Core Needle Biopsy Enhances the Activity of the CCL2/CCR2 Pathway in the Microenvironment of Invasive Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Tissue Samples
2.2. Immunohistochemistry
2.3. Evaluation of the Staining Results
2.4. Statistical Methods
3. Results
3.1. Distribution of the CCL2/CCR2 Pathway Components
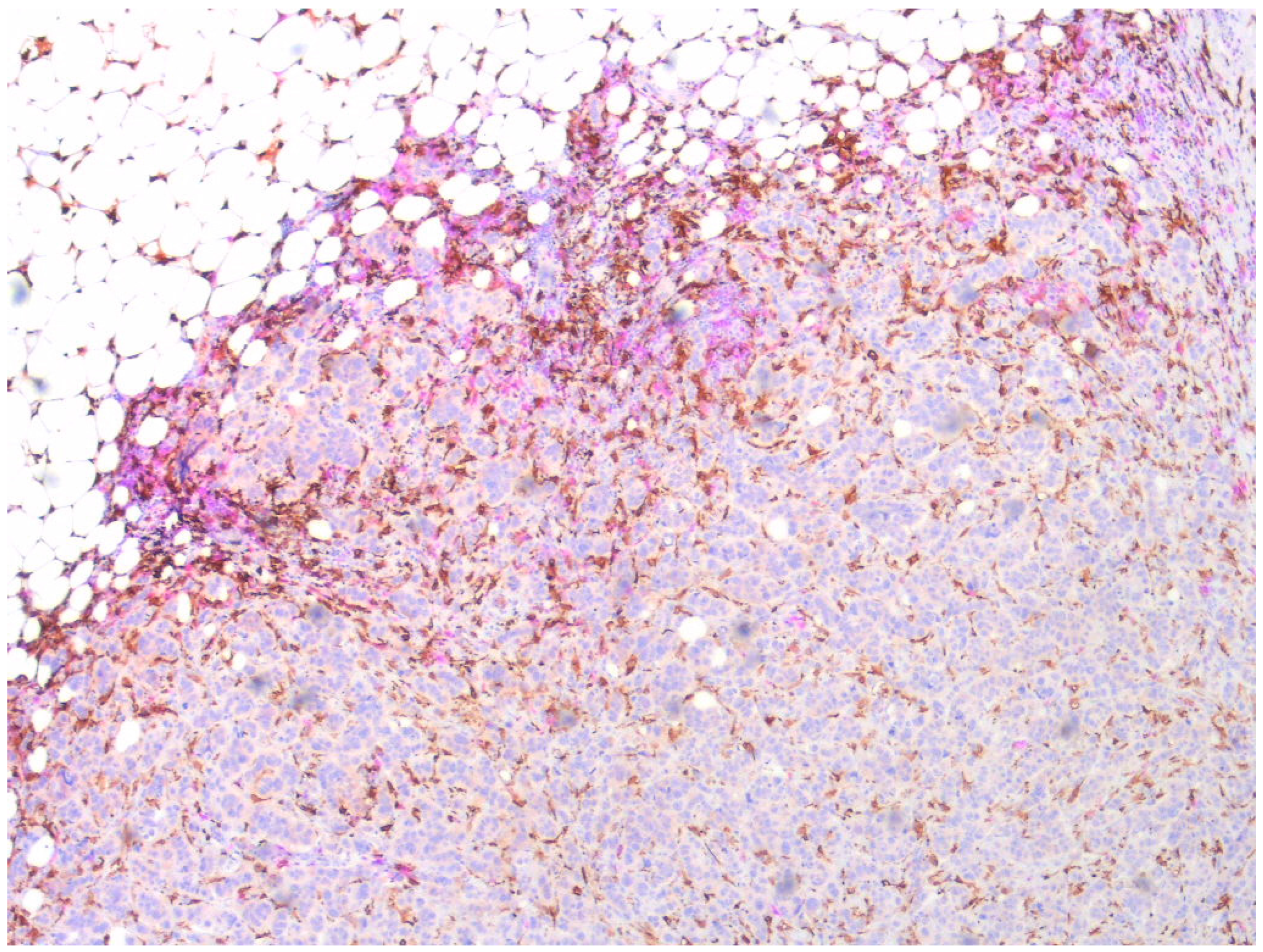
3.2. Localization of the CCL2/CCR2 Pathway Marker-Positive Monocytes/Macrophages in Tumors
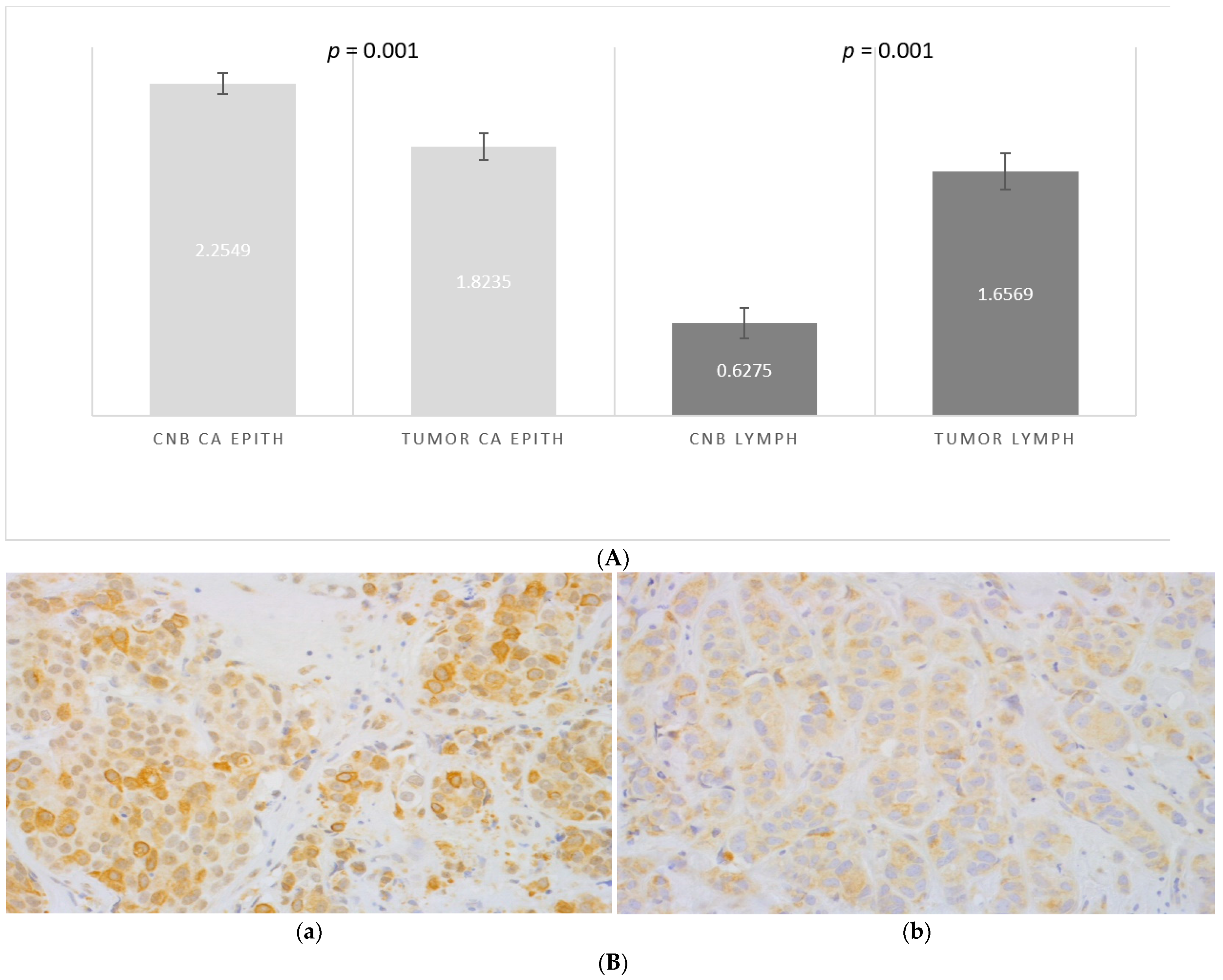
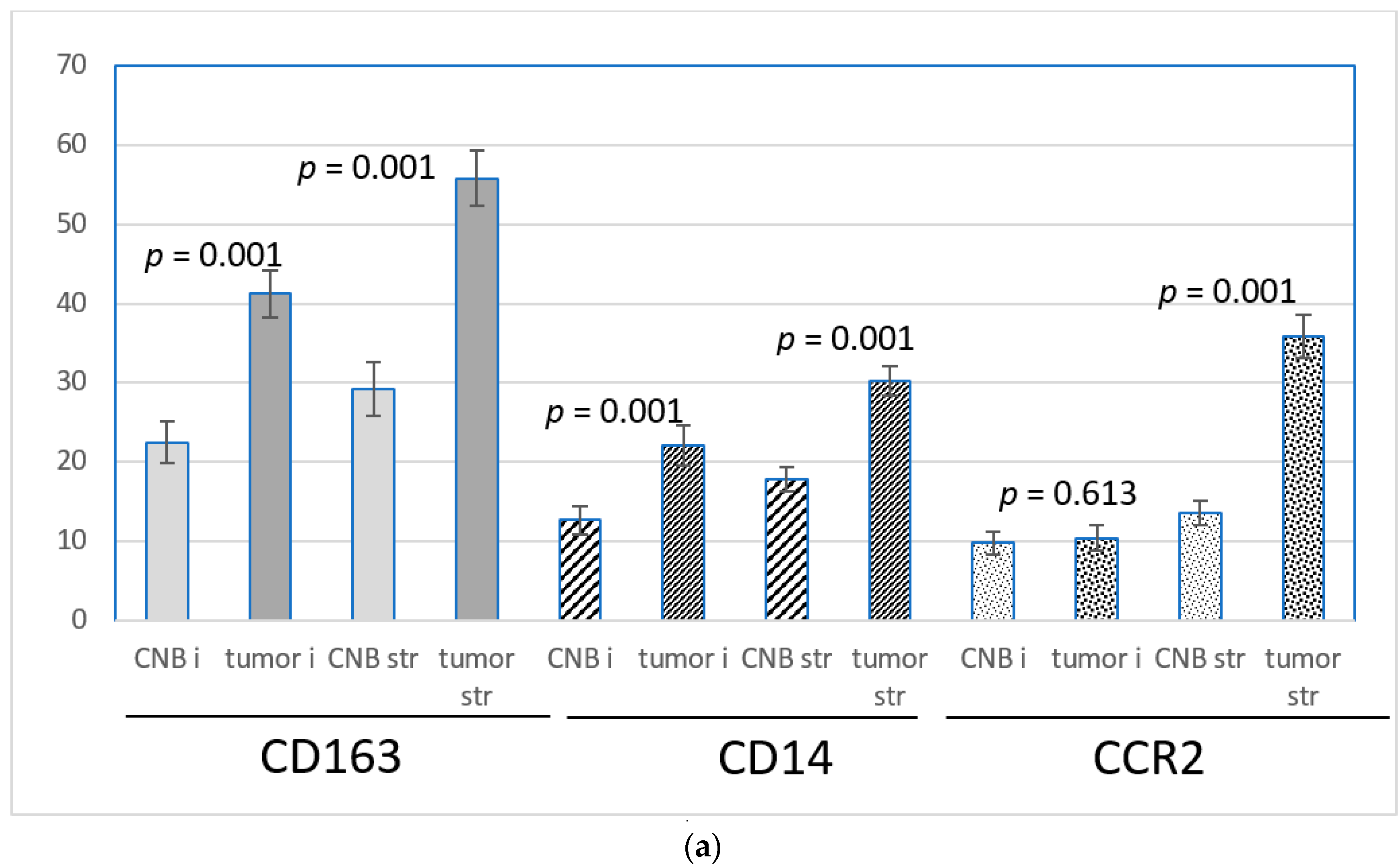
3.3. CNB-Induced Changes in the Expression of CCL2, CCR2, CD163 and CD14
3.4. Expression of CD163, CD14 and CCR2
3.5. Correlation of the CCL2/CCR2 Pathway Marker Expression with Clinicopathologic Parameters of the Tumors
4. Discussion
4.1. Aims of the Study
4.2. Expression of CCL2
4.3. Infiltration of CD163, CD14 and CCR2-Positive Monocytes/Macrophages
4.4. Impact of Clinicopathological Prognostic Markers on CNB-Induced Activation of the CCL2/CCR2 Axis
4.5. Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kasraeian, S.; Allison, D.C.; Ahlmann, E.R.; Fedenko, A.N.; Menendez, L.R. A Comparison of Fine-needle Aspiration, Core Biopsy, and Surgical Biopsy in the Diagnosis of Extremity Soft Tissue Masses. Clin. Orthop. Relat. Res. 2010, 468, 2992–3002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, J.W.; Robbins, A.G. A late look at the safety of aspiration biopsy. Cancer 1962, 15, 826–827. [Google Scholar] [CrossRef]
- Liikanen, J.; Leidenius, M.; Joensuu, H.; Vironen, J.; Heikkilä, P.; Meretoja, T. Breast cancer prognosis and isolated tumor cell findings in axillary lymph nodes after core needle biopsy and fine needle aspiration cytology: Biopsy method and breast cancer outcome. Eur. J. Surg. Oncol. 2016, 42, 64–70. [Google Scholar] [CrossRef]
- Engzell, U.; Zajicek, J. Aspiration biopsy of tumors of the neck. I. Aspiration biopsy and cytologic findings in 100 cases of congenital cysts. Acta Cytol. 1970, 14, 51–57. [Google Scholar]
- Uematsu, T.; Kasami, M. Risk of needle tract seeding of breast cancer: Cytological results derived from core wash material. Breast Cancer Res. Treat. 2007, 110, 51–55. [Google Scholar] [CrossRef]
- Hu, X.; Chow, L.W. Fine Needle Aspiration May Shed Breast Cells into Peripheral Blood as Determined by RT-PCR. Oncology 2000, 59, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Hansen, N.M.; Ye, X.; Grube, B.J.; Giuliano, A.E. Manipulation of the Primary Breast Tumor and the Incidence of Sentinel Node Metastases From Invasive Breast Cancer. Arch. Surg. 2004, 139, 634–640. [Google Scholar] [CrossRef] [Green Version]
- Sennerstam, R.B.; Franzén, B.S.H.; Wiksell, H.O.T.; Auer, G.U. Core-needle biopsy of breast cancer is associated with a higher rate of distant metastases 5 to 15 years after diagnosis than FNA biopsy. Cancer Cytopathol. 2017, 125, 748–756. [Google Scholar] [CrossRef]
