Impacts of the Green Revolution on Rhizosphere Microbiology Related to Nutrient Acquisition
Abstract
:1. Introduction
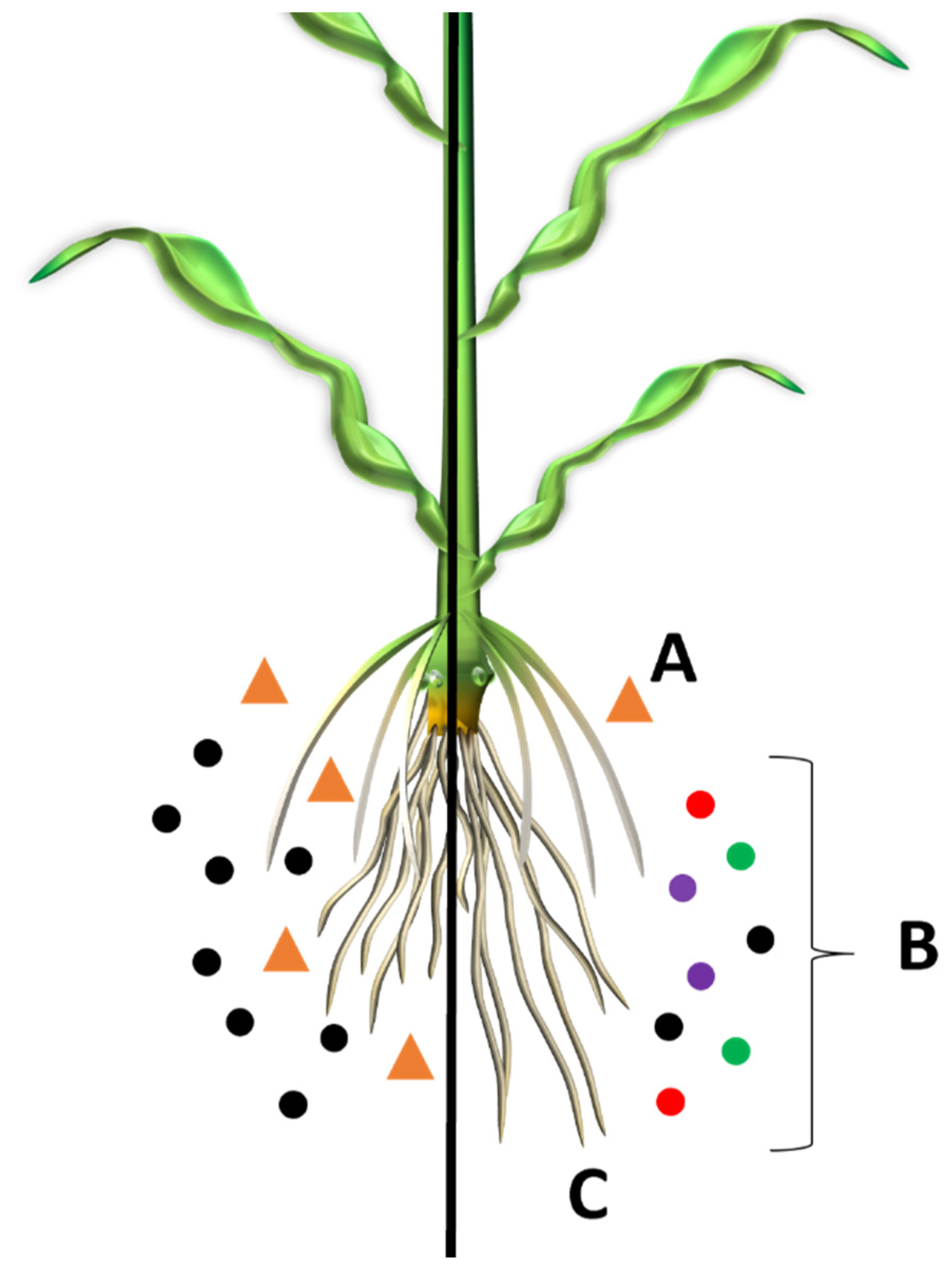
2. Microbe Facilitated Nutrient Acquisition
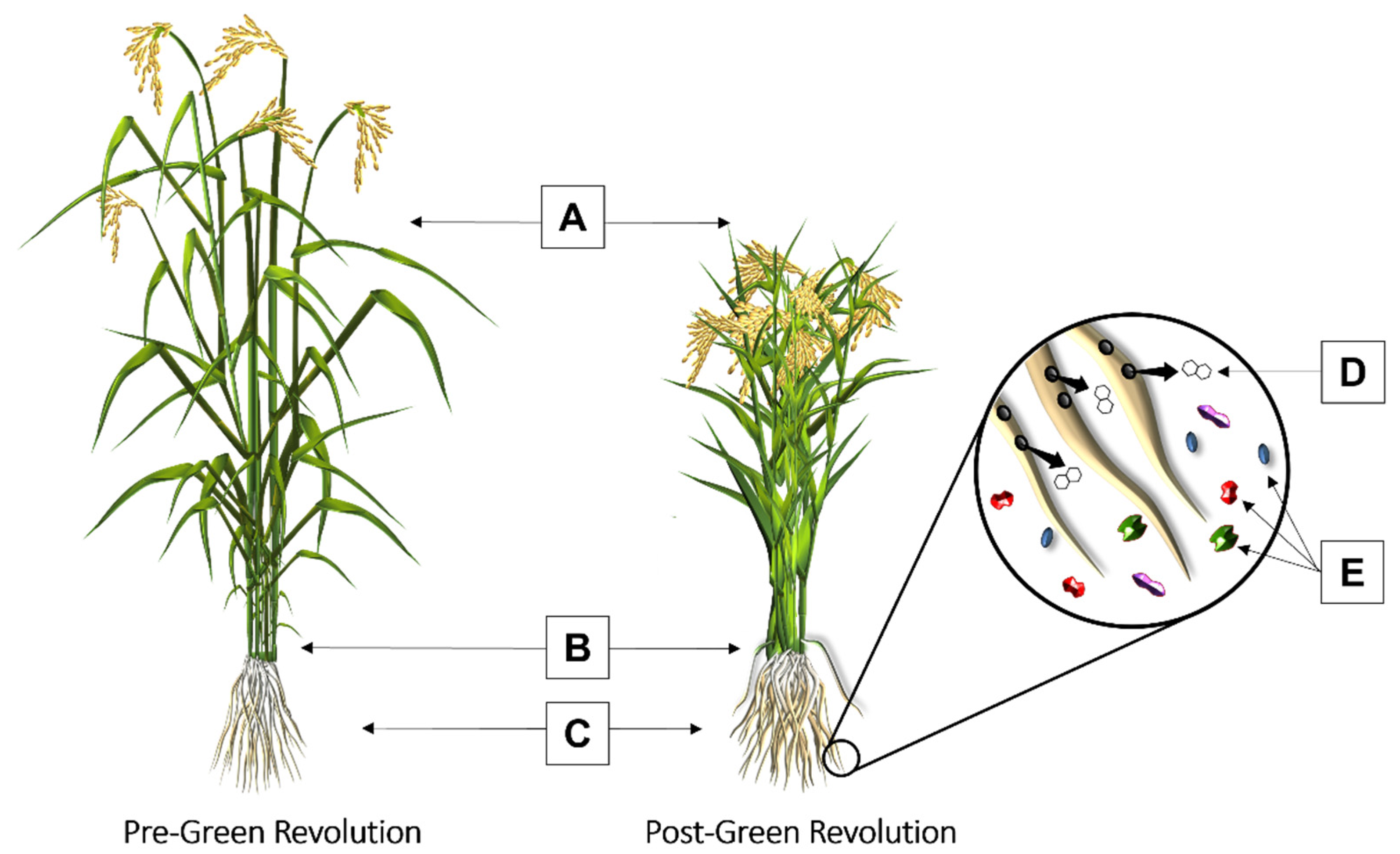
3. Influence of the Green Revolution on the Rhizosphere Microbiome of Staple Crops
3.1. Wheat
3.2. Rice
3.3. Maize
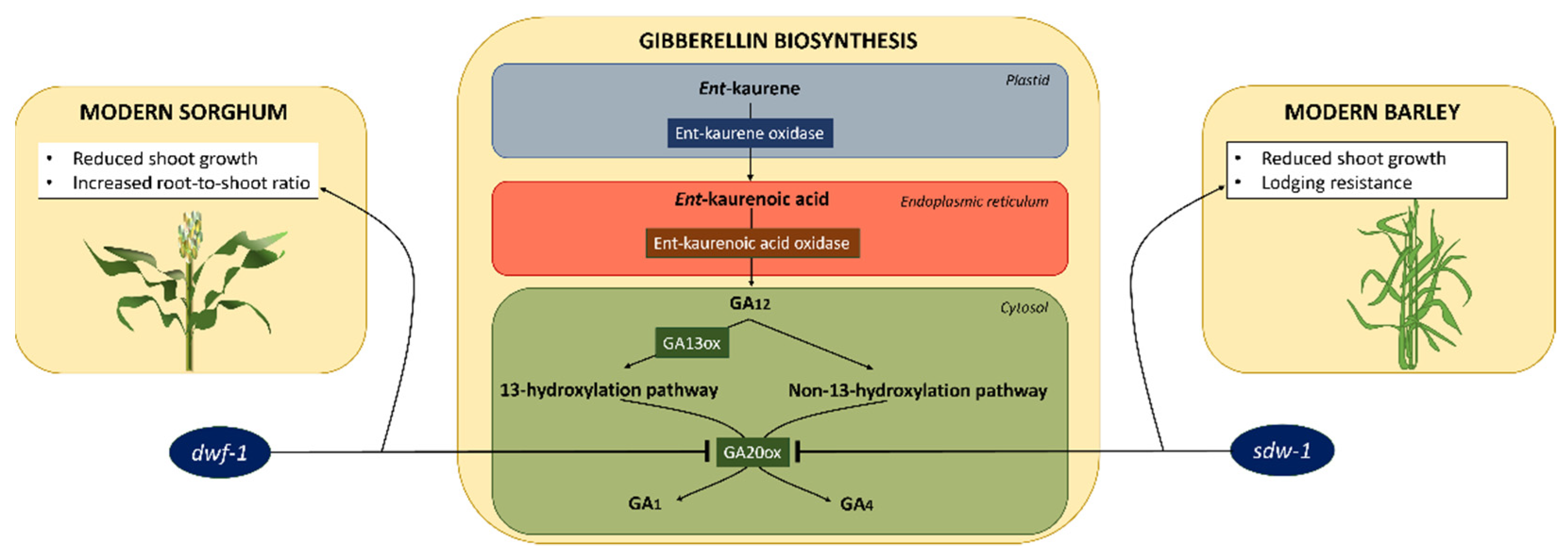
3.4. Sorghum
3.5. Barley
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- John, D.A.; Babu, G.R. Lessons from the aftermaths of green revolution on food system and health. Front. Sustain. Food Syst. 2021, 5, 644559. [Google Scholar] [CrossRef] [PubMed]
- Eliazer Nelson, A.R.L.; Ravichandran, K.; Antony, U. The impact of the Green Revolution on indigenous crops of India. J. Ethn. Foods 2019, 6, 8. [Google Scholar] [CrossRef] [Green Version]
- Gollin, D.; Morris, M.; Byerlee, D. Technology adoption in intensive post-green revolution systems. Am. J. Agric. Econ. 2005, 87, 1310–1316. [Google Scholar] [CrossRef]
- Wu, K.; Ali, I.; Xie, H.; Ullah, S.; Iqbal, A.; Wei, S.; He, L.; Huang, Q.; Wu, X.; Cheng, F. Impact of fertilization with reducing in nitrogen and phosphorous application on growth, yield and biomass accumulation of rice (Oryza sativa L.) under a dual cropping system. PeerJ 2021, 9, e11668. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Tian, Y.; Wu, K.; Ye, Y.; Yu, J.; Zhang, J.; Liu, Q.; Hu, M.; Li, H.; Tong, Y. Modulating plant growth–metabolism coordination for sustainable agriculture. Nature 2018, 560, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Spielmeyer, W.; Ellis, M.H.; Chandler, P.M. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 2002, 99, 9043–9048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asano, K.; Takashi, T.; Miura, K.; Qian, Q.; Kitano, H.; Matsuoka, M.; Ashikari, M. Genetic and molecular analysis of utility of sd1 alleles in rice breeding. Breed. Sci. 2007, 57, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Li, W.; Tan, L.; Tian, F. Harnessing knowledge from maize and rice domestication for new crop breeding. Mol. Plant 2021, 14, 9–26. [Google Scholar] [CrossRef]
- Flintham, J.; Börner, A.; Worland, A.; Gale, M. Optimizing wheat grain yield: Effects of Rht (gibberellin-insensitive) dwarfing genes. J. Agric. Sci. 1997, 128, 11–25. [Google Scholar] [CrossRef]
- Wang, Y.; Yao, Q.; Zhang, Y.; Zhang, Y.; Xing, J.; Yang, B.; Mi, G.; Li, Z.; Zhang, M. The role of gibberellins in regulation of nitrogen uptake and physiological traits in maize responding to nitrogen availability. Int. J. Mol. Sci. 2020, 21, 1824. [Google Scholar] [CrossRef]
- Hidayat, R.A.; Iskandar, J.; Gunawan, B.; Partasasmita, R. Impact of green revolution on rice cultivation practices and production system: A case study in Sindang Hamlet, Rancakalong Village, Sumedang District, West Java, Indonesia. Biodiversitas J. Biol. Divers. 2020, 21, 3. [Google Scholar]
- Tedengren, M. Eutrophication and the Disrupted Nitrogen Cycle; Springer: Berlin/Heidelberg, Germany, 2021; Volume 50, pp. 733–738. [Google Scholar]
- Bharti, C.; Mohapatra, A.; Maurya, R.; Maharana, C.; Maurya, A.; Malakar, P. Nitrate pollution with modernization of Indian agriculture. J. Pharmacogn. Phytochem. 2020, 9, 2073–2080. [Google Scholar]
