Abstract
Background: As patients interact with a healthcare service system, patterns of service utilization (PSUs) emerge. These PSUs are embedded in the sparse high-dimensional space of longitudinal cross-continuum health service encounter data. Once extracted, PSUs can provide quality assurance/quality improvement (QA/QI) efforts with the information required to optimize service system structures and functions. This may improve outcomes for complex patients with chronic diseases. Method: Working with longitudinal cross-continuum encounter data from a regional health service system, various pattern detection analyses were conducted, employing (1) graph community detection algorithms, (2) natural language processing (NLP) clustering, and (3) a hybrid NLP–graph method. Result: These approaches produced similar PSUs, as determined from a clinical perspective by clinical subject matter experts and service system operations experts. Conclusions: The similarity in the results provides validation for the methodologies. Moreover, the results stress the need to engage with clinical or service system operations experts, both in providing the taxonomies and ontologies of the service system, the cohort definitions, and determining the level of granularity that produces the most clinically meaningful results. Finally, the uniqueness of each approach provides an opportunity to take advantage of the various analytical capabilities that each approach brings, which will be further explored in our future research.
1. Introduction
1.1. Clinical Practice Guidelines, Clinical Pathways, and Services Pathways
The intent of this work is to extract useful information from data that have been accumulating in clinical information systems in order to optimize service system structure and function on behalf of patients contending with chronic/complex diseases. To achieve this, three constructs will be considered, including (1) generic clinical practice guidelines (CPGs), (2) idealized clinical pathways for those CPGs within a local service system, and (3) real-world cohort-specific service pathways located within local service system encounter data. For this study, we employ various machine learning (ML) methods to identify real-world service pathways in cross-continuum (i.e., across all services) longitudinal encounter data.
Clinical practice guidelines (CPGs) are evidence-informed recommendations intended to optimize patient care [1]. They consist of structured sequences of clinical interventions [1]. These generic guidelines are disease-class-specific but service-system-agnostic. As an example, CPGs for chronic diseases such as heart failure [2] provide evidence-based support for branching arrays of decisions that are keyed to the patient’s clinical, functional, or behavioral status. Clinical pathways, on the other hand, translate generic service system-agnostic CPGs into idealized local service-system-specific terms. As patients interact with a healthcare service system, patterns of service utilization (PSUs) emerge [3]. Hence, real-world service pathways consist of cohort-specific predictable recurring PSUs that take place within a local service system.
Perfect conformance of PSUs and of idealized service pathways is conditional upon an array of counterfactual conditions, for example an adequate supply of affordable services, and accessibility for all members of at-risk or clinically impacted populations. In the real world, pathways tracked by persons across the continuum of services are subject to the influence of at least four sets of potent factors: (1) factors attributable to the service system (e.g., limited capacity) [4]; (2) stigma associated with disease (e.g., addictions) [5,6,7]; (3) impacts arising from disease pathophysiology (e.g., difficult-to-predict emergence of comorbidities) [8]; and (4) patient factors that may impact treatment effectiveness and limit capacity to initiate and sustain requisite levels of service system engagement as a disease progresses [9,10,11]. The combined effect of these factors is real-world cohorts tracking to service pathways that may be positioned in the health service space at some distance from the idealized clinical pathways keyed to CPGs.
From this vantage point, the construct “quality” may be defined operationally in terms of the distance between idealized clinical pathways and real-world PSUs that are etched into the service terrain by cohorts of patients. If (1) PSUs are to be used to measure conformance of practice to complexly structured CPGs, and (2) those PSUs are based on machine learning methods applied to large volumes of variable-quality transactional data extracted from real-world transactional clinical information systems, then an organization using those PSUs to assure or improve quality must have confidence that they provide an accurate view of the local service system operations. This paper describes a method for supplying that assurance by applying three machine learning methods to those data and generating results that are directly comparable between methods.
1.2. Use of Graph Analytics for Healthcare Data
Numerous graph or network algorithms and methods have been applied to health-relevant data in recent years to examine diverse systems, including intracellular processes that relate clinical signs and symptoms to pathophysiological mechanisms [12], online social networks [13], biological networks [14], disease networks [15], and others. Moreover, a large body of work in the areas of disease, treatment, and health service system operations has been built upon graph analytics [16]. Examples include the following: (1) supporting medical predictive tasks such as discovering unknown disease associations for drug repositioning or comprehending disease progression [17,18]; (2) promoting drug discovery and molecular mechanism exploration in bioinformatics [18]; (3) improving critical care prediction [19]; (4) supporting diagnosis prediction, patient clinical outcome prediction, and readmission prediction [20]; (5) detecting patterns of care for patients [21]; and (6) exploring, analyzing, and understanding patterns in community referrals for elderly patients, and their use of multiple services through data visualization [22].
Node clustering is a topic that has garnered a great deal of attention in the field of graph computation [23,24]. In the context of graph analytics, clusters are often called communities. Many algorithms and methods with which to discover communities have been proposed. Some focus on the performance and some on the quality of the result. Here, quality means whether the partition of the nodes among the communities makes sense from the experts’ point of view. Well-known clustering algorithms include Fast-Greedy [25], Edge-Betweenness [25], Leading-Eigen [26], and Louvain [27].
The work conducted in [3] found that the Louvain algorithm often produces the most interpretable results. Nevertheless, while conducting some analysis in [3], we discovered issues with the Louvain method that prompted a modification to the procedure. In particular, Louvain is constrained by its resolution; given a graph, the smallest cluster or community that can be detected is bounded from below by the size of the graph; the larger the graph, the larger this minimum size.
1.3. Use of NLP in Analyzing Healthcare Data
Natural language processing (NLP) is a major branch of machine learning (ML). In recent years, NLP tools have been used extensively in healthcare as a method for extracting clinically meaningfully coded data from free text. Examples include (1) effective knowledge extraction from patient records using NLP [28,29], (2) extraction of symptoms from unstructured clinical information system data to be used in COVID-19 prognostics [30], and (3) use of NLP techniques to support clinical decisions on patients’ health outcomes [31].
NLP methods were originally designed to process texts in natural human languages. It has been known that many of the NLP methods are also applicable to many kinds of data that can be represented as strings. What is largely absent in the literature is the notion that a patient healthcare journey, consisting of a series of encounters with a large array of service entities, can also be treated as a string (after encoding the sequence of service utilization as a string of tokens). Therefore, healthcare encounters data are subject to many of the same types of analytical procedures employed with text documents or samples of human speech. Through methods such as TF-IDF (term frequency-inverse document frequency), documents can be represented as vectors in a word-space and hence can then be clustered. We-to take advantage of these NLP capabilities to extract PSUs.
1.4. Objectives
As previously stated, at a cohort level, the construct “quality” can be defined operationally as the distance between idealized clinical pathways and the real-world PSUs. With this definition, evidence-based quality assurance/quality improvement (QA/QI) requires a method for locating PSUs within sparse high-dimensional arrays of cross-continuum health service encounter data sourced from records in the clinical information systems. In the work conducted in [3], graph community detection methods were employed to detect communities of services that reflect PSUs. With graph community detection, encounter data are viewed as a bipartite graph of persons interacting with services, which is then projected to form a network of services. In this paper, a method for providing concurrent cross-validation of solutions derived from graph representations of the source data and NLP-based approaches is described.
Hence, the work in this paper is organized around the following questions:
- To what extent can NLP methods be used to extract PSUs from longitudinal heterogeneous cross-continuum healthcare data? How do the data need to be modeled and what data pre-processing needs to be conducted to generate the base data upon which the NLP methods can be applied?
- Are the results from NLP clustering for Service Classes similar to those obtained using graph community detection? Are they judged to be similar by clinical subject matter experts (SMEs) or clinical/administrative service system operations experts (SSOEs)?
- Does a hybrid NLP–graph community detection approach generate meaningful results, and how do the results compare to (a) community detection results, with simple frequency-based edge-weighted projections of service-service interactions, and (b) results obtained using NLP-based clustering approaches, employing measures of cosine similarity between vectors reflecting patient journeys?
2. Methodological Approach
2.1. Concurrent Validation via Application of Multiple Methods to the Same Body of Data, Modeled in Different Ways
The methodology reported in this document is roughly analogous to the multitrait–multimethod approach to constructing validation within a classic test and measurement paradigm, tracing back to the work of Campbell and Fiske [32]. For the work in this document, the constructs to be validated are PSUs. Our intent is to identify underlying functions expressed in terms of functions that span service system structures and emerge over the course of potentially large numbers of interactions with different services that address different needs and risks.