- Chagpar, A.B.; Scoggins, C.R.; Sahoo, S.; Martin, R.C.; Carlson, D.J.; Laidley, A.L.; El-Eid, S.E.; McGlothin, T.Q.; Noyes, R.D.; Ley, P.B.; et al. Biopsy type does not influence sentinel lymph node status. Am. J. Surg. 2005, 190, 551–556. [Google Scholar] [CrossRef]
- Liebens, F.; Carly, B.; Cusumano, P.; Van Beveren, M.; Beier, B.; Fastrez, M.; Rozenberg, S. Breast cancer seeding associated with core needle biopsies: A systematic review. Maturitas 2009, 62, 113–123. [Google Scholar] [CrossRef]
- Loughran, C.F.; Keeling, C.R. Seeding of tumour cells following breast biopsy: A literature review. Br. J. Radiol. 2011, 84, 869–874. [Google Scholar] [CrossRef]
- Shyamala, K.; Girish, H.C.; Murgod, S. Risk of tumor cell seeding through biopsy and aspiration cytology. J. Int. Soc. Prev. Community Dent. 2014, 4, 5–11. [Google Scholar] [CrossRef] [Green Version]
- Mathenge, E.G.; Dean, C.A.; Clements, D.; Vaghar-Kashani, A.; Photopoulos, S.; Coyle, K.M.; Giacomantonio, M.; Malueth, B.; Nunokawa, A.; Jordan, J.; et al. Core needle biopsy of breast cancer tumors increases distant metastases in a mouse model. Neoplasia 2014, 16, 950–960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heiskala, M.; Leidenius, M.; Joensuu, K.; Heikkilä, P. High expression of CCL2 in tumor cells and abundant infiltration with CD14 positive macrophages predict early relapse in breast cancer. Virchows Arch. 2019, 474, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, L.; Yu, C.; Yang, X.-F.; Wang, H. Monocyte and macrophage differentiation: Circulation inflammatory monocyte as biomarker for inflammatory diseases. Biomark. Res. 2014, 2, 1. [Google Scholar] [CrossRef] [Green Version]
- Yao, M.; Fang, W.; Smart, C.; Hu, Q.; Huang, S.; Alvarez, N.; Fields, P.; Cheng, N. CCR2 Chemokine Receptors Enhance Growth and Cell-Cycle Progression of Breast Cancer Cells through SRC and PKC Activation. Mol. Cancer Res. 2018, 17, 604–617. [Google Scholar] [CrossRef] [Green Version]
- Messmer, M.; Netherby, C.S.; Banik, D.; Abrams, S.I. Tumor-induced myeloid dysfunction and its implications for cancer immunotherapy. Cancer Immunol. Immunother. 2014, 64, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, W.B.; Jokar, I.; Zou, A.; Lambert, D.; Dendukuri, P.; Cheng, N. CCL2/CCR2 Chemokine Signaling Coordinates Survival and Motility of Breast Cancer Cells through Smad3 Protein- and p42/44 Mitogen-activated Protein Kinase (MAPK)-dependent Mechanisms. J. Biol. Chem. 2012, 287, 36593–36608. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Yao, W.; Yuan, Y.; Chen, P.; Li, B.; Li, J.; Chu, R.; Song, H.; Xiel, D.; Jiang, X.; et al. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut 2017, 66, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Ueno, T.; Toi, M.; Saji, H.; Muta, M.; Bando, H.; Kuroi, K.; Koike, M.; Inadera, H.; Matsushima, K. Significance of macrophage chemoattractant protein-1 in macrophage recruitment, angiogenesis, and survival in human breast cancer. Clin. Cancer Res. 2000, 6, 3282–3289. [Google Scholar]