- Medina, M.; Kaplan, D.; Milbrandt, E.C.; Tomasko, D.; Huffaker, R.; Angelini, C. Nitrogen-enriched discharges from a highly managed watershed intensify red tide (Karenia brevis) blooms in southwest Florida. Sci. Total Environ. 2022, 827, 154149. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K.; Soni, R.; Rajput, A.S. Role of microbes in organic farming for sustainable agro-ecosystem. In Microorganisms for Green Revolution; Springer: Berlin/Heidelberg, Germany, 2018; pp. 241–252. [Google Scholar]
- Bahulikar, R.A.; Chaluvadi, S.R.; Torres-Jerez, I.; Mosali, J.; Bennetzen, J.L.; Udvardi, M. Nitrogen fertilization reduces nitrogen fixation activity of diverse diazotrophs in switchgrass roots. Phytobiomes J. 2021, 5, 80–87. [Google Scholar] [CrossRef] [Green Version]
- Tian, J.; Lu, X.; Chen, Q.; Kuang, X.; Liang, C.; Deng, L.; Lin, D.; Cai, K.; Tian, J. Phosphorus fertilization affects soybean rhizosphere phosphorus dynamics and the bacterial community in karst soils. Plant Soil 2022, 475, 137–152. [Google Scholar] [CrossRef]
- Mohanram, S.; Kumar, P. Rhizosphere microbiome: Revisiting the synergy of plant-microbe interactions. Ann. Microbiol. 2019, 69, 307–320. [Google Scholar] [CrossRef]
- Pantigoso, H.A.; He, Y.; Manter, D.K.; Fonte, S.J.; Vivanco, J.M. Phosphorus-solubilizing bacteria isolated from the rhizosphere of wild potato Solanum bulbocastanum enhance growth of modern potato varieties. Bull. Natl. Res. Cent. 2022, 46, 224. [Google Scholar] [CrossRef]
- Sarkar, D.; Sankar, A.; Devika, O.S.; Singh, S.; Parihar, M.; Rakshit, A.; Sayyed, R.; Gafur, A.; Ansari, M.J.; Danish, S. Optimizing nutrient use efficiency, productivity, energetics, and economics of red cabbage following mineral fertilization and biopriming with compatible rhizosphere microbes. Sci. Rep. 2021, 11, 15680. [Google Scholar] [CrossRef]
- Lindström, K.; Mousavi, S.A. Effectiveness of nitrogen fixation in rhizobia. Microb. Biotechnol. 2020, 13, 1314–1335. [Google Scholar] [CrossRef] [Green Version]
- Roy-Bolduc, A.; Hijri, M. The use of mycorrhizae to enhance phosphorus uptake: A way out the phosphorus crisis. J. Biofertil. Biopestici. 2011, 2, 104. [Google Scholar]
- Guerrero-Galán, C.; Delteil, A.; Garcia, K.; Houdinet, G.; Conéjéro, G.; Gaillard, I.; Sentenac, H.; Zimmermann, S.D. Plant potassium nutrition in ectomycorrhizal symbiosis: Properties and roles of the three fungal TOK potassium channels in Hebeloma cylindrosporum. Environ. Microbiol. 2018, 20, 1873–1887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porter, S.S.; Sachs, J.L. Agriculture and the disruption of plant–microbial symbiosis. Trends Ecol. Evol. 2020, 35, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Favela, A.; Bohn, O.M.; Kent, D.A. Maize germplasm chronosequence shows crop breeding history impacts recruitment of the rhizosphere microbiome. ISME J. 2021, 15, 2454–2464. [Google Scholar] [CrossRef] [PubMed]
- Kavamura, V.N.; Robinson, R.J.; Hughes, D.; Clark, I.; Rossmann, M.; Melo, I.S.D.; Hirsch, P.R.; Mendes, R.; Mauchline, T.H. Wheat dwarfing influences selection of the rhizosphere microbiome. Sci. Rep. 2020, 10, 1452. [Google Scholar] [CrossRef] [Green Version]