2.2. Source Data
The source data used in this paper consist of retrospective longitudinal transactional service encounter data extracted from a single instance of a clinical information system (CIS) deployed across the continuum of services provided by one of the health authorities within Canada (hereinafter referred to as “host organization”). The host organization provides a comprehensive array of secondary and tertiary health services, for all ages, for persons contending with medical/surgical issues and/or mental health/substance use issues. This includes acute care/intensive care services, hospital- and community-based emergency response, ambulatory services, residential care services for older adults or persons contending with mental health issues, case management services, and a range of addiction harm reduction or rehab and recovery-oriented services. The encounter data accessed by this study consist of approximately 10 million encounters over 7 years for approximately 1 million patients. This represents data for all service recipients, except a few restricted services where the data are strictly prohibited (e.g., services for persons who are victims of sexual assault). To access the source data, a certificate of approval was provided by the University of Victoria Research Ethics Board (REB), following the British Columbia, Canada, ethics harmonization guideline.
2.3. Data Pre-Processing
Two main branches of analysis are presented in this paper: a graph method and an NLP method. However, before any analysis is undertaken, as part of the data preparation, we consider the granularity of the data. The services provided by the host organization are encapsulated into an array of roughly 2000 Service Units within a location built for the CIS used to support care delivery. Service unit names within the CIS are often opaque, rendering them unsuitable for supervised machine learning methods that require meaningfully labeled data. Moreover, the service units may vary widely with regard to granularity; for example, multiple beds will appear as a single unit within an acute care facility, but multiple beds in a large array of family care homes for frail elderly will show up as multiple service units. To address these issues, a clinical context coding scheme (CCCS) was developed [33].
The CCCS is organized around six sets of codes, constituting a semantic layer applied to all of the Service Units to generate clinically functional Services Classes with meaningful names. By applying the CCCS to source data as previously proposed in [3,34,35], the 2000 service units in the host organization’s CIS are converted into approximately 200 clinically functional Service Classes. This activity was conducted in collaboration with SSOES. These Service Classes are the codes that are used to conduct the various analyses in this study.
2.4. Use of Community Detection in Extracting PSUs
The work conducted in [3] proposes a methodology for extracting PSUs from cross-continuum longitudinal healthcare data using graph community detection. The data consist of encounter data, where each row is a record of a service accessed by a patient. We can view these data as a bipartite graph between patients and Service Classes.
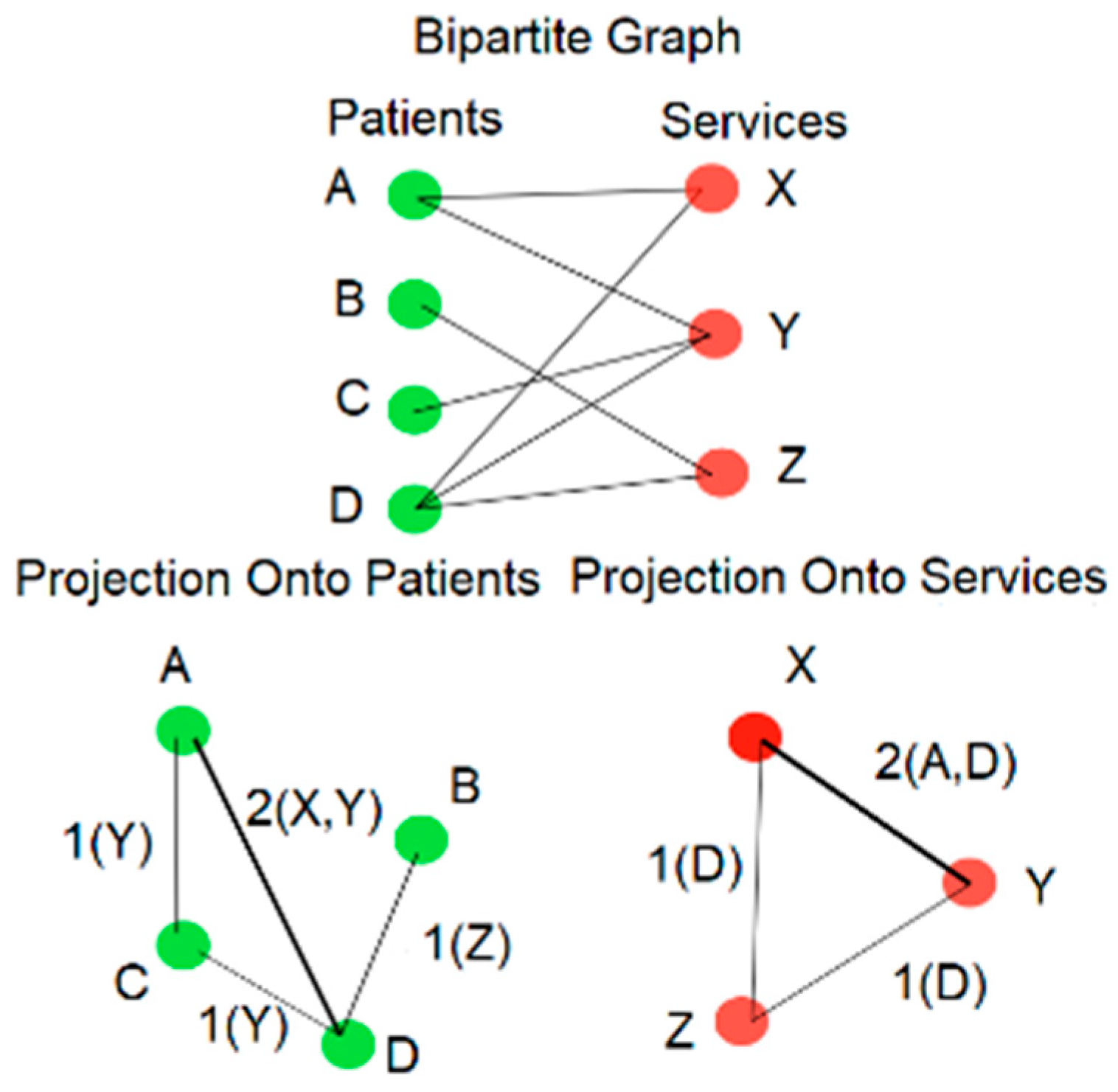
To determine which service classes cluster together based on their pattern of utilization we can perform bipartite projection onto the Service Classes, as illustrated in Figure 1. Two Service Classes are connected by an edge when there are patients who use both. The number of such patients then becomes the weight of the edge. One can then observe that if a pair of Service Classes tend to co-occur in the longitudinal encounter histories of numerous patients, they will have a strong connection between them as measured by the edge weight.

Figure 1.
Bipartite graph and bipartite projection.
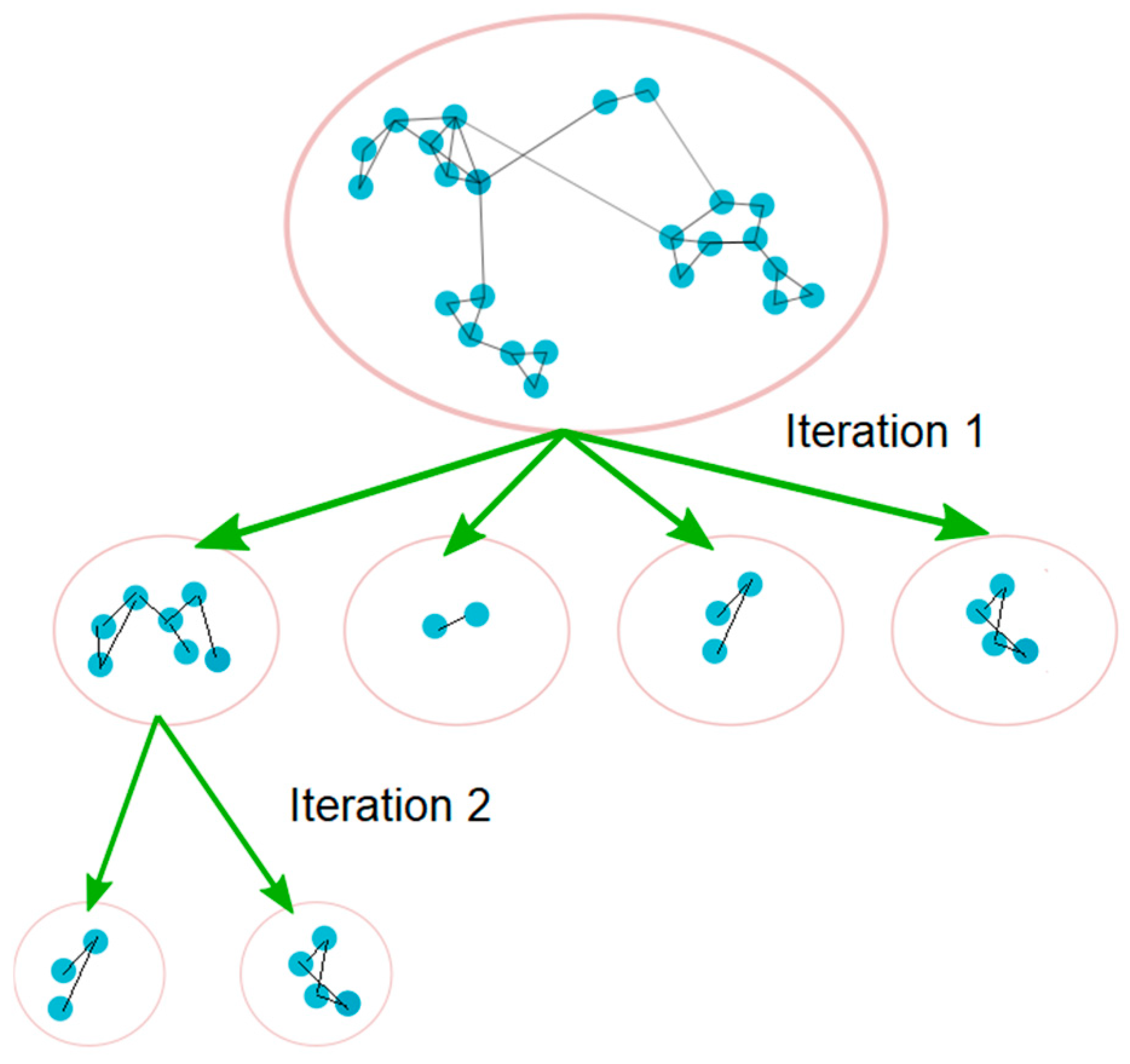
Next, the Louvain community detection [27] can be applied to the projected graph. However, while conducting the analysis in [3], it was discovered that from a clinical perspective, the results are often still too coarse, with many heterogeneous Service Classes clustered together. To solve this problem, ref. [3] proposed to iteratively apply the Louvain algorithm in a nested fashion. Note that in this case, iteration is not the same as the number of passes in the Louvain algorithm (instead, iteration refers to the level of nesting). In the approach proposed in [3], once the Louvain algorithm has generated the first set of communities, each community is isolated and treated as a new graph and the Louvain algorithm is applied again on each of the isolated graphs. The process is repeated with subsequent set of communities (see Figure 2).

