Adult B-Cell Acute Lymphoblastic Leukaemia Antigens and Enriched Pathways Identify New Targets for Therapy
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples
2.2. Antigen Identification
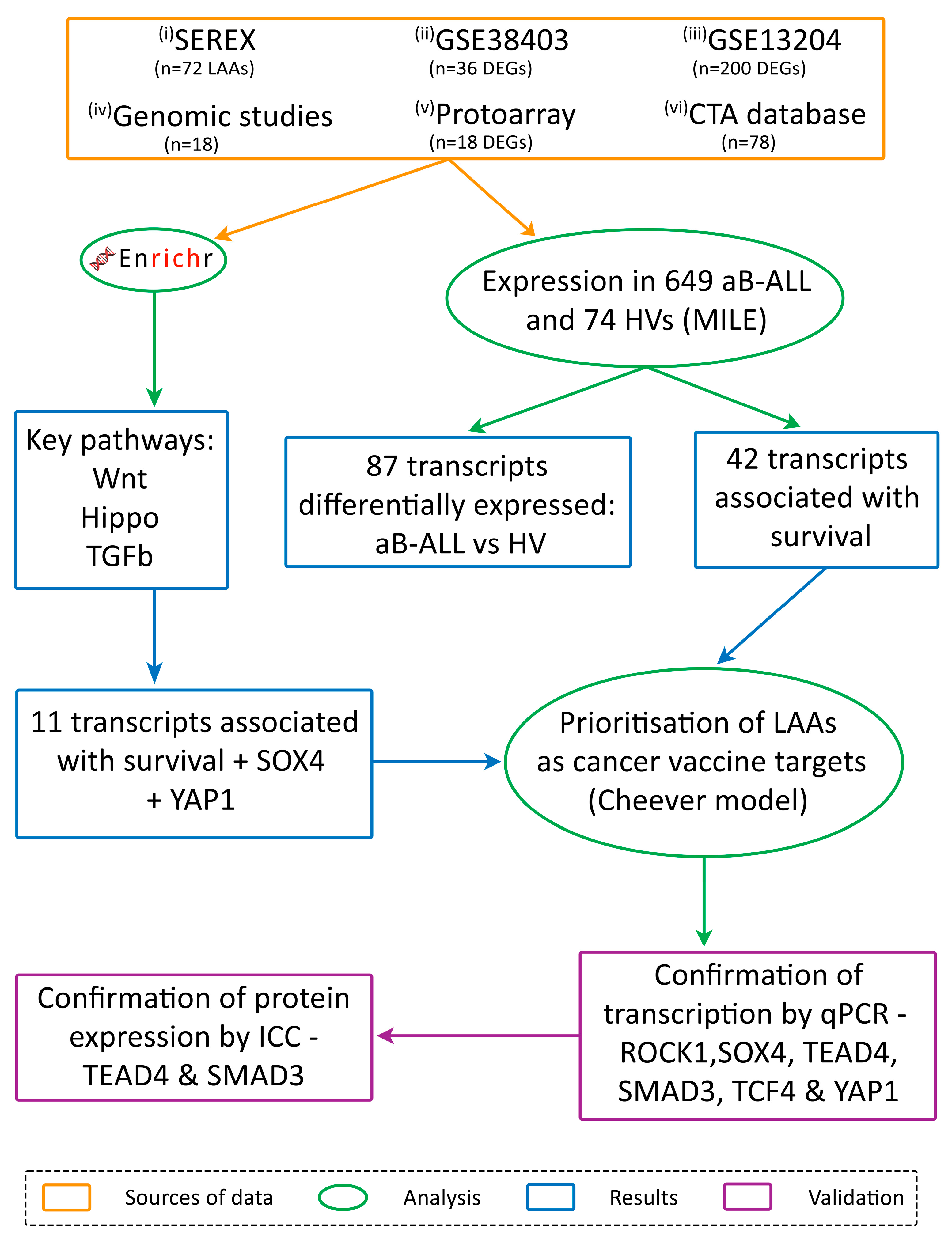
- (i)
- Serological analysis of recombinant cDNA expression libraries (SEREX)
- (ii) and (iii)
- Differentially expressed genes (DEGs) identified through the analysis of two microarray databases
- (iv)
- A review of genomic studies
- (v)
- Proto-array analysis
- (vi)
- the CTA database
2.3. Pathway Analysis and Survival
2.4. qPCR
2.5. Immunocytochemistry
3. Results
3.1. Immunoscreening Identified 72 aB-ALL Associated Antigens
3.2. Association Between Leukaemia-Associated Antigen (LAA) Expression in aB-ALL Cells and Patient Survival
3.3. TGFβ, Wnt and Hippo Pathways Were Highly Represented by the DEGs in aB-ALL Samples
3.4. Most of the aB-ALL Antigens Were Differentially Expressed in Solid Tumours
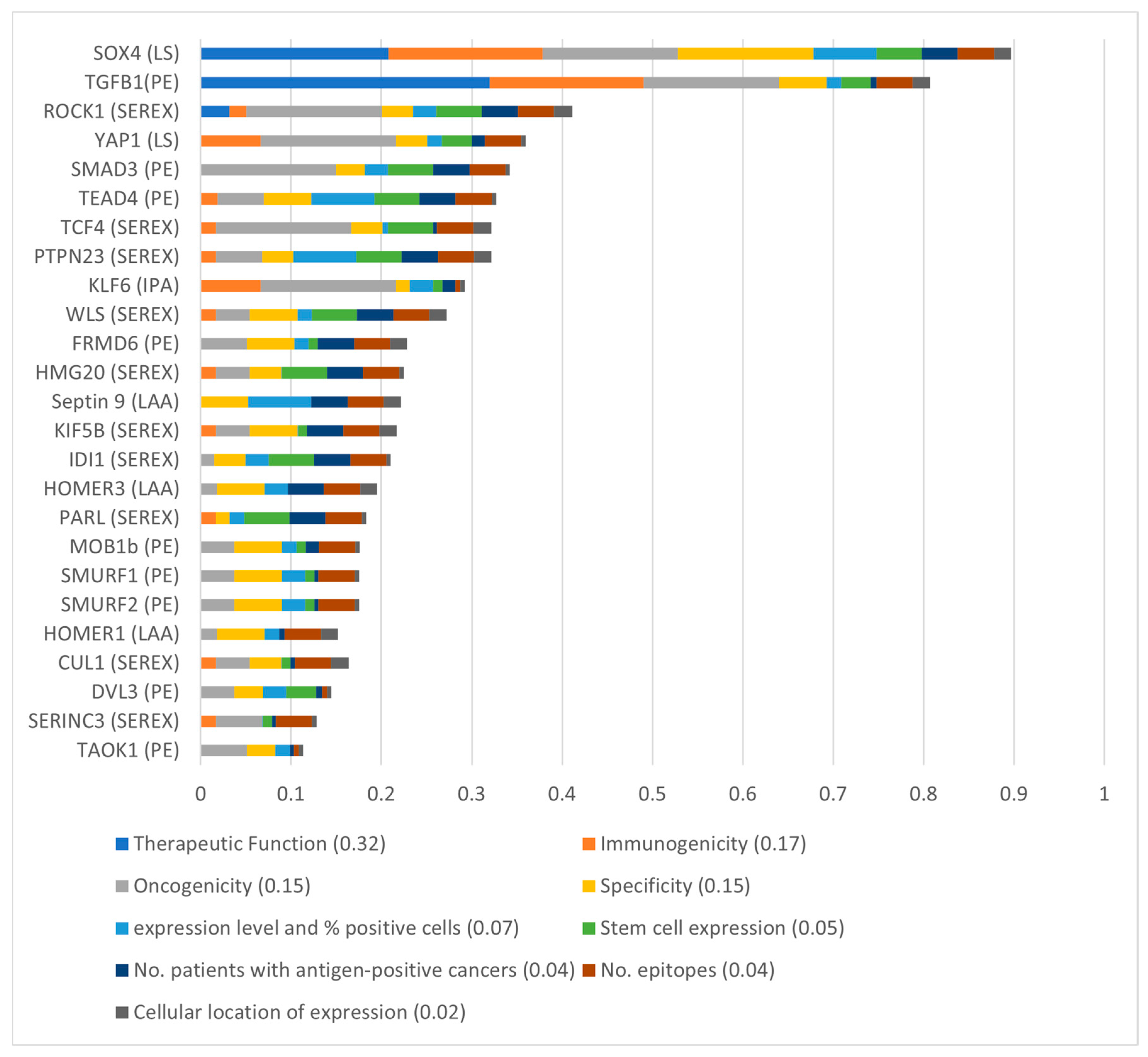
3.5. SOX4 and TGFβ1 Were the Top-Ranking aB-ALL Vaccine Targets
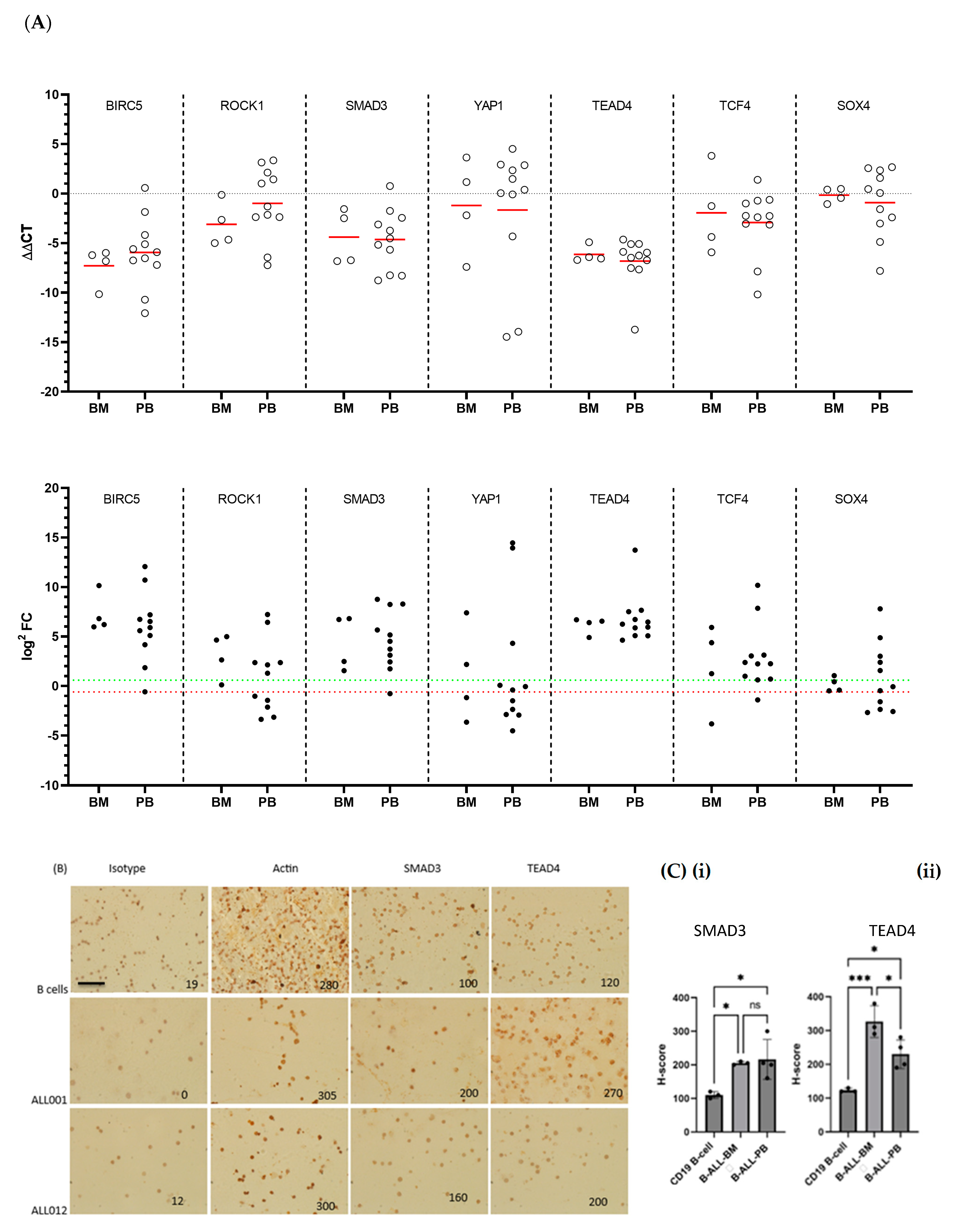
3.6. TCF4, SOX4 and SMAD3 Transcription Was Increased in Almost All aB-ALL Subtypes
3.7. TEAD4 and SMAD3 Protein Expression Was Elevated in aB-ALL Samples
4. Discussion
5. Limitations of the Study
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| aB-ALL | Adult B-cell acute lymphoblastic leukaemia |
| BIRC5 | baculoviral IAP repeat containing 5 |
| BMX | bone marrow tyrosine kinase |
| CAR | Chimeric antigen receptor |
| cB-ALL | children with B-ALL |
| CTA | cancer-testis antigen |
| DEGs | Differentially expressed genes |
| HV | Healthy volunteer |
| LAA | leukaemia-associated antigens |
| NS | Not significant |
| PB | Peripheral blood |
| PRKG1 | protein kinase cGMP-dependent 1 |
| R/R | relapsed/refractory |
| ROCK1 | Rho-associated coiled-coil containing protein kinase 1 |
| SMAD3 | SMAD family member 3 |
| SOX4 | SRY-Box Transcription Factor 4 |
| TAAs | Tumour-associated antigens |
| TBP1 | TATA-box binding protein |
| TCF4 | T cell Factor 4 |
| TEAD4 | TEA Domain Transcription Factor 4 |
| TGFβ | Transforming Growth Factor-β |
| YAP1 | Yes-associated protein1 |
References
- Samra, B.; Jabbour, E.; Ravandi, F.; Kantarjian, H.; Short, N.J. Evolving therapy of adult acute lymphoblastic leukemia: State-of-the-art treatment and future directions. J. Hematol. Oncol. 2020, 13, 70. [Google Scholar] [CrossRef]
- DeAngelo, D.J.; Jabbour, E.; Advani, A. Recent advances in managing acute lymphoblastic leukemia. Am. Soc. Clin. Oncol. Educ. Book. 2020, 40, 330–342. [Google Scholar] [CrossRef]
- Gokbuget, N.; Dombret, H.; Giebel, S.; Bruggemann, M.; Doubek, M.; Foa, R.; Hoelzer, D.; Kim, C.; Martinelli, G.; Parovichnikova, E.; et al. Minimal residual disease level predicts outcome in adults with Ph-negative B-precursor acute lymphoblastic leukemia. Hematology 2019, 24, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Kantarjian, H.M.; Vandendries, E.; Advani, A.S. Inotuzumab Ozogamicin for Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2016, 375, 2100–2101. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.A.; Ji, L.; Xu, X.; Devidas, M.; Hogan, L.E.; Borowitz, M.J.; Raetz, E.A.; Zugmaier, G.; Sharon, E.; Bernhardt, M.B.; et al. Effect of Postreinduction Therapy Consolidation With Blinatumomab vs. Chemotherapy on Disease-Free Survival in Children, Adolescents, and Young Adults With First Relapse of B-Cell Acute Lymphoblastic Leukemia: A Randomized Clinical Trial. JAMA 2021, 325, 833–842. [Google Scholar] [CrossRef]
- Stackelberg, A.V.; Jaschke, K.; Jousseaume, E.; Templin, C.; Jeratsch, U.; Kosmides, D.; Steffen, I.; Gokbuget, N.; Peters, C. Tisagenlecleucel vs. historical standard of care in children and young adult patients with relapsed/refractory B-cell precursor acute lymphoblastic leukemia. Leukemia 2023, 37, 2346–2355. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Kantarjian, H.; Jabbour, E.J. Adult acute lymphoblastic leukemia. In Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 2016; pp. 1645–1666. [Google Scholar]
- Martino, M.; Alati, C.; Canale, F.A.; Musuraca, G.; Martinelli, G.; Cerchione, C. A Review of Clinical Outcomes of CAR T-Cell Therapies for B-Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2021, 22, 2150. [Google Scholar] [CrossRef]
- Webster, J.; Luskin, M.R.; Prince, G.T.; DeZern, A.E.; DeAngelo, D.J.; Levis, M.J.; Blackford, A.; Sharon, E.; Streicher, H.; Luznik, L. Blinatumomab in combination with immune checkpoint inhibitors of PD-1 and CTLA-4 in adult patients with relapsed/refractory (R/R) CD19 positive B-cell acute lymphoblastic leukemia (ALL): Preliminary results of a phase I study. Blood 2018, 132, 557. [Google Scholar] [CrossRef]
- Namuduri, M.; Brentjens, R.J. Enhancing CAR T cell efficacy: The next step toward a clinical revolution? Expert. Rev. Hematol. 2020, 13, 533–543. [Google Scholar] [CrossRef]
- Jordaens, S.; Cooksey, L.; Freire Boullosa, L.; Van Tendeloo, V.; Smits, E.; Mills, K.I.; Orchard, K.H.; Guinn, B.A. New targets for therapy: Antigen identification in adults with B-cell acute lymphoblastic leukaemia. Cancer Immunol. Immunother. 2020, 69, 867–877. [Google Scholar] [CrossRef]
- Coutre, S.E.; Byrd, J.C.; Hillmen, P.; Barrientos, J.C.; Barr, P.M.; Devereux, S.; Robak, T.; Kipps, T.J.; Schuh, A.; Moreno, C.; et al. Long-term safety of single-agent ibrutinib in patients with chronic lymphocytic leukemia in 3 pivotal studies. Blood Adv. 2019, 3, 1799–1807. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R.; Eichhorst, B.; Hillmen, P.; Jurczak, W.; Kazmierczak, M.; Lamanna, N.; O’Brien, S.M.; Tam, C.S.; Qiu, L.; Zhou, K.; et al. Zanubrutinib or Ibrutinib in Relapsed or Refractory Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2023, 388, 319–332. [Google Scholar] [CrossRef]
- Hillmen, P.; Eichhorst, B.; Brown, J.R.; Lamanna, N.; O’Brien, S.M.; Tam, C.S.; Qiu, L.; Kazmierczak, M.; Zhou, K.; Simkovic, M.; et al. Zanubrutinib Versus Ibrutinib in Relapsed/Refractory Chronic Lymphocytic Leukemia and Small Lymphocytic Lymphoma: Interim Analysis of a Randomized Phase III Trial. J. Clin. Oncol. 2023, 41, 1035–1045. [Google Scholar] [CrossRef]
- Boullosa, L.F.; Savaliya, P.; Bonney, S.; Orchard, L.; Wickenden, H.; Lee, C.; Smits, E.; Banham, A.H.; Mills, K.I.; Orchard, K. Identification of survivin as a promising target for the immunotherapy of adult B-cell acute lymphoblastic leukemia. Oncotarget 2018, 9, 3853. [Google Scholar] [CrossRef]
- Li, Y.; He, W.; Gao, X.; Lu, X.; Xie, F.; Um, S.-W.; Kang, M.-W.; Yang, H.; Shang, Y.; Wang, Z. Cullin7 induces docetaxel resistance by regulating the protein level of the antiapoptotic protein Survivin in lung adenocarcinoma cells. J. Thorac. Dis. 2023, 15, 5006–5019. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Aljahdali, I.; Ling, X. Cancer therapeutics using survivin BIRC5 as a target: What can we do after over two decades of study? J. Exp. Clin. Cancer Res. 2019, 38, 368. [Google Scholar] [CrossRef]