- Smulders, L.; Benítez, E.; Moreno, B.; López-García, Á.; Pozo, M.J.; Ferrero, V.; de la Peña, E.; Alcalá Herrera, R. Tomato domestication affects potential functional molecular pathways of root-associated soil bacteria. Plants 2021, 10, 1942. [Google Scholar] [CrossRef]
- Pérez-Jaramillo, J.E.; Mendes, R.; Raaijmakers, J.M. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 2016, 90, 635–644. [Google Scholar] [CrossRef] [Green Version]
- Waines, J.G.; Ehdaie, B. Domestication and crop physiology: Roots of green-revolution wheat. Ann. Bot. 2007, 100, 991–998. [Google Scholar] [CrossRef] [Green Version]
- Hallama, M.; Pekrun, C.; Lambers, H.; Kandeler, E. Hidden miners–the roles of cover crops and soil microorganisms in phosphorus cycling through agroecosystems. Plant Soil 2019, 434, 7–45. [Google Scholar] [CrossRef] [Green Version]
- Prasad, M.; Chaudhary, M.; Choudhary, M.; Kumar, T.K.; Jat, L.K. Rhizosphere microorganisms towards soil sustainability and nutrient acquisition. In Agriculturally Important Microbes for Sustainable Agriculture; Springer: Berlin/Heidelberg, Germany, 2017; pp. 31–49. [Google Scholar]
- Sun, H.; Jiang, S.; Jiang, C.; Wu, C.; Gao, M.; Wang, Q. A review of root exudates and rhizosphere microbiome for crop production. Environ. Sci. Pollut. Res. 2021, 28, 54497–54510. [Google Scholar] [CrossRef]
- Xiong, J.; Lu, J.; Li, X.; Qiu, Q.; Chen, J.; Yan, C. Effect of rice (Oryza sativa L.) genotype on yield: Evidence from recruiting spatially consistent rhizosphere microbiome. Soil Biol. Biochem. 2021, 161, 108395. [Google Scholar] [CrossRef]
- Moreau, D.; Bardgett, R.D.; Finlay, R.D.; Jones, D.L.; Philippot, L. A plant perspective on nitrogen cycling in the rhizosphere. Funct. Ecol. 2019, 33, 540–552. [Google Scholar] [CrossRef] [Green Version]
- Sattar, A.; Naveed, M.; Ali, M.; Zahir, Z.A.; Nadeem, S.M.; Yaseen, M.; Meena, V.S.; Farooq, M.; Singh, R.; Rahman, M. Perspectives of potassium solubilizing microbes in sustainable food production system: A review. Appl. Soil Ecol. 2019, 133, 146–159. [Google Scholar] [CrossRef]
- Charmet, G. Wheat domestication: Lessons for the future. Comptes Rendus Biol. 2011, 334, 212–220. [Google Scholar] [CrossRef]
- Mondal, S.; Dutta, S.; Crespo-Herrera, L.; Huerta-Espino, J.; Braun, H.J.; Singh, R.P. Fifty years of semi-dwarf spring wheat breeding at CIMMYT: Grain yield progress in optimum, drought and heat stress environments. Field Crops Res. 2020, 250, 107757. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, J.; Hu, G.; Xue, H.; Xu, H.; Zhao, C.; Qin, R.; Cui, F.; Sun, H. Functional analysis of the “Green Revolution” gene Photoperiod-1 and its selection trends during bread wheat breeding. Front. Plant Sci. 2021, 12, 745411. [Google Scholar] [CrossRef] [PubMed]
- Iannucci, A.; Fragasso, M.; Beleggia, R.; Nigro, F.; Papa, R. Evolution of the crop rhizosphere: Impact of domestication on root exudates in tetraploid wheat (Triticum turgidum L.). Front. Plant Sci. 2017, 8, 2124. [Google Scholar] [CrossRef] [Green Version]
- Nunes da Rocha, U.; Plugge, C.M.; George, I.; Van Elsas, J.D.; Van Overbeek, L.S. The rhizosphere selects for particular groups of Acidobacteria and Verrucomicrobia. PLoS ONE 2013, 8, e82443. [Google Scholar] [CrossRef]