- Owen, J.L.; Lopez, D.M.; Grosso, J.F.; Guthrie, K.M.; Herbert, L.M.; Torroella-Kouri, M.; Iragavarapu-Charyulu, V. The expression of CCL2 by T lymphocytes of mammary tumor bearers: Role of tumor-derived factors. Cell. Immunol. 2005, 235, 122–135. [Google Scholar] [CrossRef]
- Hao, Q.; Vadgama, J.V.; Wang, P. CCL2/CCR2 signaling in cancer pathogenesis. Cell Commun. Signal. 2020, 18, 82. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, T. The chemokine MCP-1 (CCL2) in the host interaction with cancer: A foe or ally? Cell and Mol. Immunology 2018, 15, 335–345. [Google Scholar] [CrossRef] [Green Version]
- Bussard, K.M.; Mutkus, L.; Strumpf, K.; Comez-Manzano, C.; Marini, F.C. Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res. 2016, 18, 84. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Xu, M.; Yuan, C.; Yin, L.; Chen, X.; Zhou, X.; Li, G.; Fu, Y.; Feghali-Bostwick, C.A.; Pang, L. Transcriptional regulation of increased CCL2 expression in pulmonary fibrosis involves nuclear factor-κB and activator protein-1. Int. J. Biochem. Cell Biol. 2013, 45, 1366–1376. [Google Scholar] [CrossRef] [PubMed]
- Nakatsumi, H.; Matsumoto, M.; Nakayama, K.I. Noncanonical Pathway for Regulation of CCL2 Expression by an mTORC1-FOXK1 Axis Promotes Recruitment of Tumor-Associated Macrophages. Cell Rep. 2017, 21, 2471–2486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; A Knight, D.; A Snyder, L.; Smyth, M.J.; Stewart, T.J. A role for CCL2 in both tumor progression and immunosurveillance. OncoImmunology 2013, 2, e25474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gschwandtner, M.; Derler, R.; Midwood, K.S. More Than Just Attractive: How CCL2 Influences Myeloid Cell Behavior Beyond Chemotaxis. Front. Immunol. 2019, 10, 2759. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.N.; Campbell, J.J.; Salanga, C.L.; Ertl, L.S.; Wang, Y.; Yau, S.; Dang, T.; Zeng, Y.; McMahon, J.P.; Krasinski, A.; et al. CCR2-Mediated Uptake of Constitutively Produced CCL2: A Mechanism for Regulating Chemokine Levels in the Blood. J. Immunol. 2019, 203, 3157–3165. [Google Scholar] [CrossRef]
- Yao, M.; Brummer, G.; Acevedo, D.; Cheng, N. Cytokine Regulation of Metastasis and Tumorigenicity. Adv. Cancer Res. 2016, 132, 265–367. [Google Scholar] [CrossRef]
- O’Connor, T.; Borsig, L.; Heikenwalder, M. CCL2-CCR2 signaling in disease pathogenesis. Endocr. Metab. Immune Disord. Drug Targets 2015, 15, 105–118. [Google Scholar] [CrossRef]
- Willenborg, S.; Lucas, T.; Van Loo, G.; Knipper, J.; Krieg, T.; Haase, I.; Brachvogel, B.; Hammerschmidt, M.; Nagy, A.; Ferrara, N.; et al. CCR2 recruits an inflammatory macrophage subpopulation critical for angiogenesis in tissue repair. Blood 2012, 120, 613–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, B.-Z.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Campion, L.R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef] [Green Version]