- Valipour, B.; Abedelahi, A.; Naderali, E.; Velaei, K.; Movassaghpour, A.; Talebi, M.; Montazersaheb, S.; Karimipour, M.; Darabi, M.; Chavoshi, H.; et al. Cord blood stem cell derived CD16+ NK cells eradicated acute lymphoblastic leukemia cells using with anti-CD47 antibody. Life Sci. 2020, 242, 117223. [Google Scholar] [CrossRef]
- Haferlach, T.; Kohlmann, A.; Wieczorek, L.; Basso, G.; Kronnie, G.T.; Béné, M.-C.; De Vos, J.; Hernández, J.M.; Hofmann, W.-K.; Mills, K.I. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: Report from the International Microarray Innovations in Leukemia Study Group. J. Clin. Oncol. 2010, 28, 2529–2537. [Google Scholar] [CrossRef] [PubMed]
- Boncheva, V.B.; Linnebacher, M.; Kdimati, S.; Draper, H.; Orchard, L.; Mills, K.I.; O’Sullivan, G.; Tangney, M.; Guinn, B.-a. Identification of the Antigens Recognised by Colorectal Cancer Patients Using Sera from Patients Who Exhibit a Crohn’s-like Lymphoid Reaction. Biomolecules 2022, 12, 1058. [Google Scholar] [CrossRef]
- Liggins, A.P.; Guinn, B.A.; Banham, A.H. Identification of lymphoma-associated antigens using SEREX. Methods Mol. Med. 2005, 115, 109–128. [Google Scholar]
- Sahin, U.; Tureci, O.; Schmitt, H.; Cochlovius, B.; Johannes, T.; Schmits, R.; Stenner, F.; Luo, G.; Schobert, I.; Pfreundschuh, M. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc. Natl. Acad. Sci. USA 1995, 92, 11810–11813. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.; Brennan, S.; Milne, T.A.; Chen, W.-Y.; Li, Y.; Hurtz, C.; Kweon, S.-M.; Zickl, L.; Shojaee, S.; Neuberg, D. Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia. Cancer Discov. 2012, 2, 1004–1023. [Google Scholar] [CrossRef]
- Kohlmann, A.; Kipps, T.J.; Rassenti, L.Z.; Downing, J.R.; Shurtleff, S.A.; Mills, K.I.; Gilkes, A.F.; Hofmann, W.K.; Basso, G.; Dell’orto, M.C.; et al. An international standardization programme towards the application of gene expression profiling in routine leukaemia diagnostics: The Microarray Innovations in LEukemia study prephase. Br. J. Haematol. 2008, 142, 802–807. [Google Scholar] [CrossRef] [PubMed]
- Akhmedov, M.; Martinelli, A.; Geiger, R.; Kwee, I. Omics Playground: A comprehensive self-service platform for visualization, analytics and exploration of Big Omics Data. NAR Genom. Bioinform. 2020, 2, lqz019. [Google Scholar] [CrossRef] [PubMed]
- Iacobucci, I.; Mullighan, C.G. Genetic basis of acute lymphoblastic leukemia. J. Clin. Oncol. 2017, 35, 975. [Google Scholar] [CrossRef]
- Naik, A.; Lattab, B.; Qasem, H.; Decock, J. Cancer testis antigens: Emerging therapeutic targets leveraging genomic instability in cancer. Mol. Ther. Oncol. 2024, 32, 200768. [Google Scholar] [CrossRef]
- Almeida, L.G.; Sakabe, N.J.; deOliveira, A.R.; Silva, M.C.; Mundstein, A.S.; Cohen, T.; Chen, Y.T.; Chua, R.; Gurung, S.; Gnjatic, S.; et al. CTdatabase: A knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 2009, 37, 816–819. [Google Scholar] [CrossRef]
- Mohamed, E. Identification of Tumour Antigens That May Facilitate Effective Cancer Detection and Treatment; University of Hull: Hull, UK, 2023. [Google Scholar]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Gislason, M.H.; Demircan, G.S.; Prachar, M.; Furtwangler, B.; Schwaller, J.; Schoof, E.M.; Porse, B.T.; Rapin, N.; Bagger, F.O. BloodSpot 3.0: A database of gene and protein expression data in normal and malignant haematopoiesis. Nucleic Acids Res. 2024, 52, D1138–D1142. [Google Scholar] [CrossRef]
- Cheever, M.A.; Allison, J.P.; Ferris, A.S.; Finn, O.J.; Hastings, B.M.; Hecht, T.T.; Mellman, I.; Prindiville, S.A.; Viner, J.L.; Weiner, L.M.; et al. The prioritization of cancer antigens: A national cancer institute pilot project for the acceleration of translational research. Clin. Cancer Res. 2009, 15, 5323–5337. [Google Scholar] [CrossRef] [PubMed]
- Lossos, I.S.; Czerwinski, D.K.; Wechser, M.A.; Levy, R. Optimization of quantitative real-time RT-PCR parameters for the study of lymphoid malignancies. Leukemia 2003, 17, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Khan, G.; Brooks, S.; Mills, K.; Guinn, B. Infrequent expression of the cancer–testis antigen, PASD1, in ovarian cancer. Biomark. Cancer 2015, 7, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Biesterfeld, S.; Veuskens, U.; Schmitz, F.; Amo-Takyi, B.; Böcking, A. Interobserver reproducibility of immunocytochemical estrogen-and progesterone receptor status assessment in breast cancer. Anticancer. Res. 1996, 16, 2497–2500. [Google Scholar]
- Deng, Z.; Hasegawa, M.; Aoki, K.; Matayoshi, S.; Kiyuna, A.; Yamashita, Y.; Uehara, T.; Agena, S.; Maeda, H.; Xie, M. A comprehensive evaluation of human papillomavirus positive status and p16INK4a overexpression as a prognostic biomarker in head and neck squamous cell carcinoma. Int. J. Oncol. 2014, 45, 67–76. [Google Scholar] [CrossRef]
- Mehta, G.A.; Angus, S.P.; Khella, C.A.; Tong, K.; Khanna, P.; Dixon, S.A.; Verzi, M.P.; Johnson, G.L.; Gatza, M.L. SOX4 and SMARCA4 cooperatively regulate PI3k signaling through transcriptional activation of TGFBR2. NPJ Breast Cancer 2021, 7, 40. [Google Scholar] [CrossRef]
- Steinhardt, A.A.; Gayyed, M.F.; Klein, A.P.; Dong, J.; Maitra, A.; Pan, D.; Montgomery, E.A.; Anders, R.A. Expression of Yes-associated protein in common solid tumors. Hum. Pathol. 2008, 39, 1582–1589. [Google Scholar] [CrossRef]
- Mostufi-Zadeh-Haghighi, G.; Veratti, P.; Zodel, K.; Greve, G.; Waterhouse, M.; Zeiser, R.; Cleary, M.L.; Lübbert, M.; Duque-Afonso, J. Functional Characterization of Transforming Growth Factor-β Signaling in Dasatinib Resistance and Pre-BCR+ Acute Lymphoblastic Leukemia. Cancers 2023, 15, 4328. [Google Scholar] [CrossRef]
- Luo, X.; Ji, X.; Xie, M.; Zhang, T.; Wang, Y.; Sun, M.; Huang, W.; Xia, L. Advance of SOX transcription factors in hepatocellular carcinoma: From role, tumor immune relevance to targeted therapy. Cancers 2022, 14, 1165. [Google Scholar] [CrossRef] [PubMed]
- Mali, R.S.; Kapur, S.; Kapur, R. Role of Rho kinases in abnormal and normal hematopoiesis. Curr. Opin. Hematol. 2014, 21, 271. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gao, X.; Tian, X. High expression of long intergenic non-coding RNA LINC00662 contributes to malignant growth of acute myeloid leukemia cells by upregulating ROCK1 via sponging microRNA-340-5p. Eur. J. Pharmacol. 2019, 859, 172535. [Google Scholar] [CrossRef]
- Pan, T.; Wang, S.; Feng, H.; Xu, J.; Zhang, M.; Yao, Y.; Xu, K.; Niu, M. Preclinical evaluation of the ROCK1 inhibitor, GSK269962A, in acute myeloid leukemia. Front. Pharmacol. 2022, 13, 1064470. [Google Scholar] [CrossRef]
- Mali, R.S.; Ramdas, B.; Ma, P.; Shi, J.; Munugalavadla, V.; Sims, E.; Wei, L.; Vemula, S.; Nabinger, S.C.; Goodwin, C.B.; et al. Rho kinase regulates the survival and transformation of cells bearing oncogenic forms of KIT, FLT3, and BCR-ABL. Cancer Cell 2011, 20, 357–369. [Google Scholar] [CrossRef]
- Tamayo, E.; Alvarez, P.; Merino, R. TGFβ superfamily members as regulators of B cell development and function—Implications for autoimmunity. Int. J. Mol. Sci. 2018, 19, 3928. [Google Scholar] [CrossRef] [PubMed]