- Wang, M.; Sun, H.; Xu, L.; Xu, Z. Bacterial diversity in tea plant (Camellia sinensis) rhizosphere soil from Qinling Mountains and its relationship with environmental elements. Plant Soil 2021, 460, 403–415. [Google Scholar] [CrossRef]
- Roucou, A.; Violle, C.; Fort, F.; Roumet, P.; Ecarnot, M.; Vile, D. Shifts in plant functional strategies over the course of wheat domestication. J. Appl. Ecol. 2018, 55, 25–37. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Zhang, X.; Ni, H.; Gai, X.; Huang, Z.; Du, X.; Zhong, Z. Soil carbon and associated bacterial community shifts driven by fine root traits along a chronosequence of Moso bamboo (Phyllostachys edulis) plantations in subtropical China. Sci. Total Environ. 2021, 752, 142333. [Google Scholar] [CrossRef]
- Pervaiz, Z.H.; Contreras, J.; Hupp, B.M.; Lindenberger, J.H.; Chen, D.; Zhang, Q.; Wang, C.; Twigg, P.; Saleem, M. Root microbiome changes with root branching order and root chemistry in peach rhizosphere soil. Rhizosphere 2020, 16, 100249. [Google Scholar] [CrossRef]
- Li, P.; Chen, J.; Wu, P.; Zhang, J.; Chu, C.; See, D.; Brown-Guedira, G.; Zemetra, R.; Souza, E. Quantitative trait loci analysis for the effect of Rht-B1 dwarfing gene on coleoptile length and seedling root length and number of bread wheat. Crop Sci. 2011, 51, 2561–2568. [Google Scholar] [CrossRef]
- McGrail, R.K.; McNear Jr, D.H. Two centuries of breeding has altered root system architecture of winter wheat. Rhizosphere 2021, 19, 100411. [Google Scholar] [CrossRef]
- Nakhforoosh, A.; Nagel, K.A.; Fiorani, F.; Bodner, G. Deep soil exploration vs. topsoil exploitation: Distinctive rooting strategies between wheat landraces and wild relatives. Plant Soil 2021, 459, 397–421. [Google Scholar] [CrossRef] [PubMed]
- Kavamura, V.N.; Hayat, R.; Clark, I.M.; Rossmann, M.; Mendes, R.; Hirsch, P.R.; Mauchline, T.H. Inorganic nitrogen application affects both taxonomical and predicted functional structure of wheat rhizosphere bacterial communities. Front. Microbiol. 2018, 9, 1074. [Google Scholar] [CrossRef] [Green Version]
- Fierer, N.; Wood, S.A.; de Mesquita, C.P.B. How microbes can, and cannot, be used to assess soil health. Soil Biol. Biochem. 2021, 153, 108111. [Google Scholar] [CrossRef]
- Kavamura, V.N.; Mendes, R.; Bargaz, A.; Mauchline, T.H. Defining the wheat microbiome: Towards microbiome-facilitated crop production. Comput. Struct. Biotechnol. J. 2021, 19, 1200–1213. [Google Scholar] [CrossRef]
- Chen, S.; Waghmode, T.R.; Sun, R.; Kuramae, E.E.; Hu, C.; Liu, B. Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 2019, 7, 136. [Google Scholar] [CrossRef] [Green Version]
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 2014, 1324, 7–14. [Google Scholar] [CrossRef]
- Chen, E.; Huang, X.; Tian, Z.; Wing, R.A.; Han, B. The genomics of Oryza species provides insights into rice domestication and heterosis. Annu. Rev. Plant Biol. 2019, 70, 639–665. [Google Scholar] [CrossRef]
- Peng, Y.; Hu, Y.; Qian, Q.; Ren, D. Progress and prospect of breeding utilization of Green Revolution gene SD 1 in rice. Agriculture 2021, 11, 611. [Google Scholar] [CrossRef]