- Franklin, R.A.; Liao, W.; Sarkar, A.; Kim, M.V.; Bivona, M.R.; Liu, K.; Pamer, E.G.; Li, M.O. The cellular and molecular origin of tumor-associated macrophages. Science 2014, 344, 921–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Speigl, L.; Burow, H.; Bailur, J.K.; Janssen, N.; Walter, C.-B.; Pawelec, G.; Shipp, C. CD14+ HLA-DR-/low MDSCs are elevated in the periphery of early-stage breast cancer patients and suppress autologous T cell proliferation. Breast Cancer Res. Treat. 2018, 168, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Laborde, R.R.; Elin, Y.; Gustafson, M.P.; Bulur, P.A.; Dietz, A.B. Cancer Vaccines in the World of Immune Suppressive Monocytes (CD14+HLA-DRlo/neg Cells): The Gateway to Improved Responses. Front. Immunol. 2014, 5, 147. [Google Scholar] [CrossRef]
- Shi, C.; Pamer, E.G. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 2011, 11, 762–774. [Google Scholar] [CrossRef] [Green Version]
- Inwald, E.C.; Klinkhammer-Schalke, M.; Hofstädter, F.; Zeman, F.; Koller, M.; Gerstenhauer, M.; Ortmann, O. Ki-67 is a prognostic parameter in breast cancer patients: Results of a large population-based cohort of a cancer registry. Breast Cancer Res. Treat. 2013, 139, 539–552. [Google Scholar] [CrossRef] [Green Version]
- Tabar, L.; Fagerberg, G.; Chen, H.H.; Duffy, S.W.; Gad, A. Tumour development, histology and grade of breast cancers: Prognosis and progression. Int. J. Cancer 1996, 66, 413–419. [Google Scholar] [CrossRef]
- Patani, N.; Martin, L.-A.; Dowsett, M. Biomarkers for the clinical management of breast cancer: International perspective. Int. J. Cancer 2012, 133, 1–13. [Google Scholar] [CrossRef]
- Hammond, M.E.H.; Hayes, D.F.; Dowsett, M.; Allred, D.C.; Hagerty, K.L.; Badve, S.; Fitzgibbons, P.L.; Francis, G.; Goldstein, N.S.; Hayes, M.; et al. American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Immunohistochemical Testing of Estrogen and Progesterone Receptors in Breast Cancer. J. Clin. Oncol. 2010, 28, 2784–2795. [Google Scholar] [CrossRef] [Green Version]
- Rakha, E.A.; El-Sayed, M.E.; Lee, A.H.S.; Elston, C.W.; Grainge, M.J.; Hodi, Z.; Blamey, R.W.; Ellis, I.O. Prognostic Significance of Nottingham Histologic Grade in Invasive Breast Carcinoma. J. Clin. Oncol. 2008, 26, 3153–3158. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; He, M.; Zuo, W.-J.; Hao, S.; Wang, Z.-H.; Shao, Z.-M. Tumor Size Still Impacts Prognosis in Breast Cancer With Extensive Nodal Involvement. Front. Oncol. 2021, 11, 585613. [Google Scholar] [CrossRef] [PubMed]
- Witz, I.P. Tumor-microenvironment interactions: Dangerous liaisons. Adv. Cancer Res. 2008, 100, 203–239. [Google Scholar]
- Chun, E.; Lavoie, S.; Michaud, M.; Gallini, C.A.; Kim, J.; Soucy, G.; Odze, R.; Glickman, J.N.; Garrett, W.S. CCL2 Promotes Colorectal Carcinogenesis by Enhancing Polymorphonuclear Myeloid-Derived Suppressor Cell Population and Function. Cell Rep. 2015, 12, 244–257. [Google Scholar] [CrossRef] [Green Version]
