium Formation in Betaherpesvirus Infection. Viruses 2021, 13, 1973. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, S.; Yadav, R.P.; Kumar, S.; Yadav, S.C. Ultrastructural study confirms the formation of single and heterotypic syncytial cells in bronchoalveolar fluids of COVID-19 patients. Virol. J. 2023, 20, 97. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zheng, Y.; Niu, Z.; Zhang, B.; Wang, C.; Yao, X.; Peng, H.; Franca, D.N.; Wang, Y.; Zhu, Y.; et al. SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination. Cell Death Differ. 2021, 28, 2765–2777. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wu, X.; Mao, Q.; Gao, F.; Liu, M.; Song, Z.; Bian, L.; Liang, Z. How SARS-CoV-2 dodges immune surveillance and facilitates infection: An analytical review. Expert Rev. Anti-Infect. Ther. 2022, 20, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Nemudryi, A.; Nemudraia, A.; Wiegand, T.; Nichols, J.; Snyder, D.T.; Hedges, J.F.; Cicha, C.; Lee, H.; Vanderwood, K.K.; Bimczok, D.; et al. SARS-CoV-2 genomic surveillance identifies naturally occurring truncation of ORF7a that limits immune suppression. Cell Rep. 2021, 35, 109197. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Sun, J.; Yuan, Y.; Yao, F.; Zheng, B.; Yang, G.; Xie, W.; Ye, G.; Li, Z.; Jiao, X.; et al. Surveillance of SARS-CoV-2 antibodies of patients in the local affected area during Wuhan lockdown. BMC Infect. Dis. 2022, 22, 10. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Zheng, Y.; Zeng, X.; He, B.; Cheng, W. Structural biology of SARS-CoV-2: Open the door for novel therapies. Signal Transduct. Target. Ther. 2022, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Rao, Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 2021, 19, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Y.; Zhao, R.; Gao, L.J.; Gao, X.F.; Wang, D.P.; Cao, J.M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell Infect. Microbiol. 2020, 10, 587269. [Google Scholar] [CrossRef] [PubMed]
- Hardenbrook, N.J.; Zhang, P. A structural view of the SARS-CoV-2 virus and its assembly. Curr. Opin. Virol. 2022, 52, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Scott, Z.C.; Koning, K.; Vanderwerp, M.; Cohen, L.; Westrate, L.M.; Koslover, E.F. Endoplasmic reticulum network heterogeneity guides diffusive transport and kinetics. Biophys. J. 2023, 122, 3191–3205. [Google Scholar] [CrossRef] [PubMed]
- Malone, B.; Urakova, N.; Snijder, E.J.; Campbell, E.A. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2022, 23, 21–39. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Ma, Y.; Chang, W. SARS-CoV-2 and the Nucleus. Int. J. Biol. Sci. 2022, 18, 4731–4743. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.E.; Wang, Y.; Du, X.; Zhang, T.; Mak, H.Y.; Hancock, S.E.; McEwen, H.; Pandzic, E.; Whan, R.M.; Aw, Y.C.; et al. TMEM41B and VMP1 are scramblases and regulate the distribution of cholesterol and phosphatidylserine. J. Cell Biol. 2021, 220, e202103105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, Y.E.; Yuan, Y.; Du, X.; Wang, Y.; Dong, X.; Yang, H.; Qi, S. TMEM41B and VMP1 are phospholipid scramblases. Autophagy 2021, 17, 2048–2050. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Altan-Bonnet, N. Viral pores are everywhere. Mol. Cell 2021, 81, 2061–2063. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.R.; Yin, W.C.; Jiang, Y.; Xu, H.E. Structure genomics of SARS-CoV-2 and its Omicron variant: Drug design templates for COVID-19. Acta Pharmacol. Sin. 2022, 43, 3021–3033. [Google Scholar] [CrossRef] [PubMed]
