Yeast and Virus-like Particles: A Perfect or Imperfect Couple?
Abstract
:1. Introduction
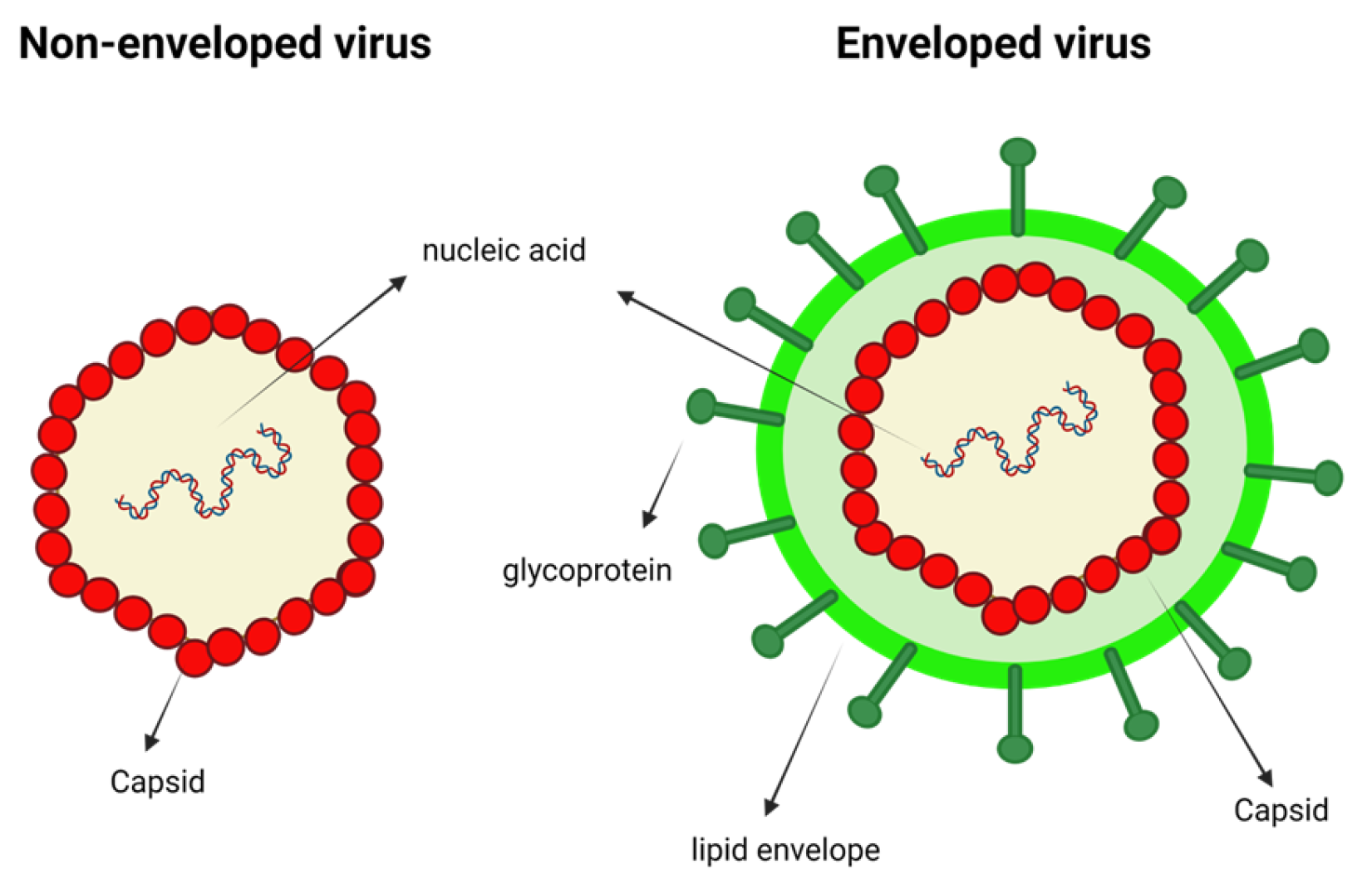
1.1. Types of VLPs
1.2. Critical Points in VLP Production
1.3. General Considerations of Yeast as Expression System
2. Saccharomyces cerevisiae
3. Pichia pastoris
4. Hansenula polymorpha
5. Comparison of VLP Production in the Three Yeast Species
6. Conclusions
- The number of proteins to be expressed. Some proteins must be in a suitable ratio to assemble in functional VLPs;
- Primary assessment of heterologous protein expression. The evaluation of the effect of the protein is crucial; toxicity should be avoided; therefore, several mutants (protease deficient, high temperature tolerant, secretory efficient) should be considered;
- Inducible promoters and episomal plasmids are preferable; usually, high efficiency of DNA transformation is required. For this reason, S. cerevisiae and P. pastoris strains could be more suitable;
- Plasmid stability. In general, in S. cerevisiae and P. pastoris, episomal or centromeric plasmids are more stable than in H. polymorpha. In H. polymorpha, plasmids must be integrated into the genome to achieve high stability. Therefore, if the expression of the heterologous proteins confers toxicity when expressed in high copy number, the use of H. polymorpha expression system which consists of one single stable integrated copy of the gene is preferable.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fraenkel-Conrat, H.; Williams, R.C. Reconstitution of Active Tobacco Mosaic Virus from Its Inactive Protein and Nucleic Acid Components. Proc. Natl. Acad. Sci. USA 1955, 41, 690–698. [Google Scholar] [CrossRef]
- Bayer, M.E.; Blumberg, B.S.; Werner, B. Particles associated with Australia antigen in the sera of patients with leukaemia, Down’s Syndrome and hepatitis. Nature 1968, 218, 1057–1059. [Google Scholar] [CrossRef] [PubMed]
- Klucis, E.; Jackson, L.; Parsons, P.G. Survey of human lymphoblastoid cell lines and primary cultures of normal and leukaemic leukocytes for oncornavirus production. Int. J. Cancer 1976, 18, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Graves, D.C.; Ferrer, J.F. In vitro transmission and propagation of the bovine leukemia virus in monolayer cell cultures. Cancer Res. 1976, 36, 4152–4159. [Google Scholar] [PubMed]
- Engelberg-Kulka, H.; Dekel, L.; Israeli-Reches, M. Streptomycin-resistant Escherichia coli mutant temperature sensitive for the production of Qbeta-infective particles. J. Virol. 1977, 21, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volkoff, O.; Walters, T. Virus-like particles in abnormal cells of Saccharomyces carlsbergensis. Can. J. Genet. Cytol. 1970, 12, 621–626. [Google Scholar] [CrossRef]
- Berry, E.A.; Bevan, E.A. A new species of double-stranded RNA from yeast. Nature 1972, 239, 279–280. [Google Scholar] [CrossRef]
- Bevan, E.A.; Herring, A.J.; Mitchell, D.J. Preliminary characterization of two species of dsRNA in yeast and their relationship to the “killer” character. Nature 1973, 245, 81–86. [Google Scholar] [CrossRef]
- Wood, H.A. Viruses with double-stranded RNA genomes. J. Gen. Virol. 1973, 20, 61–85. [Google Scholar] [CrossRef]
- Bruenn, J.A. Virus-like particles of yeast. Annu. Rev. Microbiol. 1980, 34, 49–68. [Google Scholar] [CrossRef]
- Wickner, R.B. Double-stranded and single-stranded RNA viruses of Saccharomyces cerevisiae. Annu. Rev. Microbiol. 1992, 46, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Mason, H.S.; Lam, D.M.; Arntzen, C.J. Expression of hepatitis B surface antigen in transgenic plants. Proc. Natl. Acad. Sci. USA 1992, 89, 11745–11749. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.H.; Church, D.; Koellhoffer, E.C.; Osota, E.; Shukla, S.; Rybicki, E.P.; Pokorski, J.K.; Steinmetz, N.F. Integrating plant molecular farming and materials research for next-generation vaccines. Nat. Rev. Mater. 2022, 7, 372–388. [Google Scholar] [CrossRef] [PubMed]
- Croizier, L.; Jousset, F.X.; Veyrunes, J.C.; Lopez-Ferber, M.; Bergoin, M.; Croizier, G. Protein requirements for assembly of virus-like particles of Junonia coenia densovirus in insect cells. J. Gen. Virol. 2000, 81, 1605–1613. [Google Scholar] [CrossRef]
- Tariq, H.; Batool, S.; Asif, S.; Ali, M.; Abbasi, B.H. Virus-Like Particles: Revolutionary Platforms for Developing Vaccines Against Emerging Infectious Diseases. Front. Microbiol. 2021, 12, 790121. [Google Scholar] [CrossRef]
- Qian, C.; Liu, X.; Xu, Q.; Wang, Z.; Chen, J.; Li, T.; Zheng, Q.; Yu, H.; Gu, Y.; Li, S.; et al. Recent Progress on the Versatility of Virus-Like Particles. Vaccines 2020, 8, 139. [Google Scholar] [CrossRef] [Green Version]
- Ward, B.J.; Makarkov, A.; Seguin, A.; Pillet, S.; Trepanier, S.; Dhaliwall, J.; Libman, M.D.; Vesikari, T.; Landry, N. Efficacy, immunogenicity, and safety of a plant-derived, quadrivalent, virus-like particle influenza vaccine in adults (18–64 years) and older adults (>/=65 years): Two multicentre, randomised phase 3 trials. Lancet 2020, 396, 1491–1503. [Google Scholar] [CrossRef]
- Palucha, A.; Loniewska, A.; Satheshkumar, S.; Boguszewska-Chachulska, A.M.; Umashankar, M.; Milner, M.; Haenni, A.L.; Savithri, H.S. Virus-like particles: Models for assembly studies and foreign epitope carriers. Prog. Nucleic Acid. Res. Mol. Biol. 2005, 80, 135–168. [Google Scholar] [CrossRef]
- Ikwuagwu, B.; Tullman-Ercek, D. Virus-like particles for drug delivery: A review of methods and applications. Curr. Opin. Biotechnol. 2022, 78, 102785. [Google Scholar] [CrossRef]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like particles: Preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnol. 2021, 19, 59. [Google Scholar] [CrossRef]
- Tornesello, A.L.; Tagliamonte, M.; Buonaguro, F.M.; Tornesello, M.L.; Buonaguro, L. Virus-like Particles as Preventive and Therapeutic Cancer Vaccines. Vaccines 2022, 10, 227. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Iwasaki, Y.; Tada, H.; Iwabuki, H.; Chuah, M.K.; VandenDriessche, T.; Fukuda, H.; Kondo, A.; Ueda, M.; Seno, M.; et al. Nanoparticles for the delivery of genes and drugs to human hepatocytes. Nat. Biotechnol. 2003, 21, 885–890. [Google Scholar] [CrossRef]