- Lo, S.-F.; Yang, S.-Y.; Chen, K.-T.; Hsing, Y.-I.; Zeevaart, J.A.; Chen, L.-J.; Yu, S.-M. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 2008, 20, 2603–2618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saleem, M.; Law, A.D.; Sahib, M.R.; Pervaiz, Z.H.; Zhang, Q. Impact of root system architecture on rhizosphere and root microbiome. Rhizosphere 2018, 6, 47–51. [Google Scholar] [CrossRef]
- Beltran-Garcia, M.J.; Martínez-Rodríguez, A.; Olmos-Arriaga, I.; Valdes-Salas, B.; Di Mascio, P.; White, J.F. Nitrogen fertilization and stress factors drive shifts in microbial diversity in soils and plants. Symbiosis 2021, 84, 379–390. [Google Scholar] [CrossRef]
- Xiong, Q.; Hu, J.; Wei, H.; Zhang, H.; Zhu, J. Relationship between plant roots, rhizosphere microorganisms, and nitrogen and its special focus on rice. Agriculture 2021, 11, 234. [Google Scholar] [CrossRef]
- Pérez-Jaramillo, J.E.; Carrión, V.J.; Bosse, M.; Ferrão, L.F.; De Hollander, M.; Garcia, A.A.; Ramírez, C.A.; Mendes, R.; Raaijmakers, J.M. Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J. 2017, 11, 2244–2257. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Vivanco, J.M.; Manter, D.K. Nitrogen fertilizer rate affects root exudation, the rhizosphere microbiome and nitrogen-use-efficiency of maize. Appl. Soil Ecol. 2016, 107, 324–333. [Google Scholar] [CrossRef] [Green Version]
- Briones, A.M.; Okabe, S.; Umemiya, Y.; Ramsing, N.-B.; Reichardt, W.; Okuyama, H. Influence of different cultivars on populations of ammonia-oxidizing bacteria in the root environment of rice. Appl. Environ. Microbiol. 2002, 68, 3067–3075. [Google Scholar] [CrossRef] [Green Version]
- Shenton, M.; Iwamoto, C.; Kurata, N.; Ikeo, K. Effect of wild and cultivated rice genotypes on rhizosphere bacterial community composition. Rice 2016, 9, 42. [Google Scholar] [CrossRef] [Green Version]
- Baruah, K.; Gogoi, B.; Gogoi, P. Plant physiological and soil characteristics associated with methane and nitrous oxide emission from rice paddy. Physiol. Mol. Biol. Plants 2010, 16, 79–91. [Google Scholar] [CrossRef] [Green Version]
- Warburton, M.L.; Wilkes, G.; Taba, S.; Charcosset, A.; Mir, C.; Dumas, F.; Madur, D.; Dreisigacker, S.; Bedoya, C.; Prasanna, B. Gene flow among different teosinte taxa and into the domesticated maize gene pool. Genet. Resour. Crop Evol. 2011, 58, 1243–1261. [Google Scholar] [CrossRef]
- Stitzer, M.C.; Ross-Ibarra, J. Maize domestication and gene interaction. New Phytol. 2018, 220, 395–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Nussbaum-Wagler, T.; Li, B.; Zhao, Q.; Vigouroux, Y.; Faller, M.; Bomblies, K.; Lukens, L.; Doebley, J.F. The origin of the naked grains of maize. Nature 2005, 436, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Byerlee, D. The globalization of hybrid maize, 1921–70. J. Glob. Hist. 2020, 15, 101–122. [Google Scholar] [CrossRef]
- Curry, H.A. From working collections to the World Germplasm Project: Agricultural modernization and genetic conservation at the Rockefeller Foundation. Hist. Philos. Life Sci. 2017, 39, 5. [Google Scholar] [CrossRef] [Green Version]
- Grote, U.; Fasse, A.; Nguyen, T.T.; Erenstein, O. Food security and the dynamics of wheat and maize value chains in Africa and Asia. Front. Sustain. Food Syst. 2021, 4, 617009. [Google Scholar] [CrossRef]