- Valković, T.; Lučin, K.; Krstulja, M.; Dobi-Babić, R.; Jonjić, N. Expression of monocyte chemotactic protein-1 in human invasive ductal breast cancer. Pathol. Res. Pract. 1998, 194, 335–340. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, Q.; Xu, M.; Wang, L.; Chen, X.; Feng, Y.; Li, Y.; Zhang, X.; Cui, W.; Jia, X. CCL2-CCR2 axis recruits tumor associated macrophages to induce immune evasion through PD-1 signaling in esophageal carcinogenesis. Mol. Cancer 2020, 19, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, C.-B.; Wang, A.; Meloni, D.; Zhang, K.; Kong, L.; Feng, H.; Glenn, J.; Huang, T.; Zhang, Y.; Cao, G.; et al. Discovery of INCB3344, a potent, selective and orally bioavailable antagonist of human and murine CCR2. Bioorganic Med. Chem. Lett. 2010, 20, 7473–7478. [Google Scholar] [CrossRef] [PubMed]
- Roblek, M.; Strutzmann, E.; Zankl, C.; Adage, T.; Heikenwalder, M.; Atlic, A.; Weis, R.; Kungl, A.; Borsig, L. Targeting of CCL2-CCR2-Glycosaminoglycan Axis Using a CCL2 Decoy Protein Attenuates Metastasis through Inhibition of Tumor Cell Seeding. Neoplasia 2016, 18, 49–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, S.Y.; Yuzhalin, A.; Gordon-Weeks, A.N.; Muschel, R.J. Targeting the CCL2-CCR2 signaling axis in cancer metastasis. Oncotarget 2016, 7, 28697–28710. [Google Scholar] [CrossRef] [Green Version]
- Lepique, A.P.; Daghastanli, K.R.P.; Cuccovia, I.M.; Villa, L.L. HPV16 Tumor Associated Macrophages Suppress Antitumor T Cell Responses. Clin. Cancer Res. 2009, 15, 4391–4400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allavena, P.; Mantovani, A. Immunology in the clinic review series; focus on cancer: Tumour-associated macrophages: Undisputed stars of the inflammatory tumour microenvironment. Clin. Exp. Immunol. 2012, 167, 195–205. [Google Scholar] [CrossRef]
- Predina, J.; Eruslanov, E.; Judy, B.; Kapoor, V.; Cheng, G.; Wang, L.-C.; Sun, J.; Moon, E.K.; Fridlender, Z.G.; Albelda, S.; et al. Changes in the local tumor microenvironment in recurrent cancers may explain the failure of vaccines after surgery. Proc. Natl. Acad. Sci. USA 2013, 110, E415–E424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sha, W.; Brune, B.; Weigert, A. The multi-faceted roles of € prostaglandin E2 in cancer-infiltrating mononuclear phagocyte biology. Immunobiology 2012, 217, 1225–1232. [Google Scholar] [CrossRef]
- Shurin, M.R. Dual role of immunomodulation by anticancer chemotherapy. Nat. Med. 2013, 19, 20–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| age at surgery | <50 year | ≥50 year |
| 9 (17.3%) | 43 (82.7%) | |
| nodal metastases 1 | − | + |
| 23 (44.2%) | 27 (51.9%) | |
| tumor size | <20 mm | ≥20 mm |
| 26 (50%) | 26 (50%) | |
| grade | 1 | 2–3 |
| 11 (21.2%) | 41 (78.8%) | |
| ER status | − | + |
| 4 (7.7%) | 48 (92.3%) | |
| PR status | − | + |
| 10 (19.2%) | 42 (80.8%) | |
| Ki67 expression | <30% | ≥30% |
| 31 (59.6%) | 21 (40.4%) | |
| HER2 status | − | + |
| 47 (90.4%) | 5 (9.6%) | |
| Histology 2 | ductal | lobular |
| 25 (48.1%) | 24 (46.2%) | |
| age at surgery | <50 year | ≥50 year |
| 9 (17.3%) | 43 (82.7%) | |
| nodal metastases 1 | − | + |
| 23 (44.2%) | 27 (51.9%) | |
| tumor size | <20 mm | ≥20 mm |
| 26 (50%) | 26 (50%) | |