Figure 2.
Iterative community detection.
With each iteration, the number of communities increases, and the size of each resulting community is reduced. At a certain point, Louvain no longer breaks the communities any further (i.e., the number of communities remains unchanged, with further iterations) and the algorithm stops. The output communities for each iteration are collected, and the results are compared using modularity values and by engaging with clinical SMEs. Additionally, for each node, the internal weighted degree (the weighted degree inside the community), and the external weighted degree (the weighted degree of a node to nodes outside of its community), are computed. The internal and external weighted degrees can be used to measure the strength of the bond of each node to its community.
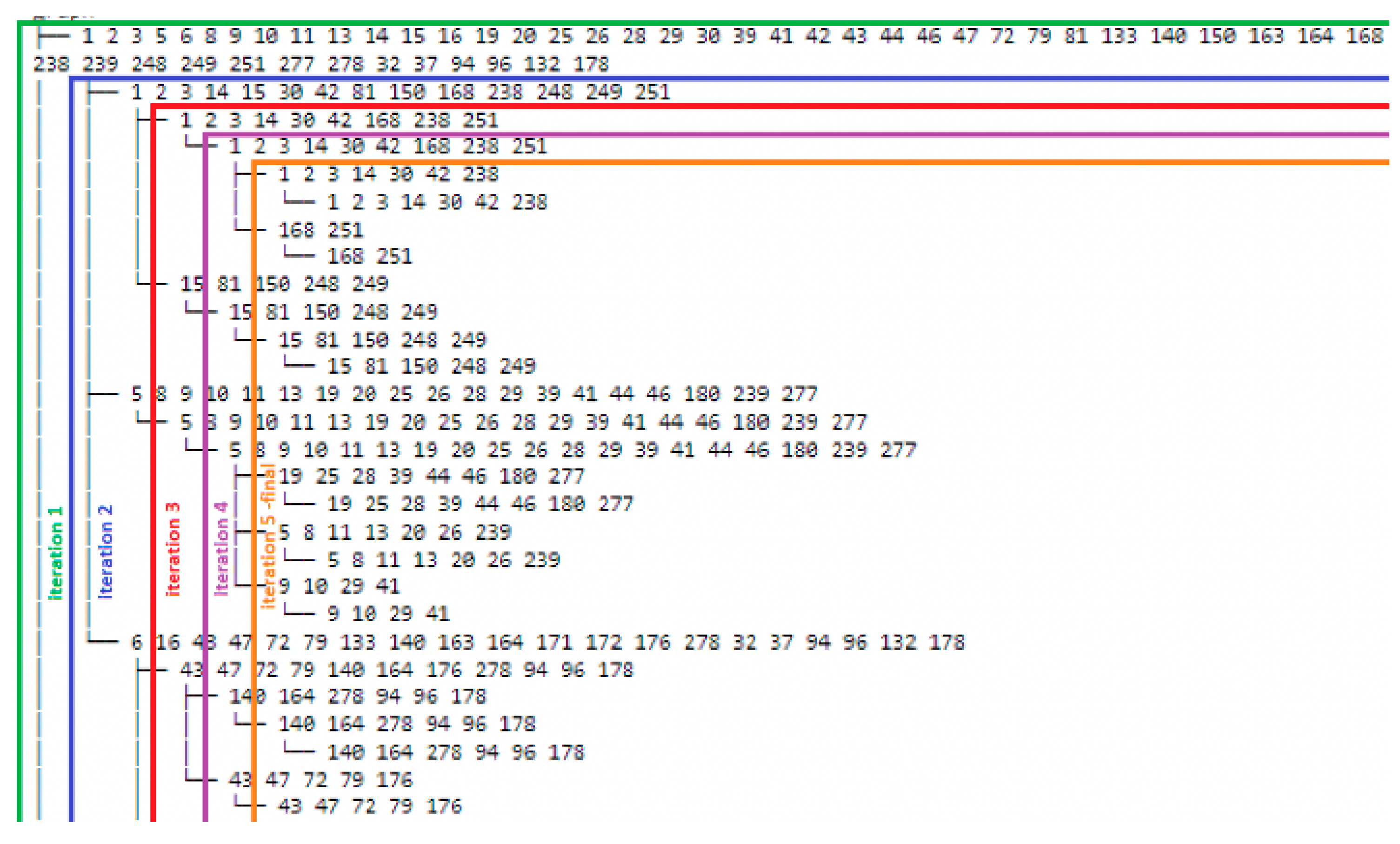
Figure 3 provides a snapshot of the results for an illustrative community detection iteration process. For this illustration, the sub-community with Service Classes 1, 2, 3, 14, 30, 42, 81, 150, 168, 238, 248, 249, and 251 is considered. This is one of the communities that were generated as the result of iteration 2. At iteration 3, this sub community will split into two, including sub-communities 1, 2, 3, 14, 30, 42, 168, 238, and 251 and sub-communities 15, 81, 150, 248, and 249. Additionally, at iteration 5, we observe that the community detection algorithm is no longer able to break the last sub-communities any further. As a result, the iteration stops at this level, and the result is reviewed with SMEs and SSOEs to determine the iteration that provides the result that is most clinically meaningful.

Figure 3.
Sample community detection iteration process.
2.5. Use of NLP in Extracting PSUs
Natural language processing (NLP) is a rapidly expanding field in computer science. NLP methods begin by systematically tokening elements within a document (e.g., words) and mapping the tokens into a vector space. Thus, every document in a collection of documents (a corpus) has a vector representation. The process of tokenization can be conducted in several ways, i.e., using frequency counts or considering the proportion of occurrences throughout the corpus (TF-IDF) [36]. First, the tokens must be defined: this can be simply the words in the document or patterns of words. Once the tokens have been defined, vectors can be created for each document in the corpus. TF-IDF is used to generate a normalized count of the tokens in a document weighted by the logarithmic ratio of the entire corpus. Hence, if a token only appears in one of the documents in the corpus, it will have a very small weighting. Conversely, if a token appears in most of the documents in the corpus, its weight will be much larger.
After each document has a TF-IDF vector representation, document vectors can be compared pairwise using a dot product. If two documents are highly similar in token weights, then their corresponding dot product (a.k.a. cosine similarity) will be close to one. Likewise, if two documents are dissimilar in their token weights, then the dot product will be close to zero. Therefore, with the vector representation, one can measure similarity between documents in a given corpus. Moreover, the similarity scores allow algorithms such as K-means to measure the closeness of documents in a corpus and generate clusters of related documents from a given corpus.