- Li, Y.; Liu, D.; Wang, Y.; Su, W.; Liu, G.; Dong, W. The Importance of Glycans of Viral and Host Proteins in Enveloped Virus Infection. Front. Immunol. 2021, 12, 638573. [Google Scholar] [CrossRef] [PubMed]
- Coombs, K.M. Stoichiometry of reovirus structural proteins in virus, ISVP, and core particles. Virology 1998, 243, 218–228. [Google Scholar] [CrossRef] [Green Version]
- Weitzman, M.D.; Linden, R.M. Adeno-associated virus biology. Methods Mol. Biol. 2011, 807, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Byrne, L.J.; O’Callaghan, K.J.; Tuite, M.F. Heterologous gene expression in yeast. Methods Mol. Biol. 2005, 308, 51–64. [Google Scholar] [CrossRef]
- Vieira Gomes, A.M.; Souza Carmo, T.; Silva Carvalho, L.; Mendonca Bahia, F.; Parachin, N.S. Comparison of Yeasts as Hosts for Recombinant Protein Production. Microorganisms 2018, 29, 38. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.; Kumar, P. Yeast-based vaccines: New perspective in vaccine development and application. FEMS Yeast Res. 2019, 19, foz007. [Google Scholar] [CrossRef] [Green Version]
- Bill, R.M. Recombinant protein subunit vaccine synthesis in microbes: A role for yeast? J. Pharm. Pharmacol. 2015, 67, 319–328. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, V.; Nand, K.N.; Ahmad, A.; Kumar, R. Yeast-Based Virus-like Particles as an Emerging Platform for Vaccine Development and Delivery. Vaccines 2023, 11, 479. [Google Scholar] [CrossRef]
- Gassler, T.; Heistinger, L.; Mattanovich, D.; Gasser, B.; Prielhofer, R. CRISPR/Cas9-Mediated Homology-Directed Genome Editing in Pichia pastoris. Methods Mol. Biol. 2019, 1923, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Laughery, M.F.; Wyrick, J.J. Simple CRISPR-Cas9 Genome Editing in Saccharomyces cerevisiae. Curr. Protoc. Mol. Biol. 2019, 129, e110. [Google Scholar] [CrossRef] [PubMed]
- Numamoto, M.; Maekawa, H.; Kaneko, Y. Efficient genome editing by CRISPR/Cas9 with a tRNA-sgRNA fusion in the methylotrophic yeast Ogataea polymorpha. J. Biosci. Bioeng. 2017, 124, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, A.; Arun, K.B.; Sindhu, R.; Krishnamoorthy, J.; Reshmy, R.; Sirohi, R.; Pugazhendi, A.; Awasthi, M.K.; Szakacs, G.; Binod, P. Customized yeast cell factories for biopharmaceuticals: From cell engineering to process scale up. Microb. Cell Fact. 2021, 20, 124. [Google Scholar] [CrossRef] [PubMed]
- Gnugge, R.; Rudolf, F. Saccharomyces cerevisiae Shuttle vectors. Yeast 2017, 34, 205–221. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Shao, Z.; Zhao, H.; Nair, N.; Wen, F.; Xu, J.H. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2012, 109, 2082–2092. [Google Scholar] [CrossRef]
- Weinhandl, K.; Winkler, M.; Glieder, A.; Camattari, A. Carbon source dependent promoters in yeasts. Microb. Cell Fact. 2014, 13, 5. [Google Scholar] [CrossRef] [Green Version]
- Partow, S.; Siewers, V.; Bjorn, S.; Nielsen, J.; Maury, J. Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast 2010, 27, 955–964. [Google Scholar] [CrossRef]
- West, R.W., Jr.; Chen, S.M.; Putz, H.; Butler, G.; Banerjee, M. GAL1-GAL10 divergent promoter region of Saccharomyces cerevisiae contains negative control elements in addition to functionally separate and possibly overlapping upstream activating sequences. Genes. Dev. 1987, 1, 1118–1131. [Google Scholar] [CrossRef] [Green Version]
- Butt, T.R.; Ecker, D.J. Yeast metallothionein and applications in biotechnology. Microbiol. Rev. 1987, 51, 351–364. [Google Scholar] [CrossRef]
- Hauf, J.; Zimmermann, F.K.; Muller, S. Simultaneous genomic overexpression of seven glycolytic enzymes in the yeast Saccharomyces cerevisiae. Enzyme Microb. Technol. 2000, 26, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; DaSilva, N.A. Evaluation of the Saccharomyces cerevisiae ADH2 promoter for protein synthesis. Yeast 2005, 22, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Tian, Y.; Liu, H.; Kan, Y.; Zhou, Y.; Wang, Y.; Luo, Y. Harnessing the Endogenous 2mu Plasmid of Saccharomyces cerevisiae for Pathway Construction. Front. Microbiol. 2021, 12, 679665. [Google Scholar] [CrossRef]
- Valenzuela, P.; Medina, A.; Rutter, W.J.; Ammerer, G.; Hall, B.D. Synthesis and assembly of hepatitis B virus surface antigen particles in yeast. Nature 1982, 298, 347–350. [Google Scholar] [CrossRef]
- Hofmann, K.J.; Cook, J.C.; Joyce, J.G.; Brown, D.R.; Schultz, L.D.; George, H.A.; Rosolowsky, M.; Fife, K.H.; Jansen, K.U. Sequence determination of human papillomavirus type 6a and assembly of virus-like particles in Saccharomyces cerevisiae. Virology 1995, 209, 506–518. [Google Scholar] [CrossRef]
- Sapp, M.; Volpers, C.; Streeck, R.E. Synthesis, properties and applications of papillomavirus-like particles. Intervirology 1996, 39, 49–53. [Google Scholar] [CrossRef]
- Sasnauskas, K.; Bulavaite, A.; Hale, A.; Jin, L.; Knowles, W.A.; Gedvilaite, A.; Dargeviciute, A.; Bartkeviciute, D.; Zvirbliene, A.; Staniulis, J.; et al. Generation of recombinant virus-like particles of human and non-human polyomaviruses in yeast Saccharomyces cerevisiae. Intervirology 2002, 45, 308–317. [Google Scholar] [CrossRef]
- Zeltins, A. Construction and characterization of virus-like particles: A review. Mol. Biotechnol. 2013, 53, 92–107. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, H.J. Yeast as an expression system for producing virus-like particles: What factors do we need to consider? Lett. Appl. Microbiol. 2017, 64, 111–123. [Google Scholar] [CrossRef]
- Sakuragi, S.; Goto, T.; Sano, K.; Morikawa, Y. HIV type 1 Gag virus-like particle budding from spheroplasts of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2002, 99, 7956–7961. [Google Scholar] [CrossRef]
- Mazumder, S.; Rastogi, R.; Undale, A.; Arora, K.; Arora, N.M.; Pratim, B.; Kumar, D.; Joseph, A.; Mali, B.; Arya, V.B.; et al. PRAK-03202: A triple antigen virus-like particle vaccine candidate against SARS CoV-2. Heliyon 2021, 7, e08124. [Google Scholar] [CrossRef] [PubMed]