- Vicioso, Y.; Gram, H.; Beck, R.; Asthana, A.; Zhang, K.; Wong, D.P.; Letterio, J.; Parameswaran, R. Combination Therapy for Treating Advanced Drug-Resistant Acute Lymphoblastic Leukemia. Cancer Immunol. Res. 2019, 7, 1106–1119. [Google Scholar] [CrossRef]
- Rouce, R.H.; Shaim, H.; Sekine, T.; Weber, G.; Ballard, B.; Ku, S.; Barese, C.; Murali, V.; Wu, M.F.; Liu, H.; et al. The TGF-β/SMAD pathway is an important mechanism for NK cell immune evasion in childhood B-acute lymphoblastic leukemia. Leukemia 2016, 30, 800–811. [Google Scholar] [CrossRef]
- Khoury, H.; Pinto, R.; Meng, Y.; He, R.; Wu, S.; Minden, M.D. Noggin Overexpression Enhances Leukemic Progenitors Self-Renewal in AML by Abrogating the BMP Pathway Activation. Blood 2006, 108, 2214. [Google Scholar] [CrossRef]
- Chiarini, F.; Paganelli, F.; Martelli, A.M.; Evangelisti, C. The Role Played by Wnt/β-Catenin Signaling Pathway in Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2020, 21, 1098. [Google Scholar] [CrossRef]
- Cassaro, A.; Grillo, G.; Notaro, M.; Gliozzo, J.; Esposito, I.; Reda, G.; Trojani, A.; Valentini, G.; Di Camillo, B.; Cairoli, R.; et al. FZD6 triggers Wnt–signalling driven by WNT10BIVS1 expression and highlights new targets in T-cell acute lymphoblastic leukemia. Hematol. Oncol. 2021, 39, 364–379. [Google Scholar] [CrossRef]
- Yu, J.; Zheng, Y.; Dong, J.; Klusza, S.; Deng, W.-M.; Pan, D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev. Cell 2010, 18, 288–299. [Google Scholar] [CrossRef]
- Hill, V.K.; Dunwell, T.L.; Catchpoole, D.; Krex, D.; Brini, A.T.; Griffiths, M.; Craddock, C.; Maher, E.R.; Latif, F. Frequent epigenetic inactivation of KIBRA, an upstream member of the Salvador/Warts/Hippo (SWH) tumor suppressor network, is associated with specific genetic event in B-cell acute lymphocytic leukemia. Epigenetics 2011, 6, 326–332. [Google Scholar] [CrossRef]
- Fun, X.H.; Thibault, G. Lipid bilayer stress and proteotoxic stress-induced unfolded protein response deploy divergent transcriptional and non-transcriptional programmes. Biochim. Biophys. Acta BBA-Mol. Cell Biol. Lipids 2020, 1865, 158449. [Google Scholar] [CrossRef]
- Liu, S.X.; Xiao, H.R.; Wang, G.B.; Chen, X.W.; Li, C.G.; Mai, H.R.; Yuan, X.L.; Liu, G.S.; Wen, F.Q. Preliminary investigation on the abnormal mechanism of CD4+FOXP3+CD25high regulatory T cells in pediatric B-cell acute lymphoblastic leukemia. Exp. Ther. Med. 2018, 16, 1433–1441. [Google Scholar]
- Takahashi, H.; Kajiwara, R.; Kato, M.; Hasegawa, D.; Tomizawa, D.; Noguchi, Y.; Koike, K.; Toyama, D.; Yabe, H.; Kajiwara, M.; et al. Treatment outcome of children with acute lymphoblastic leukemia: The Tokyo Children’s Cancer Study Group (TCCSG) Study L04-16. Int. J. Hematol. 2018, 108, 98–108. [Google Scholar] [CrossRef]
- Backert, L.; Kohlbacher, O. Immunoinformatics and epitope prediction in the age of genomic medicine. Genome Med. 2015, 7, 119. [Google Scholar] [CrossRef]
- Caron, E.; Aebersold, R.; Banaei-Esfahani, A.; Chong, C.; Bassani-Sternberg, M. A case for a human immuno-peptidome project consortium. Immunity 2017, 47, 203–208. [Google Scholar] [CrossRef]
- Thomas, D.A.; Massagué, J. TGF-β directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 2005, 8, 369–380. [Google Scholar] [CrossRef]
- Greiner, J.; Goetz, M.; Schuler, P.J.; Bulach, C.; Hofmann, S.; Schrezenmeier, H.; Dӧhner, H.; Schneider, V.; Guinn, B.A. Enhanced stimulation of antigen-specific immune responses against nucleophosmin 1 mutated acute myeloid leukaemia by an anti-programmed death 1 antibody. Br. J. Haematol. 2022, 198, 866–874. [Google Scholar] [CrossRef]
- Brown, G. Lessons to cancer from studies of leukemia and hematopoiesis. Front. Cell Dev. Biol. 2022, 10, 993915. [Google Scholar] [CrossRef]
- Guinn, B.A.; Bland, E.A.; Lodi, U.; Liggins, A.P.; Tobal, K.; Petters, S.; Wells, J.W.; Banham, A.H.; Mufti, G.J. Humoral detection of leukaemia-associated antigens in presentation acute myeloid leukaemia. Biochem. Biophys. Res. Commun. 2005, 335, 1293–1304. [Google Scholar] [CrossRef]
- Liggins, A.P.; Guinn, B.A.; Hatton, C.S.; Pulford, K.; Banham, A.H. Serologic detection of diffuse large B-cell lymphoma-associated antigens. Int. J. Cancer 2004, 110, 563–569, Erratum in Int. J. Cancer 2004, 110, 934. [Google Scholar] [CrossRef]
- Silva, W.A.; Gnjatic, S.; Ritter, E.; Chua, R.; Cohen, T.; Hsu, M.; Jungbluth, A.A.; Altorki, N.K.; Chen, Y.-T.; Old, L.J. PLAC1, a trophoblast-specific cell surface protein, is expressed in a range of human tumors and elicits spontaneous antibody responses. Cancer Immun. Arch. 2007, 7, 18. [Google Scholar]
- Watkins, P.A.; Maiguel, D.; Jia, Z.; Pevsner, J. Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genomes. J. Lipid Res. 2007, 48, 2736–2750. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, M.; Xu, Z.; Cai, Y.; Peng, B.; Liang, Q.; Yan, Y.; Liu, W.; Kang, F.; He, Q.; et al. Identification of ACSF gene family as therapeutic targets and immune-associated biomarkers in hepatocellular carcinoma. Aging 2022, 14, 7926–7940. [Google Scholar] [CrossRef]
- Wang, Y.; Wo, Y.; Lu, T.; Sun, X.; Liu, A.; Dong, Y.; Du, W.; Su, W.; Huang, Z.; Jiao, W. Circ-AASDH functions as the progression of early stage lung adenocarcinoma by targeting miR-140-3p to activate E2F7 expression. Transl. Lung Cancer Res. 2021, 10, 57–70. [Google Scholar] [CrossRef]
- Tan, Y.; You, H.; Wu, C.; Altomare, D.A.; Testa, J.R. Appl1 is dispensable for mouse development, and loss of Appl1 has growth factor-selective effects on Akt signaling in murine embryonic fibroblasts. J. Biol. Chem. 2010, 285, 6377–6389. [Google Scholar] [CrossRef]
- Thomas, R.M.; Nechamen, C.A.; Mazurkiewicz, J.E.; Ulloa-Aguirre, A.; Dias, J.A. The adapter protein APPL1 links FSH receptor to inositol 1,4,5-trisphosphate production and is implicated in intracellular Ca2+ mobilization. Endocrinology 2011, 152, 1691–1701. [Google Scholar] [CrossRef]
- Zhai, J.S.; Song, J.G.; Zhu, C.H.; Wu, K.; Yao, Y.; Li, N. Expression of APPL1 is correlated with clinicopathologic characteristics and poor prognosis in patients with gastric cancer. Curr. Oncol. 2016, 23, e95–e101. [Google Scholar] [CrossRef]
- Hupalowska, A.; Pyrzynska, B.; Miaczynska, M. APPL1 regulates basal NF-κB activity by stabilizing NIK. J. Cell science 2012, 125, 4090–4102. [Google Scholar] [CrossRef]
- Wang, Y.; Tong, X.; Li, G.; Li, J.; Deng, M.; Ye, X. Ankrd17 positively regulates RIG-I-like receptor (RLR)-mediated immune signaling. Eur. J. Immunol. 2012, 42, 1304–1315. [Google Scholar] [CrossRef]
- Dong, L.; Lin, F.; Wu, W.; Huang, W.; Cai, Z. Transcriptional cofactor Mask2 is required for YAP-induced cell growth and migration in bladder cancer cell. J. Cancer 2016, 7, 2132–2138. [Google Scholar] [CrossRef]
- Wilkinson, S. ER-phagy: Shaping up and destressing the endoplasmic reticulum. FEBS J. 2019, 286, 2645–2663. [Google Scholar] [CrossRef]