This method can be translated quite directly to patient encounter histories (patient journeys) embedded in vectors reflecting the history of encounters with roughly 200 Service Classes. Now, for every patient one can create “sentences” composed of the Service Class IDs.
We then tokenize the sentences with a uni-gram, bi-gram, or skip-gram vectorization. Here, a k-gram is a sequence of k words in a document, and a skip-gram is a sequence of words in a document that are separated by other intervening words. Once tokenized, we apply TF-IDF. After this tokenization, the TF-IDF vectors can be used to measure the patient’s journey similarity using various similarity metrics with regards to the other patients in the “corpus”.
Cosine similarity is a metric used to measure how similar the documents are irrespective of their sizes. Mathematically, it measures the cosine of the angle between two vectors projected in a multi-dimensional space by taking a dot product of the two vectors [37]. In the case of patients’ journey similarities, a dot product between two patient’s vectors provides a measure of similarity on a patient-to-patient level. Finally, one can apply the K-means algorithm to the patients’ dot products and generate a clustering of similar patients in the “corpus”.
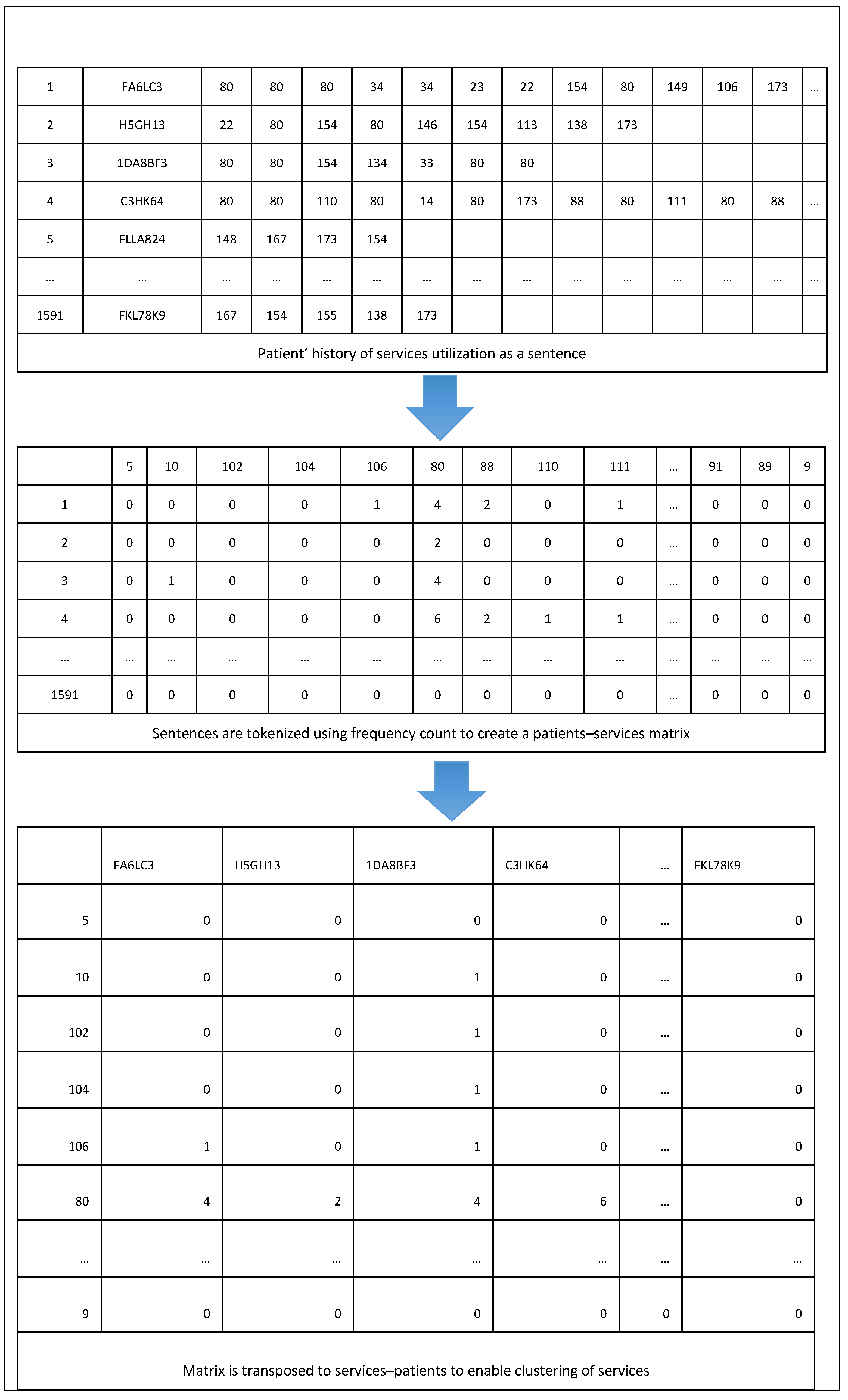
To create a cluster of services classes, a similar approach to the above is used. First, each patient’s history of service utilization is generated as a sentence. The words in such sentences are Service Class ids that a patient had interacted with. For example, if patient “A” engaged with Service Classes “X”, “Y”, “Z”, and “X” again, one would illustrate the sequence of service engagement as “X Y Z X”. Second, the sentences are tokenized using frequency counts to transform a sentence showing the history of service utilization into a vector. From this, a matrix that illustrates the frequency of each Service Class utilization for a patient is generated. In other words, each row describes a patient and each column describes a Service Class. Third, unlike the process used for patient clustering, to cluster Service Classes, the matrix is transposed such that each column corresponds to patients that had an interaction with a Service Class, and rows correspond to Service Classes that patients engaged with. Fourth, TF-IDF is applied on the resulting transposed matrix to have normalized counts. Fifth, cosine similarity is applied to the service vectors to measure the degree of similarity between the services classes. Finally, a clustering algorithm is applied to create a cluster of services classes based on degree of similarity with regards to patients’ engagement with the services.
Figure 4 provides an illustrative summary of part of the process used in using NLP to generate PSUs. Note that for purposes of privacy protection, the data in the above tables consist of simulated patient journeys.

Figure 4.
PSUs generation using NLP.
2.6. Combining Community Detection with the NLP to Extract PSUs
In this approach, both the capabilities of NLP and graph community detection are combined. Instead of creating a bipartite graph with patients and Service Classes, we used a TF-IDF matrix to create a projected graph for Service Classes. To accomplish this, first, each patient’s history of service utilization was created as a sentence. Then, using frequency counts, these sentences were tokenized. As a result, we formed a matrix in which the columns indicate the frequency count of Service Classes for each patient and the rows represent the patients. Then, to produce normalized counts, TF-IDF was applied to the resulting transposed matrix.
To create a projected graph of Service Classes, the weights between two Service Classes were calculated by computing the dot product of each service vector with other Service Classes. This results in a measure of similarity on a service-to-service level. Hence, services that are utilized by many of the same patients will have a high dot product and a correspondingly high weight. Similarly, the services that are less accessed by the same patients will have a low dot product, resulting in low weight. Once the service-to-service graph is created, the Louvain algorithm can be applied iteratively, as previously described in generating PSU using graph community detection.
3. Analysis
In collaboration with clinical SMEs, a cohort of patients who have taken an opioid overdose (OD-cohort) was considered. The data used represent anonymized cross-continuum patients’ data, extracted from the host organization’s CIS. The OD-cohort was analyzed, applying the methods described in the previous sections. The data contained 5279 patients (1606 females, 3672 males, and 1 unknown sex), aged between 14 and 92 years, with a range between 1 and 200 interactions.
For the analysis, three approaches were used. First, we performed community detection using weights from the bipartite projection. Second, we applied NLP using TF-IDF, cosine similarity, and clustering algorithms. Note that for NLP, two clustering approaches were used, including K-means and hierarchical clustering. Third, NLP, using TF-IDF and cosine similarity, was combined with the community detection algorithm.
The analyses were conducted in Python using the libraries igraph [38], scikit-learn [39], and scipy [40]. We also used the pandas and numpy libraries for data pre-processing.