- Backovic, A.; Cervelli, T.; Salvetti, A.; Zentilin, L.; Giacca, M.; Galli, A. Capsid protein expression and adeno-associated virus like particles assembly in Saccharomyces cerevisiae. Microb. Cell Fact. 2012, 11, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barajas, D.; Aponte-Ubillus, J.J.; Akeefe, H.; Cinek, T.; Peltier, J.; Gold, D. Generation of infectious recombinant Adeno-associated virus in Saccharomyces cerevisiae. PLoS ONE 2017, 12, e0173010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, S.; Sandal, T.; Kamp-Hansen, P.; Dalboge, H. Comparison of expression systems in the yeasts Saccharomyces cerevisiae, Hansenula polymorpha, Klyveromyces lactis, Schizosaccharomyces pombe and Yarrowia lipolytica. Cloning of two novel promoters from Yarrowia lipolytica. Yeast 1998, 14, 1267–1283. [Google Scholar] [CrossRef]
- Tran, A.M.; Nguyen, T.T.; Nguyen, C.T.; Huynh-Thi, X.M.; Trinh, M.T.; Tran, L.T.; Cartwright, S.P.; Bill, R.M.; Tran-Van, H. Pichia pastoris versus Saccharomyces cerevisiae: A case study on the recombinant production of human granulocyte-macrophage colony-stimulating factor. BMC Res. Notes 2017, 10, 148. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Wang, G.; Qin, J.; Petranovic, D.; Nielsen, J. Engineering the protein secretory pathway of Saccharomyces cerevisiae enables improved protein production. Proc. Natl. Acad. Sci. USA 2018, 115, E11025–E11032. [Google Scholar] [CrossRef] [Green Version]
- Turcotte, B.; Liang, X.B.; Robert, F.; Soontorngun, N. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 2010, 10, 2–13. [Google Scholar] [CrossRef] [Green Version]
- Gerngross, T. Production of complex human glycoproteins in yeast. Adv. Exp. Med. Biol. 2005, 564, 139. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, S.R.; Bobrowicz, P.; Bobrowicz, B.; Davidson, R.C.; Li, H.; Mitchell, T.; Nett, J.H.; Rausch, S.; Stadheim, T.A.; Wischnewski, H.; et al. Production of complex human glycoproteins in yeast. Science 2003, 301, 1244–1246. [Google Scholar] [CrossRef]
- Lee, H.; DeLoache, W.C.; Dueber, J.E. Spatial organization of enzymes for metabolic engineering. Metab. Eng. 2012, 14, 242–251. [Google Scholar] [CrossRef]
- Cheah, L.C.; Stark, T.; Adamson, L.S.R.; Abidin, R.S.; Lau, Y.H.; Sainsbury, F.; Vickers, C.E. Artificial Self-assembling Nanocompartment for Organizing Metabolic Pathways in Yeast. ACS Synth. Biol. 2021, 10, 3251–3263. [Google Scholar] [CrossRef] [PubMed]
- Malci, K.; Watts, E.; Roberts, T.M.; Auxillos, J.Y.; Nowrouzi, B.; Boll, H.O.; Nascimento, C.; Andreou, A.; Vegh, P.; Donovan, S.; et al. Standardization of Synthetic Biology Tools and Assembly Methods for Saccharomyces cerevisiae and Emerging Yeast Species. ACS Synth. Biol. 2022, 11, 2527–2547. [Google Scholar] [CrossRef] [PubMed]
- Buckholz, R.G.; Gleeson, M.A. Yeast systems for the commercial production of heterologous proteins. Biotechnology 1991, 9, 1067–1072. [Google Scholar] [CrossRef]
- Sturmberger, L.; Chappell, T.; Geier, M.; Krainer, F.; Day, K.J.; Vide, U.; Trstenjak, S.; Schiefer, A.; Richardson, T.; Soriaga, L.; et al. Refined Pichia pastoris reference genome sequence. J. Biotechnol. 2016, 235, 121–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cereghino, J.L.; Cregg, J.M. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol. Rev. 2000, 24, 45–66. [Google Scholar] [CrossRef] [PubMed]
- Karbalaei, M.; Rezaee, S.A.; Farsiani, H. Pichia pastoris: A highly successful expression system for optimal synthesis of heterologous proteins. J. Cell. Physiol. 2020, 235, 5867–5881. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Rajpoot, R.K.; Arora, U.; Poddar, A.; Swaminathan, S.; Khanna, N. Pichia pastoris-Expressed Bivalent Virus-Like Particulate Vaccine Induces Domain III-Focused Bivalent Neutralizing Antibodies without Antibody-Dependent Enhancement in Vivo. Front. Microbiol. 2017, 8, 2644. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Jiang, H.; Zhou, J.; Yang, X.; Tang, Y.; Fang, D.; Jiang, L. Recombinant dengue virus-like particles from Pichia pastoris: Efficient production and immunological properties. Virus Genes 2010, 40, 53–59. [Google Scholar] [CrossRef]
- Tang, Y.X.; Jiang, L.F.; Zhou, J.M.; Yin, Y.; Yang, X.M.; Liu, W.Q.; Fang, D.Y. Induction of virus-neutralizing antibodies and T cell responses by dengue virus type 1 virus-like particles prepared from Pichia pastoris. Chin. Med. J. 2012, 125, 1986–1992. [Google Scholar]
- Vogl, T.; Hartner, F.S.; Glieder, A. New opportunities by synthetic biology for biopharmaceutical production in Pichia pastoris. Curr. Opin. Biotechnol. 2013, 24, 1094–1101. [Google Scholar] [CrossRef] [Green Version]
- Garrigos-Martinez, J.; Vuoristo, K.; Nieto-Taype, M.A.; Tahtiharju, J.; Uusitalo, J.; Tukiainen, P.; Schmid, C.; Tolstorukov, I.; Madden, K.; Penttila, M.; et al. Bioprocess performance analysis of novel methanol-independent promoters for recombinant protein production with Pichia pastoris. Microb. Cell Fact. 2021, 20, 74. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Qian, J.; Yao, G.; Zhuang, Y.; Zhang, S.; Chu, J. GAP promoter library for fine-tuning of gene expression in Pichia pastoris. Appl. Environ. Microbiol. 2011, 77, 3600–3608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhou, J.; Yu, Z.; Fang, D.; Fu, C.; Zhu, X.; He, Z.; Yan, H.; Jiang, L. Tetravalent recombinant dengue virus-like particles as potential vaccine candidates: Immunological properties. BMC Microbiol. 2014, 14, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Q.; He, Y.; Lu, J. Virus-Like Particles Produced in Pichia pastoris Induce Protective Immune Responses against Coxsackievirus A16 in Mice. Med. Sci. Monit. 2016, 22, 3370–3382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eto, Y.; Saubi, N.; Ferrer, P.; Joseph-Munne, J. Expression of Chimeric HPV-HIV Protein L1P18 in Pichia pastoris; Purification and Characterization of the Virus-like Particles. Pharmaceutics 2021, 13, 1967. [Google Scholar] [CrossRef]
- Delic, M.; Valli, M.; Graf, A.B.; Pfeffer, M.; Mattanovich, D.; Gasser, B. The secretory pathway: Exploring yeast diversity. FEMS Microbiol. Rev. 2013, 37, 872–914. [Google Scholar] [CrossRef] [Green Version]