- Behrendt, L.; Hoischen, C.; Kaether, C. Disease-causing mutated ATLASTIN 3 is excluded from distal axons and reduces axonal autophagy. Neurobiol. Dis. 2021, 155, 105400. [Google Scholar] [CrossRef] [PubMed]
- Waragai, M.; Nagamitsu, S.; Xu, W.; Li, Y.J.; Lin, X.; Ashizawa, T. Ataxin 10 induces neuritogenesis via interaction with G-protein β2 subunit. J. Neurosci. Res. 2006, 83, 1170–1178. [Google Scholar] [CrossRef]
- Schäfer, M.; Oeing, C.U.; Rohm, M.; Baysal-Temel, E.; Lehmann, L.H.; Bauer, R.; Volz, H.C.; Boutros, M.; Sohn, D.; Sticht, C.; et al. Ataxin-10 is part of a cachexokine cocktail triggering cardiac metabolic dysfunction in cancer cachexia. Mol. Metab. 2016, 5, 67–78. [Google Scholar] [CrossRef]
- Garcia-Gonzalo, F.R.; Corbit, K.C.; Sirerol-Piquer, M.S.; Ramaswami, G.; Otto, E.A.; Noriega, T.R.; Seol, A.D.; Robinson, J.F.; Bennett, C.L.; Josifova, D.J. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 2011, 43, 776–784. [Google Scholar] [CrossRef]
- Hopp, K.; Heyer, C.M.; Hommerding, C.J.; Henke, S.A.; Sundsbak, J.L.; Patel, S.; Patel, P.; Consugar, M.B.; Czarnecki, P.G.; Gliem, T.J.; et al. B9D1 is revealed as a novel Meckel syndrome (MKS) gene by targeted exon-enriched next-generation sequencing and deletion analysis. Hum. Mol. Genet. 2011, 20, 2524–2534. [Google Scholar] [CrossRef] [PubMed]
- Song, C.-Q.; Li, Y.; Mou, H.; Moore, J.; Park, A.; Pomyen, Y.; Hough, S.; Kennedy, Z.; Fischer, A.; Yin, H. Genome-wide CRISPR screen identifies regulators of mitogen-activated protein kinase as suppressors of liver tumors in mice. Gastroenterology 2017, 152, 1161–1173.e1161. [Google Scholar] [CrossRef]
- Lee, Y.; Yu, Y.; Gunawardena, H.; Xie, L.; Chen, X. BCLAF1 is a radiation-induced H2AX-interacting partner involved in γH2AX-mediated regulation of apoptosis and DNA repair. Cell Death Disease 2012, 3, e359. [Google Scholar] [CrossRef] [PubMed]
- White, L.S.; Soodgupta, D.; Johnston, R.L.; Magee, J.A.; Bednarski, J.J. Bclaf1 Promotes Maintenance and Self-Renewal of Fetal Hematopoietic Stem Cells. Blood 2018, 132, 1269. [Google Scholar] [CrossRef]
- Almutairi, M.; Alrubie, T.; Alamri, A.; Almutairi, B.; Alrefaei, A.; Arafah, M.; Alanazi, M.; Semlali, A. Cancer-Testis Gene Biomarkers discovered in Colon Cancer Patients. Genes 2022, 13, 807. [Google Scholar] [CrossRef]
- Matsuda, A.; Suzuki, Y.; Honda, G.; Muramatsu, S.; Matsuzaki, O.; Nagano, Y.; Shimotohno, K.; Harada, T.; Nishida, E.; Hayashi, H. Large-scale identification and characterization of human genes that activate NF-κB and MAPK signaling pathways. Oncogene 2003, 22, 3307–3318. [Google Scholar] [CrossRef]
- Salian, S.; Guo, X.Y.; Murakami, Y.; Kinoshita, T.; Kaur, P.; Shukla, A.; Girisha, K.M.; Fujita, M.; Campeau, P.M. C18orf32 loss-of-function is associated with a neurodevelopmental disorder with hypotonia and contractures. Hum. Genet. 2022, 141, 1423–1429. [Google Scholar] [CrossRef] [PubMed]
- Malik, M.; Jividen, K.; Padmakumar, V.; Cataisson, C.; Li, L.; Lee, J.; Howard, O.Z.; Yuspa, S.H. Inducible NOS-induced chloride intracellular channel 4 (CLIC4) nuclear translocation regulates macrophage deactivation. Proc. Natl. Acad. Sci. USA 2012, 109, 6130–6135. [Google Scholar] [CrossRef]
- Huang, S.; Huang, Z.; Chen, P.; Feng, C. Aberrant Chloride Intracellular Channel 4 Expression Is Associated With Adverse Outcome in Cytogenetically Normal Acute Myeloid Leukemia. Front. Oncol. 2020, 10, 1648. [Google Scholar] [CrossRef]
- Skaar, J.R.; Pagan, J.K.; Pagano, M. SCF ubiquitin ligase-targeted therapies. Nat. Rev. Drug Discov. 2014, 13, 889–903. [Google Scholar] [CrossRef]
- Liang, K.; Liao, L.; Liu, Q.; Ouyang, Q.; Jia, L.; Wu, G. microRNA-377-3p inhibits osteosarcoma progression by targeting CUL1 and regulating Wnt/β-catenin signaling pathway. Clin. Transl. Oncol. 2021, 23, 2350–2357. [Google Scholar] [CrossRef]
- Lee, E.J.; Jo, M.; Rho, S.B.; Park, K.; Yoo, Y.N.; Park, J.; Chae, M.; Zhang, W.; Lee, J.H. Dkk3, downregulated in cervical cancer, functions as a negative regulator of β-catenin. Int. J. Cancer 2009, 124, 287–297. [Google Scholar] [CrossRef]
- Katase, N.; Nagano, K.; Fujita, S. DKK3 expression and function in head and neck squamous cell carcinoma and other cancers. J. Oral Biosci. 2020, 62, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, H.; Cheng, C.; Zheng, F.; Zhao, Z.; Chen, Q.; Zeng, W.; Zhang, P.; Huang, C.; Jiang, W.; et al. ZNF191 alters DNA methylation and activates the PI3K-AKT pathway in hepatoma cells via transcriptional regulation of DNMT1. Cancer Med. 2022, 11, 1269–1280. [Google Scholar] [CrossRef] [PubMed]
- Haque, S.; Vaiselbuh, S.R. Exosomal DNMT1 mRNA transcript is elevated in acute lymphoblastic leukemia which might reprograms leukemia progression. Cancer Genet. 2022, 260–261, 57–64. [Google Scholar] [CrossRef]
- Yin, J.-Y.; Zhang, J.-T.; Zhang, W.; Zhou, H.-H.; Liu, Z.-Q. eIF3a: A new anticancer drug target in the eIF family. Cancer Lett. 2018, 412, 81–87. [Google Scholar] [CrossRef]
- Chen, Y.-X.; Wang, C.-J.; Xiao, D.-S.; He, B.-M.; Li, M.; Yi, X.-P.; Zhang, W.; Yin, J.-Y.; Liu, Z.-Q. eIF3a R803K mutation mediates chemotherapy resistance by inducing cellular senescence in small cell lung cancer. Pharmacol. Res. 2021, 174, 105934. [Google Scholar] [CrossRef]
- Zhu, X.N.; Wei, Y.S.; Yang, Q.; Liu, H.R.; Zhi, Z.; Zhu, D.; Xia, L.; Hong, D.L.; Yu, Y.; Chen, G.Q. FBXO22 promotes leukemogenesis by targeting BACH1 in MLL-rearranged acute myeloid leukemia. J. Hematol. Oncol. 2023, 16, 9. [Google Scholar] [CrossRef]
- Greer, P. Closing in on the biological functions of Fps/Fes and Fer. Nat. Rev. Mol. Cell Biol. 2002, 3, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Wu, S.; Li, X.; Wang, Y.; Ren, R.; Lai, Y.; Ye, J. High expression of FER tyrosine kinase predicts poor prognosis in clear cell renal cell carcinoma. Oncol. Lett. 2013, 5, 473–478. [Google Scholar] [CrossRef]
- Baldassarre, M.; Razinia, Z.; Burande, C.F.; Lamsoul, I.; Lutz, P.G.; Calderwood, D.A. Filamins regulate cell spreading and initiation of cell migration. PLoS ONE 2009, 4, e7830. [Google Scholar] [CrossRef]
- del Valle-Pérez, B.; Martínez, V.G.; Lacasa-Salavert, C.; Figueras, A.; Shapiro, S.S.; Takafuta, T.; Casanovas, O.; Capellà, G.; Ventura, F.; Viñals, F. Filamin B plays a key role in vascular endothelial growth factor-induced endothelial cell motility through its interaction with Rac-1 and Vav-2. J. Biol. Chem. 2010, 285, 10748–10760. [Google Scholar] [CrossRef] [PubMed]
- Chiu, D.S.; Oram, J.F.; LeBoeuf, R.C.; Alpers, C.E.; O’Brien, K.D. High-density lipoprotein-binding protein (HBP)/vigilin is expressed in human atherosclerotic lesions and colocalizes with apolipoprotein E. Arterioscler. Thromb. Vasc. Biol. 1997, 17, 2350–2358. [Google Scholar] [CrossRef]
- Yang, W.L.; Wei, L.; Huang, W.Q.; Li, R.; Shen, W.Y.; Liu, J.Y.; Xu, J.M.; Li, B.; Qin, Y. Vigilin is overexpressed in hepatocellular carcinoma and is required for HCC cell proliferation and tumor growth. Oncol. Rep. 2014, 31, 2328–2334. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.-H.; Yi, X.; Lamb, T.; Menzl, I.; Baker, T.; Shapiro, D.J.; Chambers, S.K. Posttranscriptional suppression of proto-oncogene c-fms expression by vigilin in breast cancer. Mol. Cell. Biol. 2011, 31, 215–225. [Google Scholar] [CrossRef]