3.1. Analysis Using Community Detection
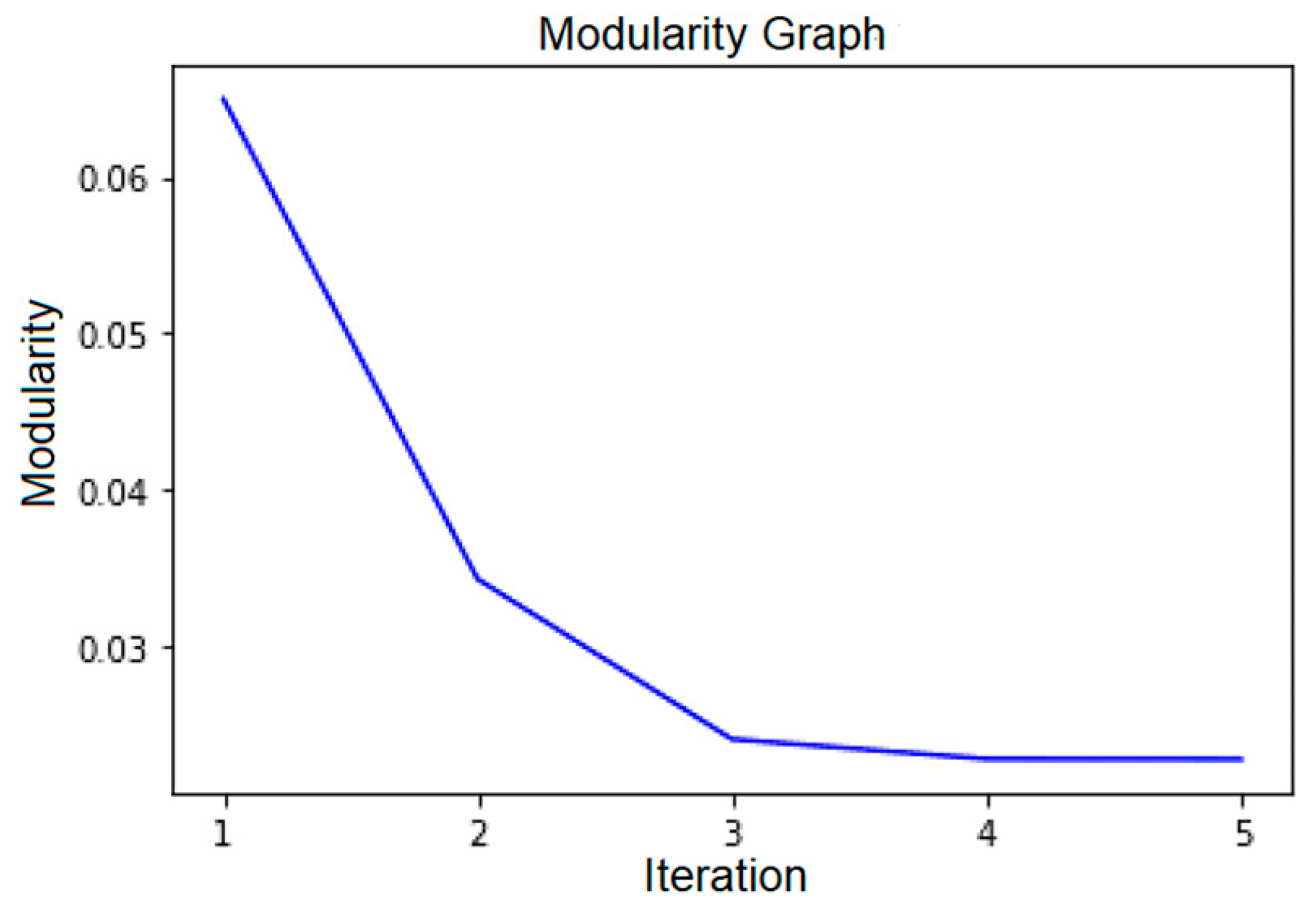
The Louvain community detection was run iteratively (as described in Section 2.4). Focusing on the OD-cohort, we found that the number of communities did not increase after five iterations. The number of communities ranged from four communities at one iteration to thirty-one at five iterations. With one iteration, the resulting communities were too functionally heterogeneous to be meaningful. On the other hand, with five iterations, the communities that were generated were “too small”. By “too small”, we mean that in an analysis of another sample of patients contending with the same cohort-defining characteristics, we are likely not to reproduce the same communities at that more granular level of resolution.
A plot of modularity value versus the number of iterations is displayed in Figure 5. Based on this figure and the elbow heuristic, the optimal number of iterations is between two and three (as the modularity plateaus after three). In consultation with clinical SMEs and SSOEs, we found that iteration three provided the most optimal clinically meaningful communities of services, in relationship to the key characteristics shared by all members of the cohort.

Figure 5.
Plot of modularity values versus the number of iterations.
Even when the number of iterations in the community detection solution has been determined, any given community may still include services that are not related in a clearly discernable way to other services in the community, or to the cohort-defining characteristics. To trim these Service Classes from the solutions, we computed the internal and external weighted degrees of the nodes. We noticed that nodes with large internal weighted degrees tended to form communities that are relatively stable and more understandable from the SMEs and SSOEs point of view. Therefore, we focused on nodes with large internal weighted degrees. In consultation with clinical SMEs and SSOEs, a cut point was drawn on the result table listing the services classes in a community to separate and discard the Service Classes with low internal weighted degrees from the others.
3.2. Analysis Using NLP Methods with K-Means and Hierarchical Clustering
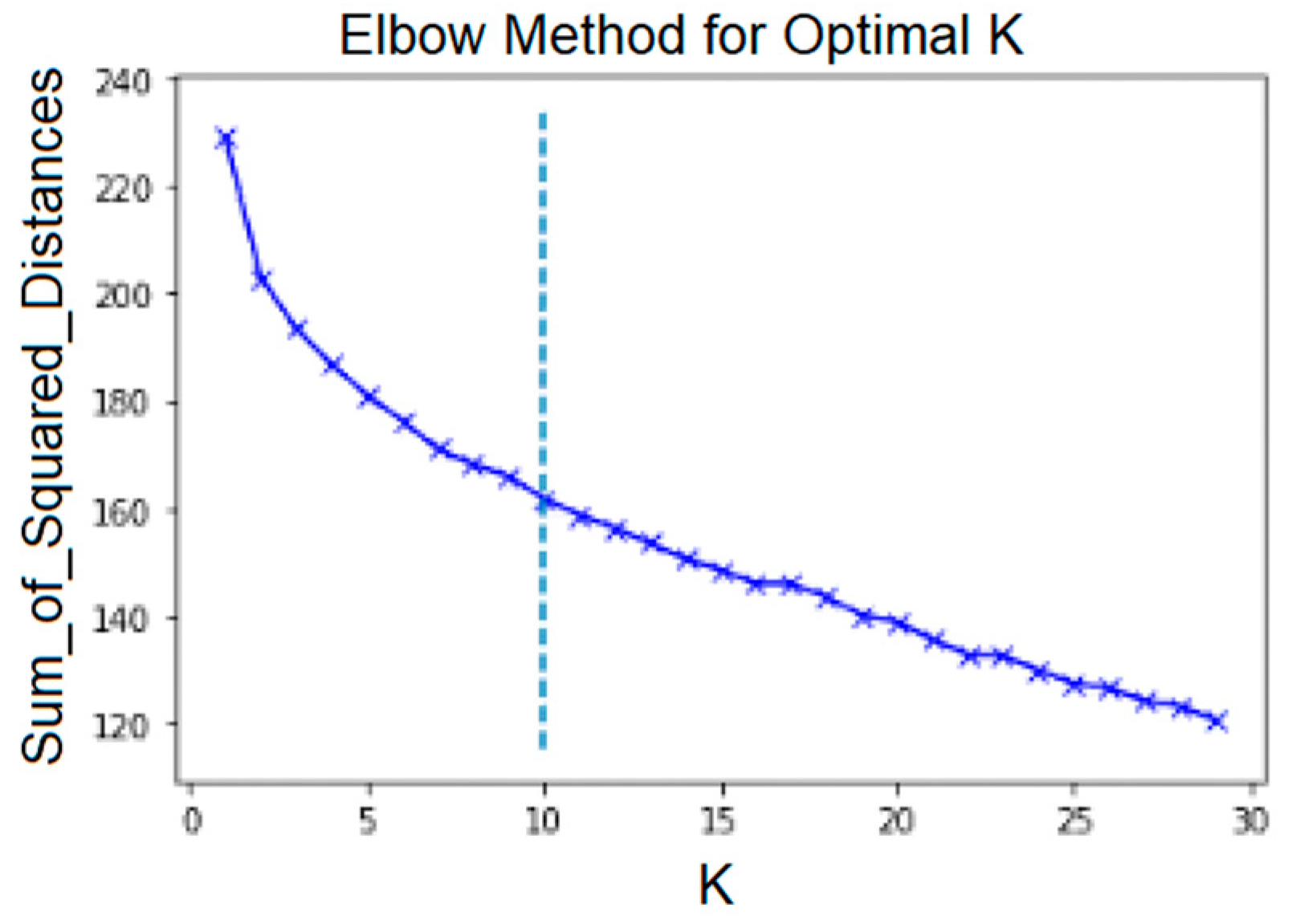
With regard to the NLP solutions, first, similarity measures among the Service Classes were built, as explained in Section 2.5. Then, various clustering algorithms were applied to the resulting matrix of similarity values. For K-means, one must choose a number k indicating the number of clusters. The elbow method provides a systematic method of determining the best k. Using this method, we plotted an average score for all clusters versus k. The score that is commonly used is the sum of square distances from each point to the centroid to which the cluster belongs. The elbow point is the inflection point on the curve [41]. The value of k for this point is regarded as the best value for k. This elbow point is not always obvious, and sometimes it is not easy to pinpoint visually. The KneeLocator function [42] was used to find the elbow point. Figure 6 shows the plot of this score. It can be noticed that for the OD-cohort, the elbow point is at around k = 9. This differs from the optimal number of communities that were generated at iteration three using the community detection approach.