| grade | 1 | 2–3 |
| 11 (21.2%) | 41 (78.8%) | |
| ER status | − | + |
| 4 (7.7%) | 48 (92.3%) | |
| PR status | − | + |
| 10 (19.2%) | 42 (80.8%) | |
| Ki67 expression | <30% | ≥30% |
| 31 (59.6%) | 21 (40.4%) | |
| HER2 status | − | + |
| 47 (90.4% | 5 (9.6%) | |
| Histology 2 | ductal | lobular |
| 25 (48.1%) | 24 (46.2%) |
| Nodal Involvement; Node Negative = n−, Node Positive = n+ | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All N | C | T | p | n− N | C | T | p | n+ N | C | T | p | |
| CCL2 tumor | 49 | 2.3 | 1.9 | 0.001 | 23 | 2.3 | 1.9 | 0.032 | 26 | 2.3 | 1.8 | 0.006 |
| CCL2 lymph | 49 | 0.63 | 1.7 | 0.001 | 23 | 0.61 | 1.7 | 0.001 | 26 | 0.65 | 1.7 | 0.001 |
| CD14 i | 49 | 12.7 | 22.1 | 0.001 | 22 | 14.7 | 28.3 | 0.008 | 27 | 11.7 | 18.5 | 0.004 |
| CD14 str | 49 | 17.8 | 30.2 | 0.001 | 22 | 20.9 | 28.9 | 0.004 | 27 | 16.3 | 32.0 | 0.001 |
| CD163 i | 50 | 22.5 | 41.3 | 0.001 | 23 | 23.5 | 44.6 | 0.001 | 27 | 22.3 | 40.0 | 0.001 |
| CD163 str | 50 | 29.2 | 55.8 | 0.001 | 23 | 32.5 | 54.8 | 0.002 | 27 | 28.0 | 57.8 | 0.001 |
| CCR2 i | 50 | 9.8 | 10.4 | 0.666 | 23 | 8.8 | 9.5 | 0.720 | 27 | 11.3 | 11.7 | 0.807 |
| CCR2 str | 50 | 13.6 | 35.8 | 0.001 | 23 | 15.7 | 37.8 | 0.001 | 27 | 12.5 | 35.2 | 0.001 |
| Tumor size <20 mm vs. ≥20 mm | ||||||||||||
| All N | C | T | p | <20 N | C | T | p | ≥20 N | C | T | p | |
| CCL2 tumor | 51 | 2.3 | 1.8 | 0.001 | 25 | 2.1 | 1.8 | 0.053 | 26 | 2.4 | 1.9 | 0.001 |
| CCL2 lymph | 51 | 0.63 | 1.7 | 0.001 | 25 | 0.56 | 1.4 | 0.001 | 26 | 0.7 | 1.9 | 0.001 |
| CD14 i | 51 | 12.7 | 22.1 | 0.001 | 25 | 11.3 | 23.2 | 0.003 | 26 | 14.0 | 21.1 | 0.026 |
| CD14 str | 51 | 17.8 | 30.2 | 0.001 | 25 | 17.4 | 29.5 | 0.001 | 26 | 18.3 | 31.0 | 0.001 |
| CD163 i | 52 | 22.5 | 41.3 | 0.001 | 26 | 19.7 | 42.5 | 0.001 | 26 | 25.3 | 40.0 | 0.001 |
| CD163 str | 52 | 29.2 | 55.8 | 0.001 | 26 | 28.2 | 54.4 | 0.001 | 26 | 30.2 | 57.1 | 0.001 |
| CCR2 i | 52 | 9.8 | 10.4 | 0.613 | 26 | 8.7 | 11.5 | 0.139 | 26 | 10.8 | 9.4 | 0.434 |
| CCR2 str | 52 | 13.6 | 35.8 | 0.001 | 26 | 13.5 | 32.7 | 0.001 | 26 | 13.7 | 38.8 | 0.001 |
| Gradus 1 vs. Gradus 2–3 | ||||||||||||
| All N | C | T | p | G1 N | C | T | p | G2–3 N | C | T | p | |
| CCL2 tumor | 51 | 2.3 | 1.8 | 0.001 | 11 | 2.2 | 2.0 | 0.506 | 40 | 2.3 | 1.8 | 0.001 |
| CCL2 lymph | 51 | 0.63 | 1.7 | 0.001 | 11 | 0.27 | 1.3 | 0.001 | 40 | 0.73 | 1.8 | 0.001 |
| CD14 i | 51 | 12.7 | 22.1 | 0.001 | 11 | 8.2 | 20.5 | 0.064 | 40 | 13.9 | 22.6 | 0.001 |
| CD14 str | 51 | 17.8 | 30.2 | 0.001 | 11 | 11.8 | 22.5 | 0.020 | 40 | 19.5 | 32.4 | 0.001 |
| CD163 i | 52 | 22.5 | 41.3 | 0.001 | 11 | 17.1 | 35.0 | 0.001 | 41 | 23.1 | 42.9 | 0.001 |
| CD163 str | 52 | 29.2 | 55.8 | 0.001 | 11 | 18.2 | 44.5 | 0.017 | 41 | 32.1 | 58.8 | 0.001 |
| CCR2 i | 52 | 9.8 | 10.4 | 0.613 | 11 | 5.4 | 6.5 | 0.319 | 41 | 11.0 | 11.5 | 0.752 |
| CCR2 str | 52 | 13.6 | 35.8 | 0.001 | 23 | 10.3 | 23.2 | 0.005 | 41 | 14.5 | 39.1 | 0.001 |
| Ductal vs. Lobular | ||||||||||||
| All N | C | T | p | Ductal N | C | T | p | Lobular N | C | T | p | |