- Han, S.P.; Tang, Y.H.; Smith, R. Functional diversity of the hnRNPs: Past, present and perspectives. Biochem. J. 2010, 430, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, K.; Sasaki, E.; Kimura, K.; Komori, K.; Shimizu, Y.; Yatabe, Y.; Aoki, M. HNRNPLL stabilizes mRNA for DNA replication proteins and promotes cell cycle progression in colorectal cancer cells. Cancer Sci. 2018, 109, 2458–2468. [Google Scholar] [CrossRef]
- Banerjee-Basu, S.; Baxevanis, A.D. Molecular evolution of the homeodomain family of transcription factors. Nucleic Acids Res. 2001, 29, 3258–3269. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, Y.; Su, X.; Lu, Y. HOXD8 inhibits the proliferation and migration of triple-negative breast cancer cells and induces apoptosis in them through regulation of AKT/mTOR pathway. Reprod. Biol. 2021, 21, 100544. [Google Scholar] [CrossRef]
- Laupitz, R.; Hecht, S.; Amslinger, S.; Zepeck, F.; Kaiser, J.; Richter, G.; Schramek, N.; Steinbacher, S.; Huber, R.; Arigoni, D. Biochemical characterization of Bacillus subtilis type II isopentenyl diphosphate isomerase, and phylogenetic distribution of isoprenoid biosynthesis pathways. Eur. J. Biochem. 2004, 271, 2658–2669. [Google Scholar] [CrossRef]
- Marona, P.; Górka, J.; Mazurek, Z.; Wilk, W.; Rys, J.; Majka, M.; Jura, J.; Miekus, K. MCPIP1 Downregulation in Clear Cell Renal Cell Carcinoma Promotes Vascularization and Metastatic Progression. Cancer Res. 2017, 77, 4905–4920. [Google Scholar] [CrossRef]
- Hirokawa, N.; Tanaka, Y. Kinesin superfamily proteins (KIFs): Various functions and their relevance for important phenomena in life and diseases. Exp. Cell Res. 2015, 334, 16–25. [Google Scholar] [CrossRef]
- Su, M.; Guo, J.; Huang, J. Meta-analysis of the correlation between the rs17401966 polymorphism in kinesin family member 1B and susceptibility to hepatitis B virus related hepatocellular carcinoma. Clin. Mol. Hepatol. 2017, 23, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Akbari Moqadam, F.; Lange-Turenhout, E.A.M.; Ariës, I.M.; Pieters, R.; den Boer, M.L. MiR-125b, miR-100 and miR-99a co-regulate vincristine resistance in childhood acute lymphoblastic leukemia. Leuk. Res. 2013, 37, 1315–1321. [Google Scholar] [CrossRef] [PubMed]
- Hansson, A.; Bloor, B.K.; Sarang, Z.; Haig, Y.; Morgan, P.R.; Stark, H.J.; Fusenig, N.E.; Ekstrand, J.; Grafström, R.C. Analysis of proliferation, apoptosis and keratin expression in cultured normal and immortalized human buccal keratinocytes. Eur. J. Oral Sci. 2003, 111, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Silveira, N.J.F.; Varuzza, L.; Machado-Lima, A.; Lauretto, M.S.; Pinheiro, D.G.; Rodrigues, R.V.; Severino, P.; Nobrega, F.G.; Silva, W.A.; de B Pereira, C.A.; et al. Searching for molecular markers in head and neck squamous cell carcinomas (HNSCC) by statistical and bioinformatic analysis of larynx-derived SAGE libraries. BMC Med. Genom. 2008, 1, 56. [Google Scholar] [CrossRef]
- Karol, S.E.; Mattano, L.A., Jr.; Yang, W.; Maloney, K.W.; Smith, C.; Liu, C.; Ramsey, L.B.; Fernandez, C.A.; Chang, T.Y.; Neale, G. Genetic risk factors for the development of osteonecrosis in children under age 10 treated for acute lymphoblastic leukemia. Blood J. Am. Soc. Hematol. 2016, 127, 558–564. [Google Scholar] [CrossRef]
- Maamari, D.; El-Khoury, H.; Saifi, O.; Muwakkit, S.A.; Zgheib, N.K. Implementation of Pharmacogenetics to Individualize Treatment Regimens for Children with Acute Lymphoblastic Leukemia. Pharmacogenomics Pers. Med. 2020, 13, 295–317. [Google Scholar] [CrossRef]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef]
- Liu, Y.J.; Hounye, A.H.; Wang, Z.; Liu, X.; Yi, J.; Qi, M. Identification and validation of three autophagy-related long noncoding RNAs as prognostic signature in cholangiocarcinoma. Front. Oncol. 2021, 11, 780601. [Google Scholar] [CrossRef]
- Yang, F.; Jing, F.; Li, Y.; Kong, S.; Zhang, S.; Huo, Y.; Huang, X.; Yu, S. Plasma lncRNA LOC338963 and mRNA AP3B2 are upregulated in paraneoplastic Lambert-Eaton myasthenic syndrome. Muscle Nerve 2022, 66, 216–222. [Google Scholar] [CrossRef]
- Czabotar, P.E.; Lessene, G.; Strasser, A.; Adams, J.M. Control of apoptosis by the BCL-2 protein family: Implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014, 15, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Wei, A.H.; Roberts, A.W.; Spencer, A.; Rosenberg, A.S.; Siegel, D.; Walter, R.B.; Caenepeel, S.; Hughes, P.; McIver, Z.; Mezzi, K.; et al. Targeting MCL-1 in hematologic malignancies: Rationale and progress. Blood Rev. 2020, 44, 100672. [Google Scholar] [CrossRef]
- Lo, W.-K.; Biswas, S.K.; Brako, L.; Shiels, A.; Gu, S.; Jiang, J.X. Aquaporin-0 targets interlocking domains to control the integrity and transparency of the eye lens. Investig. Ophthalmol. Vis. Sci. 2014, 55, 1202–1212. [Google Scholar] [CrossRef]
- Khan, S.; Ricciardelli, C.; Yool, A.J. Targeting aquaporins in novel therapies for male and female breast and reproductive cancers. Cells 2021, 10, 215. [Google Scholar] [CrossRef]
- Larance, M.; Kirkwood, K.J.; Xirodimas, D.P.; Lundberg, E.; Uhlen, M.; Lamond, A.I. Characterization of MRFAP1 turnover and interactions downstream of the NEDD8 pathway. Mol. Cell. Proteom. 2012, 11, 014407. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Bai, Z.; Ma, X.; Bai, N.; Zhang, Z. MRFAP1 plays a protective role in neddylation inhibitor MLN4924-mediated gastric cancer cell death. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8273–8280. [Google Scholar]
- Vicente-Manzanares, M.; Zareno, J.; Whitmore, L.; Choi, C.K.; Horwitz, A.F. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J. Cell Biol. 2007, 176, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Cheng, M.; Xia, X.; Han, Y.; Zhang, J.; Cao, P.; Zhou, G. Down-regulation of MYH10 driven by chromosome 17p13.1 deletion promotes hepatocellular carcinoma metastasis through activation of the EGFR pathway. J. Cell Mol. Med. 2021, 25, 11142–11156. [Google Scholar] [CrossRef]
- Milewicz, D.M.; Kwartler, C.S. Chapter 97—Genetic Variants in Smooth Muscle Contraction and Adhesion Genes Cause Thoracic Aortic Aneurysms and Dissections and Other Vascular Diseases. In Muscle; Hill, J.A., Olson, E.N., Eds.; Academic Press: Waltham, MA, USA, 2012; pp. 1291–1300. [Google Scholar] [CrossRef]
- Wang, R.J.; Wu, P.; Cai, G.X.; Wang, Z.M.; Xu, Y.; Peng, J.J.; Sheng, W.Q.; Lu, H.F.; Cai, S.J. Down-regulated MYH11 expression correlates with poor prognosis in stage II and III colorectal cancer. Asian Pac. J. Cancer Prev. 2014, 15, 7223–7228. [Google Scholar] [CrossRef]
- Wang, J.; Xu, P.; Hao, Y.; Yu, T.; Liu, L.; Song, Y.; Li, Y. Interaction between DNMT3B and MYH11 via hypermethylation regulates gastric cancer progression. BMC Cancer 2021, 21, 914. [Google Scholar] [CrossRef]