- Chahal, S.; Wei, P.; Moua, P.; Park, S.P.; Kwon, J.; Patel, A.; Vu, A.T.; Catolico, J.A.; Tsai, Y.F.; Shaheen, N.; et al. Structural characterization of the alpha-mating factor prepro-peptide for secretion of recombinant proteins in Pichia pastoris. Gene 2017, 598, 50–62. [Google Scholar] [CrossRef]
- Barrero, J.J.; Casler, J.C.; Valero, F.; Ferrer, P.; Glick, B.S. An improved secretion signal enhances the secretion of model proteins from Pichia pastoris. Microb. Cell Fact. 2018, 17, 161. [Google Scholar] [CrossRef]
- Duan, G.; Ding, L.; Wei, D.; Zhou, H.; Chu, J.; Zhang, S.; Qian, J. Screening endogenous signal peptides and protein folding factors to promote the secretory expression of heterologous proteins in Pichia pastoris. J. Biotechnol. 2019, 306, 193–202. [Google Scholar] [CrossRef]
- Obst, U.; Lu, T.K.; Sieber, V. A Modular Toolkit for Generating Pichia pastoris Secretion Libraries. ACS Synth. Biol. 2017, 6, 1016–1025. [Google Scholar] [CrossRef]
- Raschmanova, H.; Weninger, A.; Knejzlik, Z.; Melzoch, K.; Kovar, K. Engineering of the unfolded protein response pathway in Pichia pastoris: Enhancing production of secreted recombinant proteins. Appl. Microbiol. Biotechnol. 2021, 105, 4397–4414. [Google Scholar] [CrossRef] [PubMed]
- Tome-Amat, J.; Fleischer, L.; Parker, S.A.; Bardliving, C.L.; Batt, C.A. Secreted production of assembled Norovirus virus-like particles from Pichia pastoris. Microb. Cell Fact. 2014, 13, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Yang, K.; Liu, A.; Lu, X.; Yang, L.; Zhao, Q. Highly secretory expression of recombinant cowpea chlorotic mottle virus capsid proteins in Pichia pastoris and in-vitro encapsulation of ruthenium nanoparticles for catalysis. Protein Expr. Purif. 2020, 174, 105679. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.M.; Jeong, Y.E.; Wang, E.; Lee, Y.J.; Han, M.G.; Park, C.; Lee, W.J.; Choi, W. Cloning and Expression of Recombinant Tick-Borne Encephalitis Virus-like Particles in Pichia pastoris. Osong Public Health Res. Perspect. 2014, 5, 274–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bazan, S.B.; de Alencar Muniz Chaves, A.; Aires, K.A.; Cianciarullo, A.M.; Garcea, R.L.; Ho, P.L. Expression and characterization of HPV-16 L1 capsid protein in Pichia pastoris. Arch. Virol. 2009, 154, 1609–1617. [Google Scholar] [CrossRef] [Green Version]
- Mariz, F.C.; Coimbra, E.C.; Jesus, A.L.; Nascimento, L.M.; Torres, F.A.; Freitas, A.C. Development of an IP-Free Biotechnology Platform for Constitutive Production of HPV16 L1 Capsid Protein Using the Pichia pastoris PGK1 Promoter. Biomed. Res. Int. 2015, 2015, 594120. [Google Scholar] [CrossRef] [Green Version]
- Saraswat, S.; Athmaram, T.N.; Parida, M.; Agarwal, A.; Saha, A.; Dash, P.K. Expression and Characterization of Yeast Derived Chikungunya Virus Like Particles (CHIK-VLPs) and Its Evaluation as a Potential Vaccine Candidate. PLoS Negl. Trop. Dis. 2016, 10, e0004782. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.S.; Jeon, J.H.; Lee, K.J.; Ko, K. N-glycosylation modification of plant-derived virus-like particles: An application in vaccines. Biomed. Res. Int. 2014, 2014, 249519. [Google Scholar] [CrossRef] [Green Version]
- Kwon, W.T.; Lee, W.S.; Park, P.J.; Park, T.K.; Kang, H. Protective immunity of Pichia pastoris-expressed recombinant envelope protein of Japanese encephalitis virus. J. Microbiol. Biotechnol. 2012, 22, 1580–1587. [Google Scholar] [CrossRef]
- Gupta, J.; Kaul, S.; Srivastava, A.; Kaushik, N.; Ghosh, S.; Sharma, C.; Batra, G.; Banerjee, M.; Shalimar; Nayak, B.; et al. Expression, Purification and Characterization of the Hepatitis E Virus Like-Particles in the Pichia pastoris. Front. Microbiol. 2020, 11, 141. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Xie, Y.; Wu, C.; Nan, Y. The Hepatitis E Virus Open Reading Frame 2 Protein: Beyond Viral Capsid. Front. Microbiol. 2021, 12, 739124. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Zhang, J.; Chen, Z.; Pan, W.; Yan, Y.; Dai, J. Glycosylation of viral proteins: Implication in virus-host interaction and virulence. Virulence 2022, 13, 670–683. [Google Scholar] [CrossRef] [PubMed]
- Poddar, A.; Ramasamy, V.; Shukla, R.; Rajpoot, R.K.; Arora, U.; Jain, S.K.; Swaminathan, S.; Khanna, N. Virus-like particles derived from Pichia pastoris-expressed dengue virus type 1 glycoprotein elicit homotypic virus-neutralizing envelope domain III-directed antibodies. BMC Biotechnol. 2016, 16, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mani, S.; Tripathi, L.; Raut, R.; Tyagi, P.; Arora, U.; Barman, T.; Sood, R.; Galav, A.; Wahala, W.; de Silva, A.; et al. Pichia pastoris-expressed dengue 2 envelope forms virus-like particles without pre-membrane protein and induces high titer neutralizing antibodies. PLoS ONE 2013, 8, e64595. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, L.; Mani, S.; Raut, R.; Poddar, A.; Tyagi, P.; Arora, U.; de Silva, A.; Swaminathan, S.; Khanna, N. Pichia pastoris-expressed dengue 3 envelope-based virus-like particles elicit predominantly domain III-focused high titer neutralizing antibodies. Front. Microbiol. 2015, 6, 1005. [Google Scholar] [CrossRef] [Green Version]
- Khetarpal, N.; Shukla, R.; Rajpoot, R.K.; Poddar, A.; Pal, M.; Swaminathan, S.; Arora, U.; Khanna, N. Recombinant Dengue Virus 4 Envelope Glycoprotein Virus-Like Particles Derived from Pichia pastoris are Capable of Eliciting Homotypic Domain III-Directed Neutralizing Antibodies. Am. J. Trop. Med. Hyg. 2017, 96, 126–134. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, S.R.; Davidson, R.C.; Sethuraman, N.; Nett, J.H.; Jiang, Y.; Rios, S.; Bobrowicz, P.; Stadheim, T.A.; Li, H.; Choi, B.K.; et al. Humanization of yeast to produce complex terminally sialylated glycoproteins. Science 2006, 313, 1441–1443. [Google Scholar] [CrossRef] [Green Version]
- Satyanarayana, G.K. (Ed.) Yeast Biotechnology: Diversity and Applications; Springer: Dordrecht, The Netherlands, 2009. [Google Scholar]
- Manfrao-Netto, J.H.C.; Gomes, A.M.V.; Parachin, N.S. Advances in Using Hansenula polymorpha as Chassis for Recombinant Protein Production. Front. Bioeng. Biotechnol. 2019, 7, 94. [Google Scholar] [CrossRef]