Figure 6.
K-means elbow method and KneeLocator function.
Using the elbow method and setting k to 9, we generated clusters that were too big and contained Service Classes that were not related from a clinical perspective. Hence, for the purposes of the exploratory analyses reported in this paper, we decided to set the value of k to 19, which is the number of communities that was generated in the graph community detection using iteration three. (Recall that iteration three was judged overall to be the most optimal from clinical SME/SSOE perspectives; i.e., it was judged to be the array of services most clearly related to features of clinically understandable and characterizable cohorts.)
In the result comparison section, several major clusters of K-means on the OD-cohort with k = 19 can be seen. The similarity percentage column (in Table 3) shows the probability of similarity between each Service Class and other Service Classes in the cosine similarity matrix. So, the NLP clusters were ordered based on this similarity percentage to determine the most important nodes in each cluster. This approach is similar to ordering the communities based on weighted degrees inside the community to identify the most important nodes in each community.
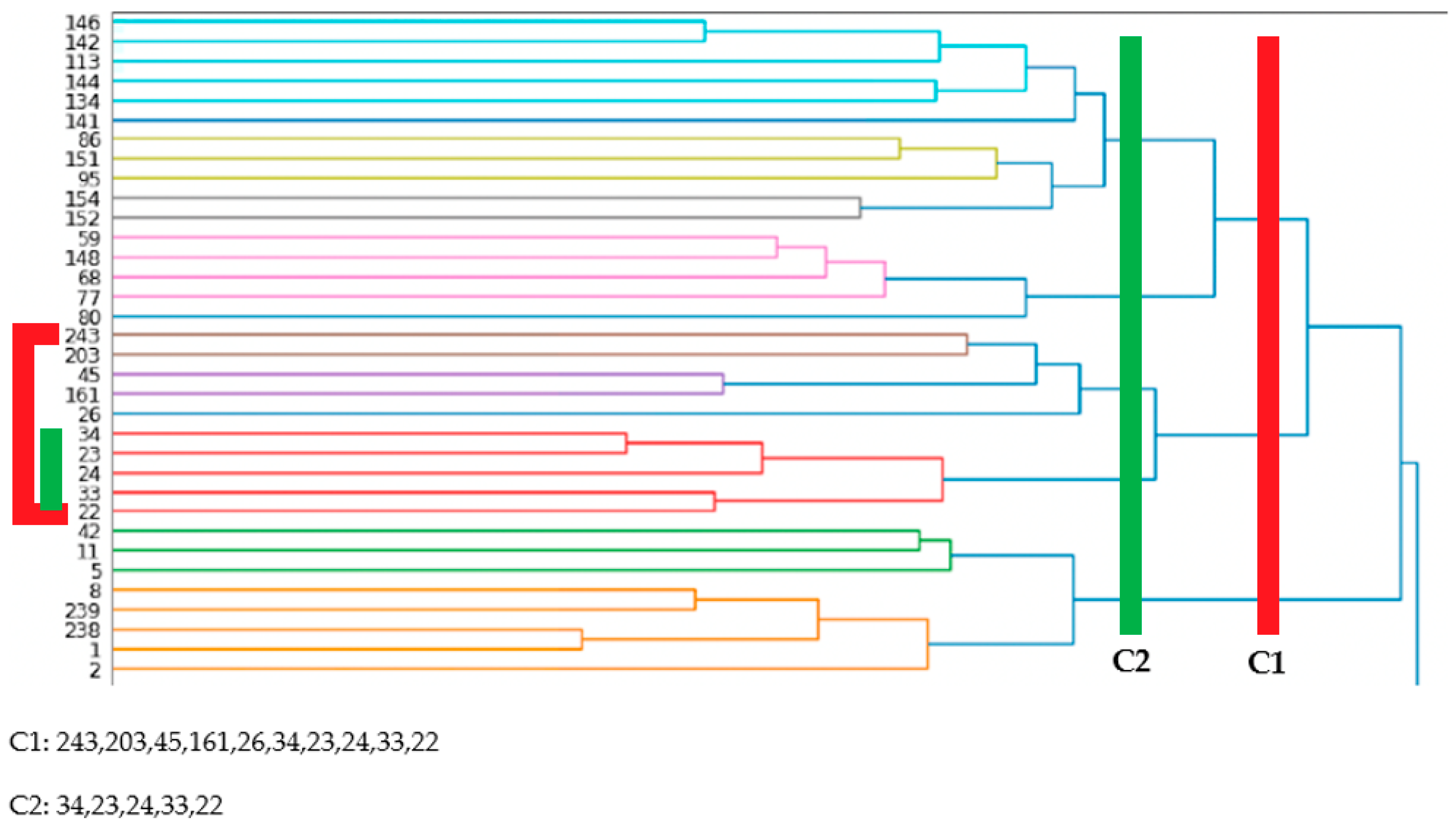
Next, a hierarchical clustering algorithm was applied. A sample of the results for the OD-cohort are shown in Section 4. With hierarchical clustering, the number of clusters was not set beforehand. This number was decided once the results were generated by choosing a cut-off line on the horizontal axis (Figure 7). This determined which services classes needed to be included in which clusters. Using this approach, several similarities with the communities from the graph community detection were observed, as well as similarities with the NLP clustering using K-means.

Figure 7.
Addiction-related services for OD-cohort—NLP and hierarchical clustering.
4. Results
Several clusters of related services were generated during the analysis of the OD-cohort. To name a few, they included emergency and acute care-related services, addictions-related services, and psychiatry-related services. For this paper, the addictions-related services were chosen to highlight the similarity of results among the different approaches. In reviewing the results of the different approaches reported in this paper, we note that none of the solutions have the status of “truth”; all contribute heuristically, together with input from SMEs and SSOEs, to a working judgment of what can be treated as “true enough”.
The Table 1, Table 2, Table 3 and Table 4 below are organized by groupings of related clusters using different approaches. A cut-off line was included to separate the strongly connected services from the weakly connected services within clusters. For the hierarchical clustering, the markings within the diagram were used to visually separate the different cut-off points and indicate the Service Classes that were used together for comparison. Two cut-off lines (C1 and C2) were added to illustrate the flexibility of interpretation that this approach provides. Finally, at the end of the cluster results, a similarity matrix was added to capture all the Service Classes that are similar across of the different approaches.

Table 1.
Addiction-related services for OD-cohort—graph community detection. The red band represents the cut-off line included to separate the strongly connected services from the weakly connected services within clusters.

Table 2.
Addiction-related services for OD-cohort—NLP and K-means clustering.

Table 3.
Addiction-related services for OD-cohort—NLP and community detection. The red band represents the cut-off line included to separate the strongly connected services from the weakly connected services within clusters.

Table 4.
Solution similarities matrix. The gray band cover cells in which a Service Class is missing from respective cluster.
In order to compare the different solutions, note that each Service Class has an associated Service Class ID. For the graph community detection results, the NLP plus K-means clustering results, and the NLP plus community detection solutions, each Service Class ID is paired with a Service Class label. For the NLP plus hierarchical clustering, as well as the similarity matrix, to make most effective use of space, only the Service Class IDs are displayed.
Note that the chosen cluster for illustration does not features all addiction services but addiction services with a rehabilitation/recovery orientation such as withdrawal management (Service Class 23) and post-withdrawal stabilization (Service Class 24).
5. Discussion
The purpose of this paper is to show the similarity of results across the different approaches for cross validation. Analogous to Campbell and Fiske’s multitrait–multimethod approach in examining construct validity [32], this paper supplies a method for validating PSUs that were generated previously using iterative graph community detection [3]. The different approaches used in this paper produced results that were similar across methods, where that similarity was manifest as overlaps in the Service Classes that, in effect, load most heavily on a given cluster. The slight differences in grouping of Service Classes that can be noticed among the approaches mostly affect the Service Classes that are not strongly connected to a given cluster regardless of method. This similarity in results provides cross validation for the PSUs, demonstrating that they are not artifacts of the method employed to produce the solutions.