- Vulchinkov, S.; Ilchovska, D.; Pavlovska, B.; Ivanova, K. Trends in productive abilities of maize hybrids from different FAO groups. Bulg. J. Agric. Sci. 2013, 19, 744–749. [Google Scholar]
- Gu, R.; Fu, J.; Guo, S.; Duan, F.; Wang, Z.; Mi, G.; Yuan, L. Comparative expression and phylogenetic analysis of maize cytokinin dehydrogenase/oxidase (CKX) gene family. J. Plant Growth Regul. 2010, 29, 428–440. [Google Scholar] [CrossRef]
- Wang, H.; Sun, H.; Xia, H.; Wu, T.; Li, P.; Xu, C.; Yang, Z. Natural variation and domestication selection of ZmCKX5 with root morphological traits at the seedling stage in maize. Plants 2020, 10, 1. [Google Scholar] [CrossRef]
- Cao, P.; Lu, C.; Yu, Z. Historical nitrogen fertilizer use in agricultural ecosystems of the contiguous United States during 1850–2015: Application rate, timing, and fertilizer types. Earth Syst. Sci. Data 2018, 10, 969–984. [Google Scholar] [CrossRef] [Green Version]
- Vyas, R.V.; Panpatte, D.G.; Jhala, Y.K.; Shelat, H.N. Wonders of microbes in agriculture for productivity and sustainability. In Microorganisms for Green Revolution; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–23. [Google Scholar]
- Egamberdiyeva, D. The effect of plant growth promoting bacteria on growth and nutrient uptake of maize in two different soils. Appl. Soil Ecol. 2007, 36, 184–189. [Google Scholar] [CrossRef]
- Fuller, D.Q.; Stevens, C.J. Sorghum domestication and diversification: A current archaeobotanical perspective. In Plants and People in the African Past; Springer: Berlin/Heidelberg, Germany, 2018; pp. 427–452. [Google Scholar]
- Chen, J.; Xin, Z.; Laza, H. Registration of BTx623dw5, a new sorghum dwarf mutant. J. Plant Regist. 2019, 13, 254–257. [Google Scholar]
- Hedden, P. The genes of the Green Revolution. TRENDS Genet. 2003, 19, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Bollam, S.; Romana, K.K.; Rayaprolu, L.; Vemula, A.; Das, R.R.; Rathore, A.; Gandham, P.; Chander, G.; Deshpande, S.P.; Gupta, R. Nitrogen use efficiency in Sorghum: Exploring native variability for traits under variable N-Regimes. Front. Plant Sci. 2021, 12, 643192. [Google Scholar] [CrossRef]
- Multani, D.S.; Briggs, S.P.; Chamberlin, M.A.; Blakeslee, J.J.; Murphy, A.S.; Johal, G.S. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 2003, 302, 81–84. [Google Scholar] [CrossRef]
- Ordonio, R.L.; Ito, Y.; Hatakeyama, A.; Ohmae-Shinohara, K.; Kasuga, S.; Tokunaga, T.; Mizuno, H.; Kitano, H.; Matsuoka, M.; Sazuka, T. Gibberellin deficiency pleiotropically induces culm bending in sorghum: An insight into sorghum semi-dwarf breeding. Sci. Rep. 2014, 4, 5287. [Google Scholar] [CrossRef] [Green Version]
- Petti, C.; Hirano, K.; Stork, J.; DeBolt, S. Mapping of a cellulose-deficient mutant named dwarf1–1 in Sorghum bicolor to the green revolution gene gibberellin20-oxidase reveals a positive regulatory association between gibberellin and cellulose biosynthesis. Plant Physiol. 2015, 169, 705–716. [Google Scholar] [CrossRef] [Green Version]