- Krendel, M.; Mooseker, M.S. Myosins: Tails (and heads) of functional diversity. Physiology 2005, 20, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ying, M.; Wu, Q.; Wang, R.; Li, Y. Overexpression of myosin VI regulates gastric cancer cell progression. Gene 2016, 593, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Placzek, W.J.; Almeida, M.S.; Wüthrich, K. NMR structure and functional characterization of a human cancer-related nucleoside triphosphatase. J. Mol. Biol. 2007, 367, 788–801. [Google Scholar] [CrossRef]
- Shang, H.; Zhang, H.; Ren, Z.; Zhao, H.; Zhang, Z.; Tong, J. Characterization of the Potential Role of NTPCR in Epithelial Ovarian Cancer by Integrating Transcriptomic and Metabolomic Analysis. Front. Genet. 2021, 12, 695245. [Google Scholar] [CrossRef]
- Meissner, C.; Lorenz, H.; Weihofen, A.; Selkoe, D.J.; Lemberg, M.K. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J. Neurochem. 2011, 117, 856–867. [Google Scholar] [CrossRef]
- Qin, C.; Wang, Y.; Zhao, B.; Li, Z.; Li, T.; Yang, X.; Zhao, Y.; Wang, W. STOML2 restricts mitophagy and increases chemosensitivity in pancreatic cancer through stabilizing PARL-induced PINK1 degradation. Cell Death Dis. 2023, 14, 191. [Google Scholar] [CrossRef]
- Du, Z.; Fei, T.; Verhaak, R.G.; Su, Z.; Zhang, Y.; Brown, M.; Chen, Y.; Liu, X.S. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol. 2013, 20, 908–913. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hong, C.; Qu, J.; Chen, J.; Qin, Z. Knockdown of lncRNA PCAT6 suppresses the growth of non-small cell lung cancer cells by inhibiting macrophages M2 polarization via miR-326/KLF1 axis. Bioengineered 2022, 13, 12834–12846. [Google Scholar] [CrossRef]
- Schagdarsurengin, U.; Paradowska, A.; Steger, K. Analysing the sperm epigenome: Roles in early embryogenesis and assisted reproduction. Nat. Rev. Urol. 2012, 9, 609–619. [Google Scholar] [CrossRef]
- Chen, Z.; Shi, C.; Gao, S.; Song, D.; Feng, Y. Impact of protamine I on colon cancer proliferation, invasion, migration, diagnosis and prognosis. Biol. Chem. 2018, 399, 265–275. [Google Scholar] [CrossRef]
- Hendriks, W.J.; Pulido, R. Protein tyrosine phosphatase variants in human hereditary disorders and disease susceptibilities. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2013, 1832, 1673–1696. [Google Scholar] [CrossRef] [PubMed]
- Karaca Atabay, E.; Mecca, C.; Wang, Q.; Ambrogio, C.; Mota, I.; Prokoph, N.; Mura, G.; Martinengo, C.; Patrucco, E.; Leonardi, G.; et al. Tyrosine phosphatases regulate resistance to ALK inhibitors in ALK+ anaplastic large cell lymphoma. Blood 2022, 139, 717–731. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S. Regulation of FYN Phosphorylation by the PTPN23 Tumor Suppressor Phosphatase in Breast Tumorigenesis. Ph.D. Thesis, State University of New York at Stony Brook, Stony Brook, NY, USA, 2017. [Google Scholar]
- Wang, T.; Hong, W. Interorganellar regulation of lysosome positioning by the Golgi apparatus through Rab34 interaction with Rab-interacting lysosomal protein. Mol. Biol. Cell 2002, 13, 4317–4332. [Google Scholar] [CrossRef]
- Wu, J.; Lu, Y.; Qin, A.; Qiao, Z.; Jiang, X. Overexpression of RAB34 correlates with poor prognosis and tumor progression in hepatocellular carcinoma. Oncol. Rep. 2017, 38, 2967–2974. [Google Scholar] [CrossRef]
- Zeigerer, A.; Gilleron, J.; Bogorad, R.L.; Marsico, G.; Nonaka, H.; Seifert, S.; Epstein-Barash, H.; Kuchimanchi, S.; Peng, C.G.; Ruda, V.M. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature 2012, 485, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.S.; Kim, M.; Kingsbury, T.J.; Civin, C.I.; Cheng, W.-C. Regulation of RAB5C is important for the growth inhibitory effects of MiR-509 in human precursor-B acute lymphoblastic leukemia. PLoS ONE 2014, 9, e111777. [Google Scholar] [CrossRef]
- Julian, L.; Olson, M.F. Rho-associated coiled-coil containing kinases (ROCK) structure, regulation, and functions. Small GTPases 2014, 5, e29846. [Google Scholar] [CrossRef]
- Warner, J.R.; McIntosh, K.B. How common are extraribosomal functions of ribosomal proteins? Mol. Cell 2009, 34, 3–11. [Google Scholar] [CrossRef]
- Labriet, A.; Lévesque, É.; Cecchin, E.; De Mattia, E.; Villeneuve, L.; Rouleau, M.; Jonker, D.; Couture, F.; Simonyan, D.; Allain, E.P.; et al. Germline variability and tumor expression level of ribosomal protein gene RPL28 are associated with survival of metastatic colorectal cancer patients. Sci. Rep. 2019, 9, 13008. [Google Scholar] [CrossRef]
- Usami, Y.; Wu, Y.; Göttlinger, H.G. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 2015, 526, 218–223. [Google Scholar] [CrossRef]
- Terasawa, K.; Sagae, S.; Toyota, M.; Tsukada, K.; Ogi, K.; Satoh, A.; Mita, H.; Imai, K.; Tokino, T.; Kudo, R. Epigenetic Inactivation of TMS1/ASC in Ovarian Cancer. Clin. Cancer Res. 2004, 10, 2000–2006. [Google Scholar] [CrossRef]
- Konyukh, M.; Delorme, R.; Chaste, P.; Leblond, C.; Lemière, N.; Nygren, G.; Anckarsäter, H.; Rastam, M.; Ståhlberg, O.; Amsellem, F.; et al. Variations of the candidate SEZ6L2 gene on Chromosome 16p11.2 in patients with autism spectrum disorders and in human populations. PLoS ONE 2011, 6, e17289. [Google Scholar] [CrossRef]
- Chen, L.; Han, S.; Li, Y.; Zheng, Y.; Zhang, Q. SEZ6L2, regulated by USF1, accelerates the growth and metastasis of breast cancer. Exp. Cell Res. 2022, 417, 113194. [Google Scholar] [CrossRef]
- Walz, A.; Ooms, A.; Gadd, S.; Gerhard, D.; Smith, M.; GuidryáAuvil, J.M.; Meerzaman, D.; Chen, Q.-R.; Hsu, C.; Yan, C. Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer Cell 2015, 27, 286–297. [Google Scholar] [CrossRef]
- Wan, Z.H.; Ma, Y.H.; Jiang, T.Y.; Lin, Y.K.; Shi, Y.Y.; Tan, Y.X.; Dong, L.W.; Wang, H.Y. Six2 is negatively correlated with prognosis and facilitates epithelial-mesenchymal transition via TGF-β/Smad signal pathway in hepatocellular carcinoma. Hepatobiliary Pancreat. Dis. Int. 2019, 18, 525–531. [Google Scholar] [CrossRef]
- Li, P.; Noegel, A.A. Inner nuclear envelope protein SUN1 plays a prominent role in mammalian mRNA export. Nucleic Acids Res. 2015, 43, 9874–9888. [Google Scholar] [CrossRef]
- Liu, L.; Li, S.W.; Yuan, W.; Tang, J.; Sang, Y. Downregulation of SUN2 promotes metastasis of colon cancer by activating BDNF/TrkB signalling by interacting with SIRT1. J. Pathol. 2021, 254, 531–542. [Google Scholar] [CrossRef]
- Lennard, A.; Gaston, K.; Fried, M. The Surf-1 and Surf-2 genes and their essential bidirectional promoter elements are conserved between mouse and human. DNA Cell Biol. 1994, 13, 1117–1126. [Google Scholar] [CrossRef]
- Jung, C.H.; Kim, E.M.; Song, J.Y.; Park, J.K.; Um, H.D. Mitochondrial superoxide dismutase 2 mediates γ-irradiation-induced cancer cell invasion. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Hatzis, P.; van der Flier, L.G.; van Driel, M.A.; Guryev, V.; Nielsen, F.; Denissov, S.; Nijman, I.c.J.; Koster, J.; Santo, E.E.; Welboren, W. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol. Cell. Biol. 2008, 28, 2732–2744. [Google Scholar] [CrossRef]