| CCL2 tumor | 51 | 2.3 | 1.8 | 0.001 | 28 | 2.2 | 1.7 | 0.010 | 23 | 2.3 | 1.9 | 0.002 |
| CCL2 lymph | 51 | 0.63 | 1.7 | 0.001 | 28 | 0.79 | 1.8 | 0.001 | 23 | 0.43 | 1.5 | 0.001 |
| CD14 i | 51 | 12.7 | 22.1 | 0.001 | 27 | 12.3 | 24.2 | 0.001 | 24 | 13.0 | 19.8 | 0.055 |
| CD14 str | 51 | 17.8 | 30.2 | 0.001 | 27 | 18.5 | 31.9 | 0.001 | 24 | 17.1 | 28.3 | 0.001 |
| CD163 i | 52 | 22.5 | 41.3 | 0.001 | 28 | 24.4 | 46.3 | 0.001 | 24 | 20.2 | 35.4 | 0.004 |
| CD163 str | 52 | 29.2 | 55.8 | 0.001 | 28 | 32.8 | 61.4 | 0.001 | 24 | 25.0 | 49.2 | 0.001 |
| CCR2 i | 52 | 9.8 | 10.4 | 0.613 | 28 | 11.8 | 11.4 | 0.860 | 24 | 7.5 | 9.3 | 0.238 |
| CCR2 str | 52 | 13.6 | 35.8 | 0.001 | 28 | 16.2 | 39.1 | 0.001 | 24 | 10.6 | 31.9 | 0.001 |
| Ki67/MIB <30% vs. ≥30% | ||||||||||||
| All N | C | T | p | MIBlow N | C | T | p | MIBhigh N | C | T | p | |
| CCL2 tumor | 51 | 2.3 | 1.8 | 0.001 | 31 | 2.2 | 1.8 | 0.001 | 20 | 2.3 | 1.9 | 0.034 |
| CCL2 lymph | 51 | 0.63 | 1.7 | 0.001 | 31 | 0.2 | 1.8 | 0.001 | 20 | 0.85 | 1.9 | 0.001 |
| CD14 i | 51 | 12.7 | 22.1 | 0.001 | 31 | 10.4 | 21.8 | 0.001 | 20 | 16.3 | 22.6 | 0.118 |
| CD14 str | 51 | 17.8 | 30.2 | 0.001 | 31 | 14.7 | 25.7 | 0.001 | 20 | 22.8 | 37.3 | 0.001 |
| CD163 i | 52 | 22.5 | 41.3 | 0.001 | 31 | 19.4 | 32.9 | 0.001 | 21 | 27.0 | 53.7 | 0.001 |
| CD163 str | 52 | 29.2 | 55.8 | 0.001 | 31 | 21.1 | 45.2 | 0.001 | 20 | 41.1 | 71.4 | 0.001 |
| CCR2 i | 52 | 9.8 | 10.4 | 0.613 | 31 | 7,2 | 9.6 | 0.052 | 21 | 13.5 | 11.7 | 0.488 |
| CCR2 str | 52 | 13.6 | 35.8 | 0.001 | 31 | 11.2 | 25.8 | 0.001 | 21 | 17.2 | 50.5 | 0.001 |
| Time lapse from biopsy to tumor resection <25 days vs. ≥25 days | ||||||||||||
| All N | C | T | p | <25 N | C | T | p | ≥25 N | C | T | p | |
| CCL2 tumor | 51 | 2.3 | 1.8 | 0.001 | 22 | 2.3 | 1.9 | 0.018 | 29 | 2.2 | 1.7 | 0.003 |
| CCL2 lymph | 51 | 0.63 | 1.7 | 0.001 | 22 | 0.60 | 1.5 | 0.001 | 29 | 0.66 | 1.7 | 0.001 |
| CD14 i | 51 | 12.7 | 22.1 | 0.001 | 23 | 12.7 | 22.9 | 0.009 | 28 | 12.6 | 21.5 | 0.009 |
| CD14 str | 51 | 17.8 | 30.2 | 0.001 | 23 | 18.0 | 30.7 | 0.001 | 28 | 17.7 | 29.9 | 0.001 |
| CD163 i | 52 | 22.5 | 41.3 | 0.001 | 23 | 21.6 | 42.4 | 0.001 | 29 | 23.1 | 40.3 | 0.001 |
| CD163 str | 52 | 29.2 | 55.8 | 0.001 | 23 | 37.0 | 57.2 | 0.001 | 29 | 23.0 | 54.7 | 0.001 |
| CCR2 i | 52 | 9.8 | 10.4 | 0.613 | 23 | 11.3 | 9.8 | 0.528 | 29 | 8.5 | 10.9 | 0.053 |
| CCR2 str | 52 | 13.6 | 35.8 | 0.001 | 23 | 13.3 | 35.7 | 0.001 | 29 | 13.9 | 35.9 | 0.001 |
| Age <50 year vs. ≥50 year | ||||||||||||
| All N | C | T | p | <50 N | C | T | p | ≥50 N | C | T | p | |
| CCL2 tumor | 51 | 2.3 | 1.8 | 0.001 | 8 | 2.4 | 2.0 | 0.25 | 43 | 2.2 | 1.8 | 0.001 |
| CCL2 lymph | 51 | 0.63 | 1.7 | 0.001 | 8 | 0.6 | 1.4 | 0.04 | 43 | 0.63 | 1.7 | 0.001 |
| CD14 i | 51 | 12.7 | 22.1 | 0.001 | 9 | 8.3 | 22.6 | 0.06 | 42 | 13.6 | 22.0 | 0.002 |
| CD14 str | 51 | 17.8 | 30.2 | 0.001 | 9 | 13.3 | 27.2 | 0.01 | 42 | 18.8 | 30.9 | 0.001 |