- Van Zutphen, T.; Baerends, R.J.; Susanna, K.A.; de Jong, A.; Kuipers, O.P.; Veenhuis, M.; van der Klei, I.J. Adaptation of Hansenula polymorpha to methanol: A transcriptome analysis. BMC Genom. 2010, 11, 1. [Google Scholar] [CrossRef] [Green Version]
- Hartner, F.S.; Glieder, A. Regulation of methanol utilisation pathway genes in yeasts. Microb. Cell Fact. 2006, 5, 39. [Google Scholar] [CrossRef] [Green Version]
- Moussa, M.; Ibrahim, M.; El Ghazaly, M.; Rohde, J.; Gnoth, S.; Anton, A.; Kensy, F.; Mueller, F. Expression of recombinant staphylokinase in the methylotrophic yeast Hansenula polymorpha. BMC Biotechnol. 2012, 12, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filyak, Y.; Finiuk, N.; Mitina, N.; Bilyk, O.; Titorenko, V.; Hrydzhuk, O.; Zaichenko, A.; Stoika, R. A novel method for genetic transformation of yeast cells using oligoelectrolyte polymeric nanoscale carriers. Biotechniques 2013, 54, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Thak, E.J.; Lee, D.J.; Agaphonov, M.O.; Kang, H.A. Hansenula polymorpha Pmt4p Plays Critical Roles in O-Mannosylation of Surface Membrane Proteins and Participates in Heteromeric Complex Formation. PLoS ONE 2015, 10, e0129914. [Google Scholar] [CrossRef] [Green Version]
- Faber, K.N.; Haima, P.; Harder, W.; Veenhuis, M.; Ab, G. Highly-efficient electrotransformation of the yeast Hansenula polymorpha. Curr. Genet. 1994, 25, 305–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sohn, J.H.; Choi, E.S.; Kim, C.H.; Agaphonov, M.O.; Ter-Avanesyan, M.D.; Rhee, J.S.; Rhee, S.K. A novel autonomously replicating sequence (ARS) for multiple integration in the yeast Hansenula polymorpha DL-1. J. Bacteriol. 1996, 178, 4420–4428. [Google Scholar] [CrossRef] [Green Version]
- Saraya, R.; Krikken, A.M.; Kiel, J.A.; Baerends, R.J.; Veenhuis, M.; van der Klei, I.J. Novel genetic tools for Hansenula polymorpha. FEMS Yeast Res. 2012, 12, 271–278. [Google Scholar] [CrossRef]
- Faber, K.N.; Westra, S.; Waterham, H.R.; Keizer-Gunnink, I.; Harder, W.; Veenhuis, G.A. Foreign gene expression in Hansenula polymorpha. A system for the synthesis of small functional peptides. Appl. Microbiol. Biotechnol. 1996, 45, 72–79. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.W.; Rhee, S.K.; Kim, J.Y.; Shimma, Y.; Chiba, Y.; Jigami, Y.; Kang, H.A. Characterization of N-linked oligosaccharides assembled on secretory recombinant glucose oxidase and cell wall mannoproteins from the methylotrophic yeast Hansenula polymorpha. Glycobiology 2004, 14, 243–251. [Google Scholar] [CrossRef] [Green Version]
- Wetzel, D.; Barbian, A.; Jenzelewski, V.; Schembecker, G.; Merz, J.; Piontek, M. Bioprocess optimization for purification of chimeric VLP displaying BVDV E2 antigens produced in yeast Hansenula polymorpha. J. Biotechnol. 2019, 306, 203–212. [Google Scholar] [CrossRef]
- McGregor, J.; Hardy, J.M.; Lay, C.S.; Boo, I.; Piontek, M.; Suckow, M.; Coulibaly, F.; Poumbourios, P.; Center, R.J.; Drummer, H.E. Virus-Like Particles Containing the E2 Core Domain of Hepatitis C Virus Generate Broadly Neutralizing Antibodies in Guinea Pigs. J. Virol. 2022, 96, e0167521. [Google Scholar] [CrossRef]
- Wetzel, D.; Chan, J.A.; Suckow, M.; Barbian, A.; Weniger, M.; Jenzelewski, V.; Reiling, L.; Richards, J.S.; Anderson, D.A.; Kouskousis, B.; et al. Display of malaria transmission-blocking antigens on chimeric duck hepatitis B virus-derived virus-like particles produced in Hansenula polymorpha. PLoS ONE 2019, 14, e0221394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheon, S.A.; Kim, H.; Oh, D.B.; Kwon, O.; Kang, H.A. Remodeling of the glycosylation pathway in the methylotrophic yeast Hansenula polymorpha to produce human hybrid-type N-glycans. J. Microbiol. 2012, 50, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Ren, S.; Chen, X.; Ge, J.; Xu, Z.; Huang, H.; Sun, H.; Gu, Y.; Zhou, T.; Li, J.; et al. Generation of hepatitis B virus PreS2-S antigen in Hansenula polymorpha. Virol. Sin. 2014, 29, 403–409. [Google Scholar] [CrossRef]
- Hardy, E.; Martinez, E.; Diago, D.; Diaz, R.; Gonzalez, D.; Herrera, L. Large-scale production of recombinant hepatitis B surface antigen from Pichia pastoris. J. Biotechnol. 2000, 77, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, D.; Rolf, T.; Suckow, M.; Kranz, A.; Barbian, A.; Chan, J.A.; Leitsch, J.; Weniger, M.; Jenzelewski, V.; Kouskousis, B.; et al. Establishment of a yeast-based VLP platform for antigen presentation. Microb. Cell Fact. 2018, 17, 17. [Google Scholar] [CrossRef]
- Shao, S.; Wang, Q.; Jin, Y.; Zhang, X.; Liu, Z.; Chen, S.; Wu, H.; Yang, S.; Tang, F.; Su, J.; et al. Recombinant Virus-like Particles of Human Parvovirus B19 with the Internal Location of VP1 Unique Region Produced by Hansenula polymorpha. Viruses 2022, 14, 2410. [Google Scholar] [CrossRef]
- Lorent, E.; Bierau, H.; Engelborghs, Y.; Verheyden, G.; Bosman, F. Structural characterisation of the hepatitis C envelope glycoprotein E1 ectodomain derived from a mammalian and a yeast expression system. Vaccine 2008, 26, 399–410. [Google Scholar] [CrossRef]
- Qian, W.; Aguilar, F.; Wang, T.; Qiu, B. Secretion of truncated recombinant rabies virus glycoprotein with preserved antigenic properties using a co-expression system in Hansenula polymorpha. J. Microbiol. 2013, 51, 234–240. [Google Scholar] [CrossRef]
- Klabunde, J.; Kleebank, S.; Piontek, M.; Hollenberg, C.P.; Hellwig, S.; Degelmann, A. Increase of calnexin gene dosage boosts the secretion of heterologous proteins by Hansenula polymorpha. FEMS Yeast Res. 2007, 7, 1168–1180. [Google Scholar] [CrossRef] [Green Version]
- Simanavicius, M.; Tamosiunas, P.L.; Petraityte-Burneikiene, R.; Johne, R.; Ulrich, R.G.; Zvirbliene, A.; Kucinskaite-Kodze, I. Generation in yeast and antigenic characterization of hepatitis E virus capsid protein virus-like particles. Appl. Microbiol. Biotechnol. 2018, 102, 185–198. [Google Scholar] [CrossRef]
- Su, C.X.; Gu, M.R.; Zhang, P.; Jin, Z.J.; Meng, F.H.; Chen, E.J.; Yang, Z.; Liu, Y.; Wang, Y.C. [Expression of ORF2 protein of HEV genotype IV in Hansenula polymorpha]. Sheng Wu Gong Cheng Xue Bao 2007, 23, 73–78. [Google Scholar]