- Saraste, J.; Prydz, K. Assembly and Cellular Exit of Coronaviruses: Hijacking an Unconventional Secretory Pathway from the Pre-Golgi Intermediate Compartment via the Golgi Ribbon to the Extracellular Space. Cells 2021, 10, 503. [Google Scholar] [CrossRef] [PubMed]
- Mironov, A.A.; Savin, M.A.; Beznoussenko, G.V. COVID-19 Biogenesis and Intracellular Transport. Int. J. Mol. Sci. 2023, 24, 4523. [Google Scholar] [CrossRef] [PubMed]
- Sergio, M.C.; Ricciardi, S.; Guarino, A.M.; Giaquinto, L.; De Matteis, M.A. Membrane remodeling and trafficking piloted by SARS-CoV-2. Trends Cell Biol. 2024; Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Saud, Z.; Tyrrell, V.J.; Zaragkoulias, A.; Protty, M.B.; Statkute, E.; Rubina, A.; Bentley, K.; White, D.A.; Rodrigues, P.D.S.; Murphy, R.C.; et al. The SARS-CoV2 envelope differs from host cells, exposes procoagulant lipids, and is disrupted in vivo by oral rinses. J. Lipid Res. 2022, 63, 100208. [Google Scholar] [CrossRef] [PubMed]
- Zandi, M.; Hosseini, P.; Soltani, S.; Rasooli, A.; Moghadami, M.; Nasimzadeh, S.; Behnezhad, F. The role of lipids in the pathophysiology of coronavirus infections. Osong Public Health Res. Perspect. 2021, 12, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Angelova, A. Coronavirus-Induced Host Cubic Membranes and Lipid-Related Antiviral Therapies: A Focus on Bioactive Plasmalogens. Front. Cell Dev. Biol. 2021, 9, 630242. [Google Scholar] [CrossRef] [PubMed]
- Hassan, Z.; Kumar, N.D.; Reggiori, F.; Khan, G. How Viruses Hijack and Modify the Secretory Transport Pathway. Cells 2021, 10, 2535. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhao, Y.G.; Zhang, H. Endomembrane remodeling in SARS-CoV-2 infection. Cell Insight 2022, 1, 100031. [Google Scholar] [CrossRef] [PubMed]
- Knupp, J.; Pletan, M.L.; Arvan, P.; Tsai, B. Autophagy of the ER: The secretome finds the lysosome. FEBS J. 2023, 290, 5656–5673. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, H.; Pei, R.; Mao, B.; Zhao, Z.; Li, H.; Lin, Y.; Lu, K. The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes. Cell Discov. 2021, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, G.R. SARS-CoV-2 spike and its adaptable furin cleavage site. Lancet Microbe 2021, 2, e488–e489. [Google Scholar] [CrossRef] [PubMed]
- Adebayo, A.; Varzideh, F.; Wilson, S.; Gambardella, J.; Eacobacci, M.; Jankauskas, S.S.; Donkor, K.; Kansakar, U.; Trimarco, V.; Mone, P.; et al. l-Arginine and COVID-19: An Update. Nutrients 2021, 13, 3951. [Google Scholar] [CrossRef] [PubMed]
- Al-Kuraishy, H.M.; Al-Gareeb, A.I.; Alexiou, A.; Batiha, G.E. COVID-19 and L-arginine Supplementations: Yet to Find the Missed Key. Curr. Protein Pept. Sci. 2022, 23, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, G.; Coppola, A.; Izzo, R.; Annunziata, A.; Bernardo, M.; Lombardi, A.; Trimarco, V.; Santulli, G.; Trimarco, B. Effects of adding L-arginine orally to standard therapy in patients with COVID-19: A randomized, double-blind, placebo-controlled, parallel-group trial. Results of the first interim analysis. EClinicalMedicine 2021, 40, 101125. [Google Scholar] [CrossRef] [PubMed]
- Trimarco, V.; Izzo, R.; Lombardi, A.; Coppola, A.; Fiorentino, G.; Santulli, G. Beneficial effects of L-Arginine in patients hospitalized for COVID-19: New insights from a randomized clinical trial. Pharmacol. Res. 2023, 191, 106702. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Chen, X.; Wu, J.; Duan, X.; Men, K. Small molecules in the treatment of COVID-19. Signal Transduct. Target. Ther. 2022, 7, 387. [Google Scholar] [CrossRef] [PubMed]