| CD163 i | 52 | 22.5 | 41.3 | 0.001 | 9 | 17.8 | 36.1 | 0.02 | 43 | 23.4 | 42.3 | 0.001 |
| CD163 str | 52 | 29.2 | 55.8 | 0.001 | 9 | 29.4 | 52.8 | 0.004 | 43 | 29.1 | 56.4 | 0.001 |
| CCR2 i | 52 | 9.8 | 10.4 | 0.613 | 9 | 13.1 | 8.4 | 0.385 | 43 | 9.07 | 10.8 | 0.118 |
| CCR2 str | 52 | 13.6 | 35.8 | 0.001 | 9 | 11.4 | 38.3 | 0.003 | 43 | 14.1 | 35.2 | 0.001 |
| p-Value | |||
| node − | node + | ||
| CD163 CNB i low | 7 (30.4%) | 16 (69.0%) | |
| CD163 CNB i high | 16 (59.3%) | 11 (40.7%) | 0.042 |
| tumor size < 20mm | tumor size ≥ 20mm | ||
| CCL2 stromal lymphocytes low | 17 (64.5%) | 9 (34.6 %) | |
| CCL2 atromal lymphocytes high | 9 (34.6%) | 17 (65.4%) | 0.027 |
| tumor grade 1 | tumor grade 2–3 | ||
| CD163 CNB str low | 8 (34.8%) | 15 (65.2%) | |
| CD163 CNB str high | 3 (10.3%) | 26 (89.7%) | 0.032 |
| CD14 tumor str low | 7 (38.9%) | 11 (61.1%) | |
| CD14 tumor str high | 4 (11.8%) | 30 (88.2%) | 0.023 |
| duktal | lobular | ||
| CCR2 CNB str low | 6 (31,6%) | 13 (68.4%) | |
| CCR2 CNB str high | 19 (63.3%) | 11 (36.7%) | 0.030 |
| CD163 tumor i low | 6 (28.6%) | 15 (71.4%) | |
| CD163 tumor i high | 19 (67.9%) | 9 (32.1%) | 0.006 |
| CD163 tumor str low | 7 (33.3%) | 14 (66.7%) | |
| CD163 tumor str high | 18 (64.3%) | 10 (35.7%) | 0.032 |
| Ki67 < 30% | Ki67 ≥ 30% | ||
| CCL2 stromal lymphocytes low | 19 (73.1%) | 7 (26.9%) | |
| CCL2 stromal lymphocytes high | 12 (46.2%) | 14 (53.8%) | 0.048 |
| CCR2 CNB str low | 21 (87.5%) | 3 (12.5%) | |
| CCR2 CNB str high | 10 (35.7%) | 18 (64.3%) | 0.001 |
| CD163 tumor i low | 19 (79.2%) | 5 (20.8%) | |
| CD163 tumor i high | 12 (42.9%) | 16 (57.1%) | 0.008 |
| CD163 tumor str low | 19 (79.2%) | 5 (20.9%) | |
| CD163 tumor str high | 12 (49.2%) | 16 (57.1%) | 0.008 |
| CD14 CNB i low | 11 (91.7%) | 1 (8.3%) | |
| CD14 CNB i high | 20 (51.3%) | 19 (48.7%) | 0.012 |
| PR − | PR + | ||
| CCR2 CNB str low | 1 (4.8%) | 20 (95.2%) | |
| CCR2 CNB str high | 9 (29.0%) | 22 (71.0%) | 0.029 |
| CCR2 tumor str low | 1 (4.2%) | 23 (95.8%) | |
| CCR2 tumor str high | 9 (32.1%) | 19 (67.9%) | 0.011 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heiskala, M.; Joensuu, K.; Heikkilä, P. Core Needle Biopsy Enhances the Activity of the CCL2/CCR2 Pathway in the Microenvironment of Invasive Breast Cancer. Onco 2022, 2, 1-18. https://doi.org/10.3390/onco2010001
Heiskala M, Joensuu K, Heikkilä P. Core Needle Biopsy Enhances the Activity of the CCL2/CCR2 Pathway in the Microenvironment of Invasive Breast Cancer. Onco. 2022; 2(1):1-18. https://doi.org/10.3390/onco2010001
Chicago/Turabian StyleHeiskala, Marja, Kristiina Joensuu, and Päivi Heikkilä. 2022. "Core Needle Biopsy Enhances the Activity of the CCL2/CCR2 Pathway in the Microenvironment of Invasive Breast Cancer" Onco 2, no. 1: 1-18. https://doi.org/10.3390/onco2010001
APA StyleHeiskala, M., Joensuu, K., & Heikkilä, P. (2022). Core Needle Biopsy Enhances the Activity of the CCL2/CCR2 Pathway in the Microenvironment of Invasive Breast Cancer. Onco, 2(1), 1-18. https://doi.org/10.3390/onco2010001