- Burden, C.S.; Jin, J.; Podgornik, A.; Bracewell, D.G. A monolith purification process for virus-like particles from yeast homogenate. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2012, 880, 82–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, J.; VanDusen, W.J.; Kolodin, D.G.; O’Keefe, D.O.; Herber, W.K.; George, H.A. Continuous culture study of the expression of hepatitis B surface antigen and its self-assembly into virus-like particles in Saccharomyces cerevisiae. Biotechnol. Bioeng. 1996, 49, 578–586. [Google Scholar] [CrossRef]
- Hadiji-Abbes, N.; Martin, M.; Benzina, W.; Karray-Hakim, H.; Gergely, C.; Gargouri, A.; Mokdad-Gargouri, R. Extraction and purification of hepatitis B virus-like M particles from a recombinant Saccharomyces cerevisiae strain using alumina powder. J. Virol. Methods 2013, 187, 132–137. [Google Scholar] [CrossRef]
- Hadiji-Abbes, N.; Mihoubi, W.; Martin, M.; Karakasyan-Dia, C.; Frikha, F.; Gergely, C.; Jouenne, T.; Gargouri, A.; Mokdad-Gargouri, R. Characterization of C69R variant HBsAg: Effect on binding to anti-HBs and the structure of virus-like particles. Arch. Virol. 2015, 160, 2427–2433. [Google Scholar] [CrossRef]
- Lunsdorf, H.; Gurramkonda, C.; Adnan, A.; Khanna, N.; Rinas, U. Virus-like particle production with yeast: Ultrastructural and immunocytochemical insights into Pichia pastoris producing high levels of the hepatitis B surface antigen. Microb. Cell Fact. 2011, 10, 48. [Google Scholar] [CrossRef] [Green Version]
- Hosseini, S.N.; Javidanbardan, A.; Alizadeh Salim, B.S.; Khatami, M. Large-scale purification of recombinant hepatitis B surface antigen from Pichia pastoris with non-affinity chromatographic methods as a substitute to immunoaffinity chromatography. Prep. Biochem. Biotechnol. 2018, 48, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Gurramkonda, C.; Adnan, A.; Gabel, T.; Lunsdorf, H.; Ross, A.; Nemani, S.K.; Swaminathan, S.; Khanna, N.; Rinas, U. Simple high-cell density fed-batch technique for high-level recombinant protein production with Pichia pastoris: Application to intracellular production of Hepatitis B surface antigen. Microb. Cell Fact. 2009, 8, 13. [Google Scholar] [CrossRef] [Green Version]
- Stephen, S.L.; Beales, L.; Peyret, H.; Roe, A.; Stonehouse, N.J.; Rowlands, D.J. Recombinant Expression of Tandem-HBc Virus-Like Particles (VLPs). Methods Mol. Biol. 2018, 1776, 97–123. [Google Scholar] [CrossRef] [PubMed]
- Rolland, D.; Gauthier, M.; Dugua, J.M.; Fournier, C.; Delpech, L.; Watelet, B.; Letourneur, O.; Arnaud, M.; Jolivet, M. Purification of recombinant HBc antigen expressed in Escherichia coli and Pichia pastoris: Comparison of size-exclusion chromatography and ultracentrifugation. J. Chromatogr. B Biomed. Sci. Appl. 2001, 753, 51–65. [Google Scholar] [CrossRef]
- Pechelyulko, A.; Andreeva-Kovalevskaya, Z.; Dmitriev, D.; Lavrov, V.; Massino, Y.; Nagel, A.; Segal, O.; Sokolova, O.S.; Solonin, A.; Tarakanova, Y.; et al. A simple method to purify recombinant HCV core protein expressed in Pichia pastoris for obtaining virus-like particles and producing monoclonal antibodies. Protein Expr. Purif. 2021, 183, 105864. [Google Scholar] [CrossRef]
- Acosta-Rivero, N.; Aguilar, J.C.; Musacchio, A.; Falcon, V.; Vina, A.; de la Rosa, M.C.; Morales, J. Characterization of the HCV core virus-like particles produced in the methylotrophic yeast Pichia pastoris. Biochem. Biophys. Res. Commun. 2001, 287, 122–125. [Google Scholar] [CrossRef]
- Fazlalipour, M.; Keyvani, H.; Monavari, S.H.; Mollaie, H.R. Expression, Purification and Immunogenic Description of a Hepatitis C Virus Recombinant CoreE1E2 Protein Expressed by Yeast Pichia pastoris. Jundishapur J. Microbiol. 2015, 8, e17157. [Google Scholar] [CrossRef] [Green Version]
- Acosta-Rivero, N.; Rodriguez, A.; Musacchio, A.; Falcon, V.; Suarez, V.M.; Martinez, G.; Guerra, I.; Paz-Lago, D.; Morera, Y.; de la Rosa, M.C.; et al. In vitro assembly into virus-like particles is an intrinsic quality of Pichia pastoris derived HCV core protein. Biochem. Biophys. Res. Commun. 2004, 325, 68–74. [Google Scholar] [CrossRef]
- Acosta-Rivero, N.; Alvarez-Obregon, J.C.; Musacchio, A.; Falcon, V.; Duenas-Carrera, S.; Marante, J.; Menendez, I.; Morales, J. In vitro self-assembled HCV core virus-like particles induce a strong antibody immune response in sheep. Biochem. Biophys. Res. Commun. 2002, 290, 300–304. [Google Scholar] [CrossRef]
- Majeau, N.; Gagne, V.; Bolduc, M.; Leclerc, D. Signal peptide peptidase promotes the formation of hepatitis C virus non-enveloped particles and is captured on the viral membrane during assembly. J. Gen. Virol. 2005, 86, 3055–3064. [Google Scholar] [CrossRef]
- Freivalds, J.; Dislers, A.; Ose, V.; Pumpens, P.; Tars, K.; Kazaks, A. Highly efficient production of phosphorylated hepatitis B core particles in yeast Pichia pastoris. Protein Expr. Purif. 2011, 75, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Cho, S.Y.; Park, M.H. Comparison of the size distributions and immunogenicity of human papillomavirus type 16 L1 virus-like particles produced in insect and yeast cells. Arch. Pharm. Res. 2018, 41, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Jin, Y. The concentration of carbon source in the medium affects the quality of virus-like particles of human papillomavirus type 16 produced in Saccharomyces cerevisiae. PLoS ONE 2014, 9, e94467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, R.S.; Brown, D.R.; Bryan, J.T.; Cook, J.C.; George, H.A.; Hofmann, K.J.; Hurni, W.M.; Joyce, J.G.; Lehman, E.D.; Markus, H.Z.; et al. Human papillomavirus type 11 (HPV-11) neutralizing antibodies in the serum and genital mucosal secretions of African green monkeys immunized with HPV-11 virus-like particles expressed in yeast. J. Infect. Dis. 1997, 176, 1141–1145. [Google Scholar] [CrossRef] [PubMed]
- Cook, J.C.; Joyce, J.G.; George, H.A.; Schultz, L.D.; Hurni, W.M.; Jansen, K.U.; Hepler, R.W.; Ip, C.; Lowe, R.S.; Keller, P.M.; et al. Purification of virus-like particles of recombinant human papillomavirus type 11 major capsid protein L1 from Saccharomyces cerevisiae. Protein Expr. Purif. 1999, 17, 477–484. [Google Scholar] [CrossRef]