- Ramon, U.; Weiss, D.; Illouz-Eliaz, N. Underground gibberellin activity: Differential gibberellin response in tomato shoots and roots. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lloret, F.; Casanovas, C.; Penuelas, J. Seedling survival of Mediterranean shrubland species in relation to root: Shoot ratio, seed size and water and nitrogen use. Funct. Ecol. 1999, 13, 210–216. [Google Scholar] [CrossRef] [Green Version]
- Niu, Y.; Chen, T.; Zhao, C.; Zhou, M. Improving Crop Lodging Resistance by Adjusting Plant Height and Stem Strength. Agronomy 2021, 11, 2421. [Google Scholar] [CrossRef]
- Binenbaum, J.; Weinstain, R.; Shani, E. Gibberellin localization and transport in plants. Trends Plant Sci. 2018, 23, 410–421. [Google Scholar] [CrossRef] [Green Version]
- Igielski, R.; Kępczyńska, E. Gene expression and metabolite profiling of gibberellin biosynthesis during induction of somatic embryogenesis in Medicago truncatula Gaertn. PLoS ONE 2017, 12, e0182055. [Google Scholar] [CrossRef] [Green Version]
- Nakayama, A.; Nakajima, M.; Yamaguchi, I. Distribution of gibberellins and expressional analysis of GA 20-oxidase genes of morning glory during fruit maturation. Biosci. Biotechnol. Biochem. 2005, 69, 334–342. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Yang, X.; Moolhuijzen, P.; Li, C.; Bellgard, M.; Lance, R.; Appels, R. Towards isolation of the barley green revolution gene. In Proceedings of the 12th Australian Barley Technical Symposium, Hobart, Tasmania, 11–14 September 2005. [Google Scholar]
- Alumira, J.; Rusike, J. The green revolution in Zimbabwe. Ejade Electron. J. Agric. Dev. Econ. 2005, 2, 50–66. [Google Scholar]
- Ouedraogo, Y.; Taonda, J.B.S.; Sermé, I.; Tychon, B.; Bielders, C.L. Factors driving cereal response to fertilizer microdosing in sub-Saharan Africa: A meta-analysis. Agron. J. 2020, 112, 2418–2431. [Google Scholar] [CrossRef]
- Cai, F.; Pang, G.; Li, R.-X.; Li, R.; Gu, X.-L.; Shen, Q.-R.; Chen, W. Bioorganic fertilizer maintains a more stable soil microbiome than chemical fertilizer for monocropping. Biol. Fertil. Soils 2017, 53, 861–872. [Google Scholar] [CrossRef]
- Rosenblueth, M.; Ormeño-Orrillo, E.; López-López, A.; Rogel, M.A.; Reyes-Hernández, B.J.; Martínez-Romero, J.C.; Reddy, P.M.; Martínez-Romero, E. Nitrogen fixation in cereals. Front. Microbiol. 2018, 9, 1794. [Google Scholar] [CrossRef] [Green Version]
- Coelho, M.R.; Marriel, I.E.; Jenkins, S.N.; Lanyon, C.V.; Seldin, L.; O’Donnell, A.G. Molecular detection and quantification of nifH gene sequences in the rhizosphere of sorghum (Sorghum bicolor) sown with two levels of nitrogen fertilizer. Appl. Soil Ecol. 2009, 42, 48–53. [Google Scholar] [CrossRef] [Green Version]
- Gaby, J.C.; Buckley, D.H. A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS ONE 2012, 7, e42149. [Google Scholar] [CrossRef] [Green Version]
- Borrell, A.; van Oosterom, E.; George-Jaeggli, B.; Rodriguez, D.; Eyre, J.; Jordan, D.J.; Mace, E.; Singh, V.; Vadez, V.; Bell, M. Sorghum. In Crop Physiology Case Histories for Major Crops; Elsevier: Amsterdam, The Netherlands, 2021; pp. 196–221. [Google Scholar]
- Haas, M.; Schreiber, M.; Mascher, M. Domestication and crop evolution of wheat and barley: Genes, genomics, and future directions. J. Integr. Plant Biol. 2019, 61, 204–225. [Google Scholar] [CrossRef] [Green Version]