- Lavie, M.; Dubuisson, J.; Belouzard, S. SARS-CoV-2 Spike Furin Cleavage Site and S2’ Basic Residues Modulate the Entry Process in a Host Cell-Dependent Manner. J. Virol. 2022, 96, e0047422. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Huang, Y.; Li, T.; Jiang, Z.; Zeng, L.; Hu, Z. The role of the Golgi apparatus in disease (Review). Int. J. Mol. Med. 2021, 47, 38. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gandy, S. The Golgi apparatus: Site for convergence of COVID-19 brain fog and Alzheimer’s disease? Mol. Neurodegener. 2022, 17, 67. [Google Scholar] [CrossRef] [PubMed]
- Choi, A.; Kots, E.D.; Singleton, D.T.; Weinstein, H.; Whittaker, G.R. Analysis of the molecular determinants for furin cleavage of the spike protein S1/S2 site in defined strains of the prototype coronavirus murine hepatitis virus (MHV). Virus Res. 2024, 340, 199283. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.A.; Zhan, S.H. The Emergence of the Spike Furin Cleavage Site in SARS-CoV-2. Mol. Biol. Evol. 2022, 39, msab327. [Google Scholar] [CrossRef] [PubMed]
- Papa, G.; Mallery, D.L.; Albecka, A.; Welch, L.G.; Cattin-Ortolá, J.; Luptak, J.; Paul, D.; McMahon, H.T.; Goodfellow, I.G.; Carter, A.; et al. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog. 2021, 17, e1009246. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Toba, S.; Itakura, Y.; Chambaro, H.M.; Kishimoto, M.; Tabata, K.; Intaruck, K.; Uemura, K.; Sanaki, T.; Sato, A.; et al. SARS-CoV-2 Bearing a Mutation at the S1/S2 Cleavage Site Exhibits Attenuated Virulence and Confers Protective Immunity. mBio 2021, 12, e0141521. [Google Scholar] [CrossRef] [PubMed]
- Lubinski, B.; Whittaker, G.R. The SARS-CoV-2 furin cleavage site: Natural selection or smoking gun? Lancet Microbe 2023, 4, e570. [Google Scholar] [CrossRef] [PubMed]
- Carabelli, A.M.; Peacock, T.P.; Thorne, L.G.; Harvey, W.T.; Hughes, J.; COVID-19 Genomics UK Consortium; Peacock, S.J.; Barclay, W.S.; de Silva, T.I.; Towers, G.J.; et al. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Bakhshandeh, B.; Jahanafrooz, Z.; Abbasi, A.; Goli, M.B.; Sadeghi, M.; Mottaqi, M.S.; Zamani, M. Mutations in SARS-CoV-2; Consequences in structure, function, and pathogenicity of the virus. Microb. Pathog. 2021, 154, 104831. [Google Scholar] [CrossRef] [PubMed]
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef] [PubMed]
- Sabanovic, B.; Piva, F.; Cecati, M.; Giulietti, M. Promising Extracellular Vesicle-Based Vaccines against Viruses, Including SARS-CoV-2. Biology 2021, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Popowski, K.D.; Zhu, D.; de Juan Abad, B.L.; Wang, X.; Liu, M.; Lutz, H.; De Naeyer, N.; DeMarco, C.T.; Denny, T.N.; et al. Exosomes decorated with a recombinant SARS-CoV-2 receptor-binding domain as an inhalable COVID-19 vaccine. Nat. Biomed. Eng. 2022, 6, 791–805. [Google Scholar] [CrossRef] [PubMed]
- Taborska, P.; Lastovicka, J.; Stakheev, D.; Strizova, Z.; Bartunkova, J.; Smrz, D. SARS-CoV-2 spike glycoprotein-reactive T cells can be readily expanded from COVID-19 vaccinated donors. Immun. Inflamm. Dis. 2021, 9, 1452–1467. [Google Scholar] [CrossRef] [PubMed]
- Rashid, F.; Xie, Z.; Suleman, M.; Shah, A.; Khan, S.; Luo, S. Roles and functions of SARS-CoV-2 proteins in host immune evasion. Front. Immunol. 2022, 13, 940756. [Google Scholar] [CrossRef] [PubMed]