- Mach, H.; Volkin, D.B.; Troutman, R.D.; Wang, B.; Luo, Z.; Jansen, K.U.; Shi, L. Disassembly and reassembly of yeast-derived recombinant human papillomavirus virus-like particles (HPV VLPs). J. Pharm. Sci. 2006, 95, 2195–2206. [Google Scholar] [CrossRef]
- Kwag, H.L.; Kim, H.J.; Chang, D.Y. The production and immunogenicity of human papillomavirus type 58 virus-like particles produced in Saccharomyces cerevisiae. J. Microbiol. 2012, 50, 813–820. [Google Scholar] [CrossRef]
- Gupta, G.; Glueck, R.; Rishi, N. Physicochemical characterization and immunological properties of Pichia pastoris based HPV16L1 and 18L1 virus like particles. Biologicals 2017, 46, 11–22. [Google Scholar] [CrossRef]
- Jiang, Z.; Tong, G.; Cai, B.; Xu, Y.; Lou, J. Purification and immunogenicity study of human papillomavirus 58 virus-like particles expressed in Pichia pastoris. Protein Expr. Purif. 2011, 80, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Hanumantha Rao, N.; Baji Babu, P.; Rajendra, L.; Sriraman, R.; Pang, Y.Y.; Schiller, J.T.; Srinivasan, V.A. Expression of codon optimized major capsid protein (L1) of human papillomavirus type 16 and 18 in Pichia pastoris; purification and characterization of the virus-like particles. Vaccine 2011, 29, 7326–7334. [Google Scholar] [CrossRef] [Green Version]
- Coimbra, E.C.; Gomes, F.B.; Campos, J.F.; D’Arc, M.; Carvalho, J.C.; Mariz, F.C.; Jesus, A.L.; Stocco, R.C.; Becak, W.; Freitas, A.C. Production of L1 protein from different types of HPV in Pichia pastoris using an integrative vector. Braz. J. Med. Biol. Res. 2011, 44, 1209–1214. [Google Scholar] [CrossRef] [Green Version]
- Dewi, K.S.; Chairunnisa, S.; Swasthikawati, S.; Yuliawati; Agustiyanti, D.F.; Mustopa, A.Z.; Kusharyoto, W.; Ningrum, R.A. Production of codon-optimized Human papillomavirus type 52 L1 virus-like particles in Pichia pastoris BG10 expression system. Prep. Biochem. Biotechnol. 2023, 53, 148–156. [Google Scholar] [CrossRef]
- Zhao, H.; Li, H.Y.; Han, J.F.; Deng, Y.Q.; Li, Y.X.; Zhu, S.Y.; He, Y.L.; Qin, E.D.; Chen, R.; Qin, C.F. Virus-like particles produced in Saccharomyces cerevisiae elicit protective immunity against Coxsackievirus A16 in mice. Appl. Microbiol. Biotechnol. 2013, 97, 10445–10452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, Q.; Ku, Z.; Hu, Y.; Ye, X.; Zhang, Y.; Huang, Z. Coxsackievirus A16-like particles produced in Pichia pastoris elicit high-titer neutralizing antibodies and confer protection against lethal viral challenge in mice. Antiviral. Res. 2016, 129, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, C.; Liu, Q.; Gong, S.; Geng, L.; Huang, Z. A virus-like particle vaccine protects mice against coxsackievirus A10 lethal infection. Antiviral. Res. 2018, 152, 124–130. [Google Scholar] [CrossRef]
- Wang, X.W.; Sheng, W.; Zeng, Y. [Formation and identification of virus-like particles of poliovirus type I]. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 2013, 27, 373–375. [Google Scholar]
- Sherry, L.; Grehan, K.; Snowden, J.S.; Knight, M.L.; Adeyemi, O.O.; Rowlands, D.J.; Stonehouse, N.J. Comparative Molecular Biology Approaches for the Production of Poliovirus Virus-like Particles Using Pichia pastoris. mSphere 2020, 5, e00838-19. [Google Scholar] [CrossRef] [Green Version]
- Sherry, L.; Grehan, K.; Swanson, J.J.; Bahar, M.W.; Porta, C.; Fry, E.E.; Stuart, D.I.; Rowlands, D.J.; Stonehouse, N.J. Production and Characterisation of Stabilised PV-3 Virus-like Particles Using Pichia pastoris. Viruses 2022, 14, 2159. [Google Scholar] [CrossRef]
- Choi, Y.R.; Kim, H.J.; Lee, J.Y.; Kang, H.A. Chromatographically-purified capsid proteins of red-spotted grouper nervous necrosis virus expressed in Saccharomyces cerevisiae form virus-like particles. Protein Expr. Purif. 2013, 89, 162–168. [Google Scholar] [CrossRef]
- Barsoe, S.; Toffan, A.; Pascoli, F.; Stratmann, A.; Pretto, T.; Marsella, A.; Er-Rafik, M.; Vendramin, N.; Olesen, N.J.; Sepulveda, D.; et al. Long-Term Protection and Serologic Response of European Sea Bass Vaccinated with a Betanodavirus Virus-Like Particle Produced in Pichia pastoris. Vaccines 2021, 2, 447. [Google Scholar] [CrossRef]
- Lowin, T.; Raab, U.; Schroeder, J.; Franssila, R.; Modrow, S. Parvovirus B19 VP2-proteins produced in Saccharomyces cerevisiae: Comparison with VP2-particles produced by baculovirus-derived vectors. J. Vet. Med. B Infect. Dis. Vet. Public Health 2005, 52, 348–352. [Google Scholar] [CrossRef]
- Penkert, R.R.; Young, N.S.; Surman, S.L.; Sealy, R.E.; Rosch, J.; Dormitzer, P.R.; Settembre, E.C.; Chandramouli, S.; Wong, S.; Hankins, J.S.; et al. Saccharomyces cerevisiae-derived virus-like particle parvovirus B19 vaccine elicits binding and neutralizing antibodies in a mouse model for sickle cell disease. Vaccine 2017, 35, 3615–3620. [Google Scholar] [CrossRef] [PubMed]
- Chandramouli, S.; Medina-Selby, A.; Coit, D.; Schaefer, M.; Spencer, T.; Brito, L.A.; Zhang, P.; Otten, G.; Mandl, C.W.; Mason, P.W.; et al. Generation of a parvovirus B19 vaccine candidate. Vaccine 2013, 31, 3872–3878. [Google Scholar] [CrossRef] [PubMed]
- Freivalds, J.; Dislers, A.; Ose, V.; Skrastina, D.; Cielens, I.; Pumpens, P.; Sasnauskas, K.; Kazaks, A. Assembly of bacteriophage Qbeta virus-like particles in yeast Saccharomyces cerevisiae and Pichia pastoris. J. Biotechnol. 2006, 123, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Bredell, H.; Smith, J.J.; Prins, W.A.; Gorgens, J.F.; van Zyl, W.H. Expression of rotavirus VP6 protein: A comparison amongst Escherichia coli, Pichia pastoris and Hansenula polymorpha. FEMS Yeast Res. 2016, 16, fow001. [Google Scholar] [CrossRef] [Green Version]
- Firdaus, M.E.R.; Mustopa, A.Z.; Ekawati, N.; Chairunnisa, S.; Arifah, R.K.; Hertati, A.; Irawan, S.; Prastyowati, A.; Kusumawati, A.; Nurfatwa, M. Optimization, characterization, comparison of self-assembly VLP of capsid protein L1 in yeast and reverse vaccinology design against human papillomavirus type 52. J. Genet. Eng. Biotechnol. 2023, 21, 68. [Google Scholar] [CrossRef]
- Weber, J.; Cheinsong-Popov, R.; Callow, D.; Adams, S.; Patou, G.; Hodgkin, K.; Martin, S.; Gotch, F.; Kingsman, A. Immunogenicity of the yeast recombinant p17/p24:Ty virus-like particles (p24-VLP) in healthy volunteers. Vaccine 1995, 13, 831–834. [Google Scholar] [CrossRef]