- Desterke, C.; Hugues, P.; Hwang, J.W.; Bennaceur-Griscelli, A.; Turhan, A.G. Embryonic Program Activated during Blast Crisis of Chronic Myelogenous Leukemia (CML) Implicates a TCF7L2 and MYC Cooperative Chromatin Binding. Int. J. Mol. Sci. 2020, 21, 4057. [Google Scholar] [CrossRef]
- Egeblad, M.; Werb, Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2002, 2, 161–174. [Google Scholar] [CrossRef]
- Saleh, M.; Khalil, M.; Abdellateif, M.S.; Ebeid, E.; Madney, Y.; Kandeel, E.Z. Role of matrix metalloproteinase MMP-2, MMP-9 and tissue inhibitor of metalloproteinase (TIMP-1) in the clinical progression of pediatric acute lymphoblastic leukemia. Hematology 2021, 26, 758–768. [Google Scholar] [CrossRef]
- Yang, W.B.; Hao, F.; Song, Z.Q.; Yang, X.C.; Ni, B. Apoptosis of the dermal papilla cells of hair follicle associated with the expression of gene HSPCO16 in vitro. Exp. Dermatol. 2005, 14, 209–214. [Google Scholar] [CrossRef]
- Song, Z.; Zhou, C.; Wang, J.; Yang, W.; Hao, F. Effect of HSPC016 gene expression on the aggregative growth of dermal papillae cells. Australas. J. Dermatol. 2012, 53, e26–e29. [Google Scholar] [CrossRef]
- Van Everdink, W.; Baranova, A.; Lummen, C.; Tyazhelova, T.; Looman, M.; Ivanov, D.; Verlind, E.; Pestova, A.; Faber, H.; van der Veen, A. RFP2, c13ORF1, and FAM10A4 are the most likely tumor suppressor gene candidates for B-cell chronic lymphocytic leukemia. Cancer Genet. Cytogenet. 2003, 146, 48–57. [Google Scholar] [CrossRef]
- Dai, T.; Ye, L.; Deng, M.; Lin, G.; Liu, R.; Yu, H.; Liu, W.; Yang, Y.; Wang, G. Upregulation of TMCO3 Promoting Tumor Progression and Contributing to the Poor Prognosis of Hepatocellular Carcinoma. J. Clin. Transl. Hepatol. 2022, 10, 913–924. [Google Scholar] [CrossRef]
- Taylor, M.R.; Slavov, D.; Gajewski, A.; Vlcek, S.; Ku, L.; Fain, P.R.; Carniel, E.; Di Lenarda, A.; Sinagra, G.; Boucek, M.M. Thymopoietin (lamina-associated polypeptide 2) gene mutation associated with dilated cardiomyopathy. Human. Mutat. 2005, 26, 566–574. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, G.; Chen, S.; Ding, J.; Ju, S.; Cao, H.; Tian, H. Depletion of thymopoietin inhibits proliferation and induces cell cycle arrest/apoptosis in glioblastoma cells. World J. Surg. Oncol. 2016, 14, 267. [Google Scholar] [CrossRef]
- Chacinska, A.; Koehler, C.M.; Milenkovic, D.; Lithgow, T.; Pfanner, N. Importing mitochondrial proteins: Machineries and mechanisms. Cell 2009, 138, 628–644. [Google Scholar] [CrossRef]
- Park, S.H.; Lee, A.R.; Choi, K.; Joung, S.; Yoon, J.B.; Kim, S. TOMM20 as a potential therapeutic target of colorectal cancer. BMB Rep. 2019, 52, 712–717. [Google Scholar] [CrossRef] [PubMed]
- De Paula, A.M.; Franques, J.; Fernandez, C.; Monnier, N.; Lunardi, J.; Pellissier, J.-F.; Figarella-Branger, D.; Pouget, J. A TPM3 mutation causing cap myopathy. Neuromuscul. Disord. 2009, 19, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wang, Y.; Bryce, N.S.; Tang, K.; Meagher, N.S.; Kang, E.Y.; Kelemen, L.E.; Köbel, M.; Ramus, S.J.; Friedlander, M. Targeting the actin/tropomyosin cytoskeleton in epithelial ovarian cancer reveals multiple mechanisms of synergy with anti-microtubule agents. Br. J. Cancer 2021, 125, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Yan, P.; Zhang, J.; Cui, Y.; Zheng, M.; Cheng, Y.; Guo, Y.; Yang, X.; Guo, X.; Zhu, H. Deficiency of TPPP2, a factor linked to oligoasthenozoospermia, causes subfertility in male mice. J. Cell Mol. Med. 2019, 23, 2583–2594. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, X.; Ni, X.; Pan, J.; Chen, M.; Lin, Y.; Zhao, Z.; Zhang, L.; Ge, N.; Song, G. Gene-based cancer-testis antigens as prognostic indicators in hepatocellular carcinoma. Heliyon 2023, 9, e13269. [Google Scholar] [CrossRef]
- Rho, S.B.; Lee, J.H.; Park, M.S.; Byun, H.-J.; Kang, S.; Seo, S.-S.; Kim, J.-Y.; Park, S.-Y. Anti-apoptotic protein TCTP controls the stability of the tumor suppressor p53. FEBS Lett. 2011, 585, 29–35. [Google Scholar] [CrossRef]
- He, S.; Huang, Y.; Wang, Y.; Tang, J.; Song, Y.; Yu, X.; Ma, J.; Wang, S.; Yin, H.; Li, Q. Histamine-releasing factor/translationally controlled tumor protein plays a role in induced cell adhesion, apoptosis resistance and chemoresistance in non-Hodgkin lymphomas. Leuk. Lymphoma 2015, 56, 2153–2161. [Google Scholar] [CrossRef]
- Kang, B.H.; Plescia, J.; Dohi, T.; Rosa, J.; Doxsey, S.J.; Altieri, D.C. Regulation of tumor cell mitochondrial homeostasis by an organelle-specific Hsp90 chaperone network. Cell 2007, 131, 257–270. [Google Scholar] [CrossRef]
- Chen, R.; Pan, S.; Lai, K.; Lai, L.A.; Crispin, D.A.; Bronner, M.P.; Brentnall, T.A. Up-regulation of mitochondrial chaperone TRAP1 in ulcerative colitis associated colorectal cancer. World J. Gastroenterol. 2014, 20, 17037–17048. [Google Scholar] [CrossRef]
- Ariës, I.M.; Bodaar, K.; Karim, S.A.; Chonghaile, T.N.; Hinze, L.; Burns, M.A.; Pfirrmann, M.; Degar, J.; Landrigan, J.T.; Balbach, S.; et al. PRC2 loss induces chemoresistance by repressing apoptosis in T cell acute lymphoblastic leukemia. J. Exp. Med. 2018, 215, 3094–3114. [Google Scholar] [CrossRef]
- Jordan, M. Mechanism of action of antitumor drugs that interact with microtubules and tubulin. Curr. Med. Chem.-Anti-Cancer Agents 2002, 2, 1–17. [Google Scholar] [CrossRef]
- Nami, B.; Wang, Z. Genetics and expression profile of the tubulin gene superfamily in breast cancer subtypes and its relation to taxane resistance. Cancers 2018, 10, 274. [Google Scholar] [CrossRef] [PubMed]
- Djureinovic, D.; Hallström, B.M.; Horie, M.; Mattsson, J.S.M.; La Fleur, L.; Fagerberg, L.; Brunnström, H.; Lindskog, C.; Madjar, K.; Rahnenführer, J.; et al. Profiling cancer testis antigens in non-small-cell lung cancer. JCI Insight 2016, 1, e86837. [Google Scholar] [CrossRef] [PubMed]
- Marinovic, A.C.; Mitch, W.E.; Price, S.R. Tools for evaluating ubiquitin (UbC) gene expression: Characterization of the rat UbC promoter and use of an unique 3’ mRNA sequence. Biochem. Biophys. Res. Commun. 2000, 274, 537–541. [Google Scholar] [CrossRef]
- Kim, J.; Kim, Y.; Choi, H.; Kwon, A.; Jekarl, D.W.; Lee, S.; Jang, W.; Chae, H.; Kim, J.R.; Kim, J.M.; et al. Ubiquitin C decrement plays a pivotal role in replicative senescence of bone marrow mesenchymal stromal cells. Cell Death Dis. 2018, 9, 139. [Google Scholar] [CrossRef] [PubMed]
- Alchahin, A.M.; Mei, S.; Tsea, I.; Hirz, T.; Kfoury, Y.; Dahl, D.; Wu, C.-L.; Subtelny, A.O.; Wu, S.; Scadden, D.T. A transcriptional metastatic signature predicts survival in clear cell renal cell carcinoma. Nat. Commun. 2022, 13, 1–15. [Google Scholar] [CrossRef]
- Metzger, M.B.; Hristova, V.A.; Weissman, A.M. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell science 2012, 125, 531–537. [Google Scholar] [CrossRef]
- Hu, K.; Liu, X.; Li, Y.; Li, Q.; Xu, Y.; Zeng, W.; Zhong, G.; Yu, C. Exosomes Mediated Transfer of Circ_UBE2D2 Enhances the Resistance of Breast Cancer to Tamoxifen by Binding to MiR-200a-3p. Med. Sci. Monit. 2020, 26, e922253. [Google Scholar] [CrossRef]