In addition to graph community detection methods, the methodology in this paper explores how we can take advantage of NLP capabilities to extract PSUs. To do this, spurious or variable granularity of the services needs to be reduced. This was accomplished by using a clinical context coding scheme (CCCS) [33]. This is a semantic layer that groups Service Units into a reduced set of equivalence classes (Service Classes) that are relatively homogeneous with regard to their clinical functions. Then, a patient journey can be viewed as a sentence, or a string of words, in which the words are made of series of encounters with the CCCS-based Service Classes, arranged in the order in which they occurred. One can then apply the TF-IDF method [36] and cosine similarity to identify similarities among patients in a chosen cohort. Based on these similarity measures, one can cluster the Service Classes that are commonly used by similar patients. These clusters can then qualify as PSUs upon review by clinical SMEs and SSOEs. In the analysis conducted in this paper, several clustering algorithms were employed, and the results were compared. These include K-means [43] and hierarchical clustering [44]. To our knowledge, focusing on longitudinal heterogeneous cross-continuum healthcare data to extract PSUs, this is the first time NLP has been used in this manner.
Furthermore, the uniqueness of each approach provides an opportunity to take advantage of the various other capabilities that each approach brings. In future work, as the concepts of predictions using patient journeys similarities are expanded, both NLP capabilities and graph various metrics, or a combination of both, will be used.
The methods outlined in this paper are applied to around 200 equivalence classes (i.e., Service Classes) generated by applying the six sets of CCCS codes to the source data, represented as patients’ encounters with roughly 2000 Service Units. Because some significant portion of the granularity of the data at the Service Unit level is not related to clinical purpose or function, given the sparseness and high dimensionality of the data, it is unlikely that the methods used in this paper would generate meaningful or usable results without prior aggregation via the use of the CCCS scheme.
The methodology outlined in our previous work [3] stresses the need to engage with clinical SMEs and SSOEs in (1) providing the taxonomies and ontologies of the Service Classes, as well as the cohort definitions, and (2) determining the level of granularity that produces the most clinically meaningful result. This still holds true for the proposed NLP method. Both the CCCS scheme and the cohort definition preceded the application of any NLP methods. Additionally, it was demonstrated that the optimal value for k in K-means, as computed via the elbow method, was not sufficient for the purposes of generating the most clinically interpretable clusters of service. In other words, the application of purely objective/quantitative criteria will not enable these methods to converge on the “best” solutions. They provide information of heuristic value that can be used by SMEs and SSOEs to arrive at solutions that are “true enough” to employ the results for other purposes, e.g., prediction models.
In support of the above, the capabilities of visually assessing the results and picking the appropriate iteration for community detection or cut-off point for NLP plus hierarchical clustering can demonstrate the power of visual analytics. This provides a platform that makes the collaboration between data scientists, ML specialists, and SME/SSOE easier, more efficient, and transparent.
Though K-means and hierarchical clustering represent the most frequently used algorithms [45], there are various other existing clustering algorithms. Moreover, there are other types of approaches to accessing vectors similarities using NLP. We have described a popular protocol in NLP that provides a document-level view of a corpus; however, NLP offers finer views of text as well. The algorithm Word2Vec [46] takes a corpus of documents and produces a vectorization of each word in the vocabulary of the corpus. Besides the ones described in this paper, other clustering algorithms or NLP approaches were not used. This is a limitation of this study.
In future studies we plan to use Word2Vec combined with other dimensionality reduction techniques such as tSNE, and we will apply other types of clustering algorithm, including Gaussian mixture models and structural clustering, to extract PSUs.
6. Conclusions
Using a cohort of patients contending with addictions, a set of analyses using anonymized cross-continuum patients’ data, extracted from the host organization’s clinical information system (CIS), was performed. The analysis consisted of three different approaches: (1) graph community detection; (2) NLP using TF-IDF (term frequency inverse document frequency), cosine similarity, and clustering algorithms; and (3) a combination of both approaches. The analyses produced comparable results from a clinical perspective, especially for services that are strongly connected. Hence, this paper begins to take on the challenge of providing what is, in effect, construct validation [32,47] for ML-derived entities.
Moreover, with the rapid advancement of transformer-based models, such as ChatGPT, used for processing NLP tasks, the work outlined in this paper provides a first step in, and a basis for, generating cohort-specific simulated data. This has the potential to provide high-quality synthetic data that still maintain the statistical characteristics of the real data. Additionally, the work outlined in this paper opens the door for combining the capabilities of NLP with prediction using patients’ encounters data. Using PSUs, algorithms such as RNN (recurrent neural networks) and random forest can be combined with NLP to predict patients who are likely to experience a certain outcome, such as an overdose, based on their pattern of engagement with the healthcare services. Such an endeavor can help address the challenge of proactivity in preventing a certain outcome, e.g., overdose, as well as potential demand estimate, in looking after patients at risk of a certain outcome. These works are currently underway.
Author Contributions
Conceptualization, J.B., K.M., A.R. and A.K.; Data curation, J.B. and H.S.; Formal analysis, J.B., K.M. and E.C.; Investigation, J.B.; Methodology, J.B. and E.C.; Project administration, J.B.; Resources, J.B. and K.M.; Software, J.B., H.S., J.H. and Y.S.; Supervision, J.B., K.M., A.R. and A.K.; Validation, J.B.; Visualization, J.B.; Writing—original draft, J.B. and L.T.E.; Writing—review and editing, J.B., Y.S., H.S., K.M., A.R., A.K., J.H. and L.T.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
A certificate of approval was provided by the University of Victoria Research Ethics Board (REB), following the British Columbia, Canada, Ethics harmonization guideline. The REB number is H21-02817.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets presented in this article are unavailable because of privacy or ethical restrictions. Requests to access the datasets require a certificate of approval by the University of Victoria Research Ethics Board, following the British Columbia, Canada, Ethics harmonization guideline.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Panteli, D.; Legido-Quigley, H.; Reichebner, C.; Ollenschläger, G.; Schäfer, C.; Busse, R. Clinical practice guidelines as a quality strategy. In Improving Healthcare Quality in Europe; OECD Publishing: Paris, France, 2019; p. 233. [Google Scholar]
- Howlett, J.G.; McKelvie, R.S.; Costigan, J.; Ducharme, A.; Estrella-Holder, E.; Ezekowitz, J.A.; Giannetti, N.; Haddad, H.; Heckman, G.A.; Herd, A.M. The 2010 Canadian Cardiovascular Society guidelines for the diagnosis and management of heart failure update: Heart failure in ethnic minority populations, heart failure and pregnancy, disease management, and quality improvement/assurance programs. Can. J. Cardiol. 2010, 26, 185–202. [Google Scholar] [CrossRef]
- Bambi, J.; Santoso, Y.; Sadri, H.; Moselle, K.; Rudnick, A.; Robertson, S.; Chang, E.; Kuo, A.; Howie, J.; Dong, G.Y. A methodological approach to extracting patterns of service utilization from a cross-continuum high dimensional Healthcare Dataset to Support Care Delivery Optimization for Patients with Complex Problems. BioMedInformatics 2024, 4, 946–965. [Google Scholar] [CrossRef]
- Dawkins, B.; Renwick, C.; Ensor, T.; Shinkins, B.; Jayne, D.; Meads, D. What factors affect patients’ ability to access healthcare? An overview of systematic reviews. Trop. Med. Int. Health 2021, 26, 1177–1188. [Google Scholar] [CrossRef] [PubMed]
- Stangl, A.L.; Earnshaw, V.A.; Logie, C.H.; Van Brakel, W.C.; Simbayi, L.; Barré, I.; Dovidio, J.F. The Health Stigma and Discrimination Framework: A global, crosscutting framework to inform research, intervention development, and policy on health-related stigmas. BMC Med. 2019, 17, 31. [Google Scholar] [CrossRef]