- Martins, S.A.; Santos, J.; Silva, R.D.M.; Rosa, C.; Cabo Verde, S.; Correia, J.D.G.; Melo, R. How promising are HIV-1-based virus-like particles for medical applications. Front. Cell Infect. Microbiol. 2022, 12, 997875. [Google Scholar] [CrossRef] [PubMed]



| Bacteria | Yeast | Mammalian Cells | Insect Cells | Plant | |
|---|---|---|---|---|---|
| Production cost | Low | Low | High | High | Moderate |
| Type of growth media | Simple | Simple | Complex | Complex | Simple |
| Growth speed | Very high | High | Moderate | Moderate | Very slow |
| Production time | Low | Low | High | High | Very high |
| Expression yield | High | High | Low | Very high | Low |
| Secretion | No | Yes | Yes | Yes | Not applicable |
| Enveloped/ non-enveloped VLPs | Non-enveloped | Non-enveloped (enveloped only in some cases) | Enveloped/ Non-enveloped | Enveloped/ non-enveloped | Not applicable |
| Scalability | Easy | Easy | Very difficult | Very difficult | Difficult |
| Safety | Safe (possibility of endotoxins) | Very safe | Safe (possible contamination with human viruses) | Very safe | Very safe |
| Genetic manipulation | Very easy | Easy | Difficult | Difficult | Difficult |
| Glycosylation of proteins | Very different from humans | Different from humans (can be optimized) | Reproducing human glycosylation | Simpler N-glycosylation pattern compared to mammalian cells | Human-like glycosylation |
| PTMs | Lack of PTM system | Lack of complex PTM pathway | Complex PTM pathway | Complex PTM pathway | Complex PTM pathway |
| Family | Virus Species |
|---|---|
| Hepadnaviridae | Hepatitis B Virus, Hepatitis E Virus, |
| Flaviviridae | Hepatitis C Virus, Japanese Encephalitis Virus, Bovine Viral diarrhea virus, Tick-borne encephalitis virus, Zika virus |
| Papillomaviridae | Human Papilloma Virus 1, 6, 11, 16, 52, 58, Cottontail rabbit Papillomavirus, bovine papilloma virus 1,2, 4 |
| Picornaviridae | Enterovirus D68, Enterovirus 71 and Coxsackievirus A6, A10 and A16, Poliovirus type I |
| Nodaviridae | Redspotted grouper nervous necrosis virus, Nervous necrosis virus |
| Parvoviridae | Porcine parvovirus, Adeno associated virus, Human Parvovirus 4, B19, Human bocaviruses |
| Paramyxoviridae | Sendai virus, Tioman virus, Human parainfluenza virus 2 and 4, Menangle virus, Nipah virus |
| Circoviridae | Porcine circovirus |
| Retroviridae | HIV |
| Kolmioviridae | Hepatitis Delta Virus |
| Fiersviridae | Cacteriophage Qbeta virus |
| Sedoreoviridae | Rotavirus |
| Potyviridae | Johnsongrass mosaic virus |
| Polyomaviridae | Human polyoma virus, hamster polyoma virus, bird polyomavirus, Goose hemorrhagic, Polyomavirus |
| Caliciviridae | Norovirus, Rabbit hemorrhagic disease virus |
| Bromoviridae | Cowpea chlorotic mottle virus |
| Birnaviridae | Infectious bursal disease virus |
| Secoviridae | Grapevine fanleaf virus |
| Togaviridae | Chikungunya virus |
| Iridoviridae | Chinese Giant Salamander iridovirus |
| Expression Promoters | |||
|---|---|---|---|
| Type of Vectors | Saccharomyces cerevisiae | Pichia pastoris | Hansenula polymorpha |
| Replicative, Inducible | GAL1, GAL10, GAL7, GAL1/10, hybrid GAL10-PYK11, hybrid GAL10-CYC1 | ||
| Replicative, Constitutive | GAD, ADH/GAPDH, PGK1, TEF1, ADC1 | ||
| Integrative, Inducible | AOX1 | MOX, FMD | |
| Integrative, Constitutive | GAP, PGK1 | ||
| Type of Antigens | |||
|---|---|---|---|
| Virus | S. cerevisiae | P. pastoris | H. polymorpha |
| Hepatitis E virus | -HEV-3: full length capsid protein (ORF2, aa 1–660); 5′ and 3′ terminally truncated capsid protein (ORF2, aa 112–608) -Rat HEV: full length capsid protein (ORF2, aa 1–645); 5′ and 3′ terminally truncated capsid protein (ORF2, aa 112–608) [121] | ORF2, aa 112–608 [90] | ORF2, aa 112–607 (HEV genotype IV) [122] |
| Hepatitis B virus | -Surface protein (HBsAg) [123,124] -Viral M [preS2 + S] [125] -Wild-type and mutant C69R S genes [126] | -Surface protein (HBsAg) [115,127,128,129] -Core protein (HBc) [130,131] | -26 amino acids of PreS2 C-terminus (aa 120–145) [114] |
| Hepatitis C virus | -Core protein (HBc) [132,133] -CoreE1E2 protein, which consists of Core (269 nt–841nt) E1 (842 nt–1417nt), and E2 (1418 nt–2506nt) [134,135,136,137,138] | E2 Core Domain [111] | |
| HPV 16, 11, 6, 58 | Human papillomavirus major capsid protein L1 [139,140,141,142,143,144] | -Human papillomavirus major capsid protein L1 [85,86,145,146,147,148,149] | |
| Coxsackievirus | Capsid protein precursor P1 and the protease 3CD of Coxsackievirus A16 [150] | -Capsid protein precursor P1 and the protease 3CD Coxsackievirus A16 [74,151] -Capsid protein precursor P1 and the protease 3CD of Coxsackievirus A10 [152] | |
| Poliovirus | Capsid precursor protein P1 and the protease 3CD [153] | Capsid precursor protein P1 and the protease 3CD [154,155] | |
| Red-spotted grouper nervous necrosis virus | Capsid protein [156] | Capsid protein [157] | |
| B19 parvovirus | Minor-capsid protein VP1 and the major-capsid protein VP2 [158,159,160] | Minor-capsid protein VP1 and the major-capsid protein VP2 [117] | |
| Bacteriophage Qβ | Coat protein (CP) [161] | Coat protein (CP) [161] | |
| Phage PP7 φCb5; Phages fr Phage SP | Coat proteins [161] | Coat proteins [161] | |
| Rotavirus | Capsid protein VP6 [162] | Capsid protein VP6 [162] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brachelente, S.; Galli, A.; Cervelli, T. Yeast and Virus-like Particles: A Perfect or Imperfect Couple? Appl. Microbiol. 2023, 3, 805-825. https://doi.org/10.3390/applmicrobiol3030056
Brachelente S, Galli A, Cervelli T. Yeast and Virus-like Particles: A Perfect or Imperfect Couple? Applied Microbiology. 2023; 3(3):805-825. https://doi.org/10.3390/applmicrobiol3030056
Chicago/Turabian StyleBrachelente, Sara, Alvaro Galli, and Tiziana Cervelli. 2023. "Yeast and Virus-like Particles: A Perfect or Imperfect Couple?" Applied Microbiology 3, no. 3: 805-825. https://doi.org/10.3390/applmicrobiol3030056
APA StyleBrachelente, S., Galli, A., & Cervelli, T. (2023). Yeast and Virus-like Particles: A Perfect or Imperfect Couple? Applied Microbiology, 3(3), 805-825. https://doi.org/10.3390/applmicrobiol3030056