- Sahni, C.; Basu Roy Chowdhury, P.; Devadas, D.; Ashish, A.; Singh, N.K.; Yadav, A.; Kaur, M.; Mishra, S.; Vishwakarma, S.; Singh, R.; et al. SARS-CoV-2 Mutations Responsible for Immune Evasion Leading to Breakthrough Infection. Cureus. 2022, 14, e29544. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, S.; Anraku, Y.; Taminishi, S.; Adachi, Y.; Kuroda, D.; Kita, S.; Higuchi, Y.; Kirita, Y.; Kotaki, R.; Tonouchi, K.; et al. Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants. Nat. Commun. 2023, 14, 4198. [Google Scholar] [CrossRef] [PubMed]
- Soh, S.M.; Kim, Y.; Kim, C.; Jang, U.S.; Lee, H.R. The rapid adaptation of SARS-CoV-2-rise of the variants: Transmission and resistance. J. Microbiol. 2021, 59, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Wang, L.; Misasi, J.; Pegu, A.; Zhang, Y.; Harris, D.R.; Olia, A.S.; Talana, C.A.; Yang, E.S.; Chen, M.; et al. Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529. Science 2022, 376, eabn8897. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Casillas, A.; Redwan, E.M.; Uversky, V.N. SARS-CoV-2: A Master of Immune Evasion. Biomedicines 2022, 10, 1339. [Google Scholar] [CrossRef] [PubMed]
- Minkoff, J.M.; tenOever, B. Innate immune evasion strategies of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 178–194. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Li, T.; Chen, G.; Zhu, Y.; Xu, L.; Lin, Y.; Sun, H.; Zhang, H.; Fang, Q.; Hong, J.; et al. Characterization and immunogenicity of SARS-CoV-2 spike proteins with varied glycosylation. Vaccine 2022, 40, 6839–6848. [Google Scholar] [CrossRef] [PubMed]
| Pharmaceutical Company | Products | Platform | Design |
|---|---|---|---|
| Pfizer-BioNTech | BNT162b2 | mRNA | Full-length S protein |
| (Mainz, Germany) | Two proline substitutions (K986P and V987P) | ||
| Lipid nanoparticle | |||
| Moderna | mRNA-1273 | mRNA | Full-length S protein |
| (Cambridge, MA, USA) | Two proline substitutions (K986P and V987P) | ||
| Lipid nanoparticle | |||
| Novavax | NVX-CoV2373 | Protein subunit | Full-length S protein |
| ( Gaithersburg, MD, USA) | Two proline substitutions (K986P and V987P) | ||
| 682-QQAQ-685 mutation at the S1/S2 cleavage site | |||
| Matrix-M adjuvant | |||
| Janssen | JNJ-78436735 | Adenovirus | Full-length S protein |
| (New Brunswick, NJ, USA) | Two proline substitutions (K986P and V987P) | ||
| Oxford-AstraZeneca | AZD1222 | Adenovirus | Full-length S protein |
| (Cambridge, UK) | The human tissue plasminogen activator gene leader |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsuzaka, Y.; Yashiro, R. Understanding the Molecular Actions of Spike Glycoprotein in SARS-CoV-2 and Issues of a Novel Therapeutic Strategy for the COVID-19 Vaccine. BioMedInformatics 2024, 4, 1531-1555. https://doi.org/10.3390/biomedinformatics4020084
Matsuzaka Y, Yashiro R. Understanding the Molecular Actions of Spike Glycoprotein in SARS-CoV-2 and Issues of a Novel Therapeutic Strategy for the COVID-19 Vaccine. BioMedInformatics. 2024; 4(2):1531-1555. https://doi.org/10.3390/biomedinformatics4020084
Chicago/Turabian StyleMatsuzaka, Yasunari, and Ryu Yashiro. 2024. "Understanding the Molecular Actions of Spike Glycoprotein in SARS-CoV-2 and Issues of a Novel Therapeutic Strategy for the COVID-19 Vaccine" BioMedInformatics 4, no. 2: 1531-1555. https://doi.org/10.3390/biomedinformatics4020084
APA StyleMatsuzaka, Y., & Yashiro, R. (2024). Understanding the Molecular Actions of Spike Glycoprotein in SARS-CoV-2 and Issues of a Novel Therapeutic Strategy for the COVID-19 Vaccine. BioMedInformatics, 4(2), 1531-1555. https://doi.org/10.3390/biomedinformatics4020084