- Wen, J.L.; Wen, X.F.; Li, R.B.; Jin, Y.C.; Wang, X.L.; Zhou, L.; Chen, H.X. UBE3C promotes growth and metastasis of renal cell carcinoma via activating Wnt/β-catenin pathway. PLoS ONE 2015, 10, e0115622. [Google Scholar] [CrossRef]
- Huang, L.Z.; Li, Y.J.; Xie, X.F.; Zhang, J.J.; Cheng, C.Y.; Yamashiro, K.; Chen, L.J.; Ma, X.Y.; Cheung, C.M.; Wang, Y.S.; et al. Whole-exome sequencing implicates UBE3D in age-related macular degeneration in East Asian populations. Nat. Commun. 2015, 6, 6687. [Google Scholar] [CrossRef]
- Liu, H.; Heller-Trulli, D.; Moore, C.L. Targeting the mRNA endonuclease CPSF73 inhibits breast cancer cell migration, invasion, and self-renewal. iScience 2022, 25, 104804. [Google Scholar] [CrossRef]
- Kimura, K.; Wakamatsu, A.; Suzuki, Y.; Ota, T.; Nishikawa, T.; Yamashita, R.; Yamamoto, J.; Sekine, M.; Tsuritani, K.; Wakaguri, H.; et al. Diversification of transcriptional modulation: Large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006, 16, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wu, S.; Wu, X.; Zhou, X.; Jin, S.; Jiang, H. Silencing circular RNA UVRAG inhibits bladder cancer growth and metastasis by targeting the microRNA-223/fibroblast growth factor receptor 2 axis. Cancer Sci. 2019, 110, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Bänziger, C.; Soldini, D.; Schütt, C.; Zipperlen, P.; Hausmann, G.; Basler, K. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell 2006, 125, 509–522. [Google Scholar] [CrossRef]
- Zhang, W.; Tao, H.; Chen, X.; Sugimura, H.; Wang, J.; Zhou, P. High expression of Wls is associated with lymph node metastasis and advanced TNM stage in gastric carcinomas. Pathol. Int. 2017, 67, 141–146. [Google Scholar] [CrossRef]
- Chiou, S.-S.; Wang, L.-T.; Huang, S.-B.; Chai, C.-Y.; Wang, S.-N.; Liao, Y.-M.; Lin, P.-C.; Liu, K.-Y.; Hsu, S.-H. Wntless (GPR177) expression correlates with poor prognosis in B-cell precursor acute lymphoblastic leukemia via Wnt signaling. Carcinogenesis 2014, 35, 2357–2364. [Google Scholar] [CrossRef]
- Memarian, A.; Vosough, P.; Asgarian-Omran, H.; Tabrizi, M.; Shabani, M.; Shokri, F. Differential WNT gene expression in various subtypes of acute lymphoblastic leukemia. Iran. J. Immunol. 2012, 9, 61–71. [Google Scholar]
- Mangino, M.; Hwang, S.-J.; Spector, T.D.; Hunt, S.C.; Kimura, M.; Fitzpatrick, A.L.; Christiansen, L.; Petersen, I.; Elbers, C.C.; Harris, T. Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Human Mol. Genet. 2012, 21, 5385–5394. [Google Scholar] [CrossRef]
- Peculis, R.; Niedra, H.; Rovite, V. Large Scale Molecular Studies of Pituitary Neuroendocrine Tumors: Novel Markers, Mechanisms and Translational Perspectives. Cancers 2021, 13, 1395. [Google Scholar] [CrossRef]


| ID | Disease Stage | Cytogenetics | WCC (109/L) | BM Blast % | Relapse | Survival Post-Sample (mo) ¶ | Age ≠ | Sex | Sample Type |
|---|---|---|---|---|---|---|---|---|---|
| ALL001 | Diagnosis Ph+ ALL | Ph+ ALL: t(9;22) | 4.9 | 40 | NK | NK | 39 | M | PB |
| ALL002 | Diagnosis T-ALL | 46,XY,t(1;7)(p36;p15) | 232 | 88 | No | Alive (post allo) | 19 | M | PB |
| ALL003 | Diagnosis B-ALL | t(1;19) | 28.6 | 91 | No | Alive | 26 | F | PB |
| ALL004 * | Diagnosis T-ALL | Complex karyotype | 591 | NK | No | Died 19 mo (post allo) | 19 | M | PB |
| ALL005 * | ¶ Post allo T-ALL | No result | NK | NK | No | Died 3.5 mo (post allo) | 46 | M | PB |
| ALL007 | Diagnosis Pre-B-ALL | Loss of one copy of ETV6 (12p13) and gain of one copy of ABL1 (9q34) by FISH | 8.2 | 92 | No | Alive | 24 | M | PB |
| ALL008 | Diagnosis Pre-B-ALL | 46XY, 5,del(5)(q15q33),dic(9;16)(p11;q11),del(13)(q12q14) | 10.4 | 72 | No | Alive | 19 | M | PB |
| ALL009 | Diagnosis Pre-B-ALL | 46,XY,t(1;7)(q25;q3?5),add(3)(p1?3) | 48.1 | 89.6 | Yes | Died 94 mo (post allo, CART) | 19 | M | BM |
| ALL010 * | Diagnosis Pre-B-ALL | Complex including t(4;11) | 229.1 | 85 | Yes | Died 3 mo | 64 | M | PB |
| ALL011 | Diagnosis Pre-B-ALL | No result—external referral | NK | NK | NK | NK | 19 | F | PB |
| ALL012 | Diagnosis Pre-B-ALL | t(11;14)(q24;q32) | 4.3 | 83 | No | Alive (post allo) | 33 | M | BM |
| ALL014 | Diagnosis Pre-B-ALL | 47,XY,+2,add(2)(p1)[3]/46,XY [47]. nucish (CRLF2)x2[100] | 4.9 | 53 | Yes | Alive (post allo & CART) | 56 | M | BM |
| ALL015 | Diagnosis Pre-B-ALL | Gain of one copy of CRLF2 (Xp22.3/Yp11.3) and loss of one copy of CSFR1 (5q32) and EBF1 (5q33.3) detected by FISH | 68.0 | 94 | No | Alive | 20 | F | PB |
| ALL016 | Diagnosis Pre-B-ALL | Hyperdiploid; 56–57 XX +X,+4,+6,+9,+10,+14,+17,+18,+21,+marker | 1.9 | 96 | No | Alive | 27 | F | BM |
| ALL017 † | Diagnosis, Pre-B-ALL | No cytogenetics available, no FISH | 3.0 | 96 | No | Alive | 19 | F | PB |
| ALL018 † | Diagnosis, T-ALL | Normal cytogenetics, SET/CAN fusion detected by FISH | 58.8 | 74 | No | Alive | 22 | M | PB |
| ALL020 | Diagnosis Pre-B-ALL | 46,XY, t(1;7)(q25;q3?5), add(3)(p1?3) TCF3 ex16-PBX1 ex3 fusion transcript detected | 31.9 | 92 | Yes | Died 34.5 mo (Post allo, relapse, salvage chemo then CART & relapse) | 56 | F | PB/BM |
| HV Control | Age ≠ | Sex | Sample Type |
|---|---|---|---|
| HV008 | 40 | F | PB |
| HV010 | 22 | M | PB |
| HV012 | 46 | F | PB |
| HV021 | 34 | M | PB |
| HV043 | NK | M | PB |
| CD19+ cells | 18–66 | F | PB |
| Pathway | p-Value | q-Value | SEREX-Identified Genes Involved in This Pathway | Database Used |
|---|---|---|---|---|
| Envelope proteins and their potential roles in EDMD physiopathology | 0.0007 | 0.0892 | TCF7L2, WNT2B | Wiki Pathway |
| Hematopoietic stem cell gene regulation | 0.001 | 0.064 | DNMT1, MCL1 | |
| TGFβ signalling pathway | 0.015 | 0.229 | TRAP1, ROCK1, CUL1 | |
| Mitotic spindle | 0.001 | 0.038 | ROCK1, KIF5B, FLNB, KIF1B, MYH10 | MSigDB Hallmark 2020 |
| Notch signalling | 0.007 | 0.117 | TCF7L2, CUL1 | |
| Apoptosis | 0.026 | 0.286 | ROCK1, TIMP1, MCL1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamed, E.; Goodman, S.; Cooksey, L.; Fletcher, D.M.; Dean, O.; Boncheva, V.B.; Mills, K.I.; Orchard, K.H.; Guinn, B.-a. Adult B-Cell Acute Lymphoblastic Leukaemia Antigens and Enriched Pathways Identify New Targets for Therapy. Onco 2025, 5, 19. https://doi.org/10.3390/onco5020019
Mohamed E, Goodman S, Cooksey L, Fletcher DM, Dean O, Boncheva VB, Mills KI, Orchard KH, Guinn B-a. Adult B-Cell Acute Lymphoblastic Leukaemia Antigens and Enriched Pathways Identify New Targets for Therapy. Onco. 2025; 5(2):19. https://doi.org/10.3390/onco5020019
Chicago/Turabian StyleMohamed, Eithar, Sara Goodman, Leah Cooksey, Daniel M. Fletcher, Olivia Dean, Viktoriya B. Boncheva, Ken I. Mills, Kim H. Orchard, and Barbara-ann Guinn. 2025. "Adult B-Cell Acute Lymphoblastic Leukaemia Antigens and Enriched Pathways Identify New Targets for Therapy" Onco 5, no. 2: 19. https://doi.org/10.3390/onco5020019
APA StyleMohamed, E., Goodman, S., Cooksey, L., Fletcher, D. M., Dean, O., Boncheva, V. B., Mills, K. I., Orchard, K. H., & Guinn, B.-a. (2025). Adult B-Cell Acute Lymphoblastic Leukaemia Antigens and Enriched Pathways Identify New Targets for Therapy. Onco, 5(2), 19. https://doi.org/10.3390/onco5020019