- Cradock-O’Leary, J.; Young, A.S.; Yano, E.M.; Wang, M.; Lee, M.L. Use of general medical Services by VA patients with psychiatric disorders. Psychiatr. Serv. 2002, 53, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Christiani, A.; Hudson, A.L.; Nyamathi, A.; Mutere, M.; Sweat, J. Attitudes of homeless and drug-using youth regarding barriers and facilitators in delivery of quality and culturally sensitive health care. J. Child Adolesc. Psychiatr. Nurs. 2008, 21, 154–163. [Google Scholar] [CrossRef] [PubMed]
- De Groot, V.; Beckerman, H.; Lankhorst, G.J.; Bouter, L.M. How to measure comorbidity: A critical review of available methods. J. Clin. Epidemiol. 2003, 56, 221–229. [Google Scholar] [CrossRef]
- UNAIDS: Joint United Nations Programme on HIV/AIDS. Protocol for the identification of discrimination against people living with HIV. In Protocol for the Identification of Discrimination against People Living with HIV; UNAIDS: Genava, Switzerland, 2000; p. 40. [Google Scholar]
- Nyblade, L.; Stockton, M.A.; Giger, K.; Bond, V.; Ekstrand, M.L.; Lean, R.M.; Mitchell, E.M.H.; Nelson, L.R.E.; Sapag, J.C.; Siraprapasiri, T. Stigma in health facilities: Why it matters and how we can change it. BMC Med. 2019, 17, 25. [Google Scholar] [CrossRef]
- Iezzoni, L.I.; McCarthy, E.P.; Davis, R.B.; Siebens, H. Mobility impairments and use of screening and preventive services. Am. J. Public Health 2000, 90, 955–961. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Loscalzo, J.; Silverman, E.K. Network Medicine: Complex Systems in Human Disease and Therapeutics; Harvard University Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Mislove, A.; Marcon, M.; Gummadi, K.P.; Druschel, P.; Bhattacharjee, B. Measurement and analysis of online social networks. In Proceedings of the 7th ACM SIGCOMM Conference on Internet Measurement, San Diego, CA, USA, 24–26 October 2007; pp. 29–42. [Google Scholar]
- Pavlopoulos, G.A.; Secrier, M.; Moschopoulos, C.N.; Soldatos, T.G.; Kossida, S.; Aerts, J.; Schneider, R.; Bagos, P.G. Using graph theory to analyze biological networks. BioData Min. 2011, 4, 1–27. [Google Scholar] [CrossRef]
- Wysocki, K.; Ritter, L. Diseasome. Annu. Rev. Nurs. Res. 2011, 29, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Rostami, M.; Oussalah, M.; Berahmand, K.; Farrahi, V. Community detection algorithms in healthcare applications: A systematic review. IEEE Access 2023, 11, 30247–30272. [Google Scholar] [CrossRef]
- Toor, R.; Chana, I. Network Analysis as a Computational technique and its benefaction for predictive analysis of healthcare data: A systematic review. Arch. Comput. Methods Eng. 2021, 28, 1689–1711. [Google Scholar] [CrossRef]
- Yi, H.-C.; You, Z.-H.; Huang, D.-S.; Kwoh, C.K. Graph representation learning in bioinformatics: Trends, methods and applications. Brief. Bioinform. 2021, 23, bbab340. [Google Scholar] [CrossRef]
- Wanyan, T.; Kang, M.; Badgeley, M.A.; Johnson, K.W.; De Freitas, J.K.; Chaudhry, F.F.; Vaid, A.; Zhao, S.; Miotto, R.; Nadkarni, G.N. Heterogeneous graph embeddings of electronic health records improve critical care disease predictions. In Proceedings of the Artificial Intelligence in Medicine: 18th International Conference on Artificial Intelligence in Medicine, AIME 2020, Minneapolis, MN, USA, 25–28 August 2020; pp. 14–25. [Google Scholar]
- Wu, T.; Wang, Y.; Wang, Y.; Zhao, E.; Yuan, Y. Leveraging graph-based hierarchical medical entity embedding for healthcare applications. Sci. Rep. 2021, 11, 5858. [Google Scholar] [CrossRef] [PubMed]
- Niyirora, J.; Aragones, O. Network analysis of medical care services. Health Inform. J. 2020, 26, 1631–1658. [Google Scholar] [CrossRef] [PubMed]
- Palmer, R.; Utley, M.; Fulop, N.J.; O’Connor, S. Using visualisation methods to analyse referral networks within community health care among patients aged 65 years and over. Health Inform. J. 2020, 26, 354–375. [Google Scholar] [CrossRef] [PubMed]
- Fortunato, S. Community detection in graphs. Phys. Rep. 2010, 486, 75–174. [Google Scholar] [CrossRef]
- Yin, H.; Benson, A.R.; Leskovec, J.; Gleich, D.F. Local higher-order graph clustering. In Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Halifax, NS, Canada, 13–17 August 2017; pp. 555–564. [Google Scholar]
- Clauset, A.; Newman, M.E.J.; Moore, C. Finding community structure in very large networks. Phys. Rev. E 2004, 70, 066111. [Google Scholar] [CrossRef]
- Newman, M.E.J. Finding community structure in networks using the eigenvectors of matrices. Phys. Rev. E 2006, 74, 036104. [Google Scholar] [CrossRef] [PubMed]
- Blondel, V.D.; Guillaume, J.-L.; Lambiotte, R.; Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008, 2008, P10008. [Google Scholar] [CrossRef]
- Stewart, R.; Velupillai, S. Applied natural language processing in mental health big data. Neuropsychopharmacology 2021, 46, 252. [Google Scholar] [CrossRef] [PubMed]
- Souili, A.; Cavallucci, D.; Rousselot, F. Natural Language Processing (NLP)—A Solution for Knowledge Extraction from Patent Unstructured Data. Procedia Eng. 2015, 131, 635–643. [Google Scholar] [CrossRef][Green Version]
- Silverman, G.M.; Sahoo, H.S.; Ingraham, N.E.; Lupei, M.; Puskarich, M.A.; Usher, M.; Dries, J.; Finzel, R.L.; Murray, E.; Sartori, J. NLP methods for extraction of symptoms from unstructured data for use in prognostic covid-19 analytic models. J. Artif. Intell. Res. 2021, 72, 429–474. [Google Scholar] [CrossRef]
- Reyes-Ortiz, J.A.; González-Beltrán, B.A.; Gallardo-López, L. Clinical decision support systems: A survey of NLP-based approaches from unstructured data. In Proceedings of the 2015 26th International Workshop on Database and Expert Systems Applications (DEXA), Valencia, Spain, 1–4 September 2015; pp. 163–167. [Google Scholar]
- Campbell, D.T.; Fiske, D.W. Convergent and discriminant validation by the multitrait-multimethod matrix. Psychol. Bull. 1959, 56, 81. [Google Scholar] [CrossRef] [PubMed]
- Koval, A.; Moselle, K. Clinical Context Coding Scheme—Describing utilisation of services of Island Health between 2007–2017. In Proceedings of the Conference of the International Population Data Linkage Association, Banff, AB, Canada, 12–14 September 2018. [Google Scholar]
- Bambi, J.; Santoso, Y.; Moselle, K.; Robertson, S.; Rudnick, A.; Chang, E.; Kuo, A. Analyzing patterns of service utilization using graph topology to understand the dynamic of the engagement of patients with complex problems with health services. BioMedInformatics 2024, 4, 1071–1084. [Google Scholar] [CrossRef]
- Bambi, J.; Dong, G.Y.; Santoso, Y.; Moselle, K.; Dugas, S.; Olobatuyi, K.; Rudnick, A.; Chang, E.; Kuo, A. Patterns of service utilization across the full continuum of care: Using patient journeys to assess disparities in access to health services. Knowledge 2024, 4, 252–264. [Google Scholar] [CrossRef]
- Ramos, J. Using TF-IDF to determine word relevance in document queries. In Proceedings of the First Instructional Conference on Machine Learning, Los Angeles, CA, USA, 23–24 June 2003; Volume 242, pp. 29–48. [Google Scholar]
- Lahitani, A.R.; Permanasari, A.E.; Setiawan, N.A. Cosine similarity to determine similarity measure: Study case in online essay assessment. In Proceedings of the 2016 4th International Conference on Cyber and IT Service Management, Bandung, Indonesia, 26–27 April 2016; pp. 1–6. [Google Scholar]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. InterJournal Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Nunez-Iglesias, J.; Van Der Walt, S.; Dashnow, H. Elegant SciPy: The Art of Scientific Python; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2017. [Google Scholar]
- Cui, M. Introduction to the K-means clustering algorithm based on the elbow method. Account. Audit. Financ. 2020, 1, 5–8. [Google Scholar]
- Satopaa, V.; Albrecht, J.; Irwin, D.; Raghavan, B. Finding a “kneedle” in a haystack: Detecting knee points in system behavior. In Proceedings of the 2011 31st International Conference on Distributed Computing Systems Workshops, Minneapolis, MN, USA, 20–24 June 2011; pp. 166–171. [Google Scholar]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A K-means clustering algorithm. J. R. Stat. Society. Ser. C (Appl. Stat.) 1979, 28, 100–108. [Google Scholar] [CrossRef]
- Johnson, S.C. Hierarchical clustering schemes. Psychometrika 1967, 32, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, B.; George, D.J.; Manikandan, G.; Thomas, T. A comparative study on K-means clustering and agglomerative hierarchical clustering. Int. J. Emerg. Trends Eng. Res. 2020, 8, 1600–1604. [Google Scholar]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient estimation of word representations in vector space. arXiv 2013, arXiv:1301.3781. [Google Scholar]
- Cronbach, L.J.; Meehl, P.E. Construct validity in psychological tests. Psychol. Bull. 1955, 52, 281. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).