- Badr, A.; Rabey, H.E.; Effgen, S.; Ibrahim, H.; Pozzi, C.; Rohde, W.; Salamini, F. On the origin and domestication history of barley (Hordeum vulgare). Mol. Biol. Evol. 2000, 17, 499–510. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Ye, H.; Liu, L.; Wu, J.; Ru, W.; Sun, G. Molecular insights on the domestication of barley (Hordeum vulgare L.). Crit. Rev. Plant Sci. 2019, 38, 280–294. [Google Scholar] [CrossRef]
- Drosse, B.; Campoli, C.; Mulki, A.; Korff, M.V. Genetic control of reproductive development. Biotechnol. Approaches Barley Improv. 2014, 173, 81–99. [Google Scholar]
- Fernández-Calleja, M.; Casas, A.M.; Igartua, E. Major flowering time genes of barley: Allelic diversity, effects, and comparison with wheat. Theor. Appl. Genet. 2021, 134, 1867–1897. [Google Scholar] [CrossRef]
- Chloupek, O.; Forster, B.P.; Thomas, W.T. The effect of semi-dwarf genes on root system size in field-grown barley. Theor. Appl. Genet. 2006, 112, 779–786. [Google Scholar] [CrossRef]
- Robertson-Albertyn, S.; Alegria Terrazas, R.; Balbirnie, K.; Blank, M.; Janiak, A.; Szarejko, I.; Chmielewska, B.; Karcz, J.; Morris, J.; Hedley, P.E. Root hair mutations displace the barley rhizosphere microbiota. Front. Plant Sci. 2017, 8, 1094. [Google Scholar] [CrossRef] [Green Version]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef]
- Cordell, D.; White, S. Tracking phosphorus security: Indicators of phosphorus vulnerability in the global food system. Food Secur. 2015, 7, 337–350. [Google Scholar] [CrossRef]
- Nanda, M.; Kansal, A.; Cordell, D. Managing agricultural vulnerability to phosphorus scarcity through bottom-up assessment of regional-scale opportunities. Agric. Syst. 2020, 184, 102910. [Google Scholar] [CrossRef]
- Yadav, V.; Karak, T.; Singh, S.; Singh, A.K.; Khare, P. Benefits of biochar over other organic amendments: Responses for plant productivity (Pelargonium graveolens L.) and nitrogen and phosphorus losses. Ind. Crops Prod. 2019, 131, 96–105. [Google Scholar] [CrossRef]
- Luo, G.; Li, L.; Friman, V.-P.; Guo, J.; Guo, S.; Shen, Q.; Ling, N. Organic amendments increase crop yields by improving microbe-mediated soil functioning of agroecosystems: A meta-analysis. Soil Biol. Biochem. 2018, 124, 105–115. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dixon, M.; Rohrbaugh, C.; Afkairin, A.; Vivanco, J. Impacts of the Green Revolution on Rhizosphere Microbiology Related to Nutrient Acquisition. Appl. Microbiol. 2022, 2, 992-1003. https://doi.org/10.3390/applmicrobiol2040076
Dixon M, Rohrbaugh C, Afkairin A, Vivanco J. Impacts of the Green Revolution on Rhizosphere Microbiology Related to Nutrient Acquisition. Applied Microbiology. 2022; 2(4):992-1003. https://doi.org/10.3390/applmicrobiol2040076
Chicago/Turabian StyleDixon, Mary, Carley Rohrbaugh, Antisar Afkairin, and Jorge Vivanco. 2022. "Impacts of the Green Revolution on Rhizosphere Microbiology Related to Nutrient Acquisition" Applied Microbiology 2, no. 4: 992-1003. https://doi.org/10.3390/applmicrobiol2040076
APA StyleDixon, M., Rohrbaugh, C., Afkairin, A., & Vivanco, J. (2022). Impacts of the Green Revolution on Rhizosphere Microbiology Related to Nutrient Acquisition. Applied Microbiology, 2(4), 992-1003. https://doi.org/10.3390/applmicrobiol2040076





