Conversion of Unsaturated Short- to Medium-Chain Fatty Acids by Unspecific Peroxygenases (UPOs)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Enzyme Preparations
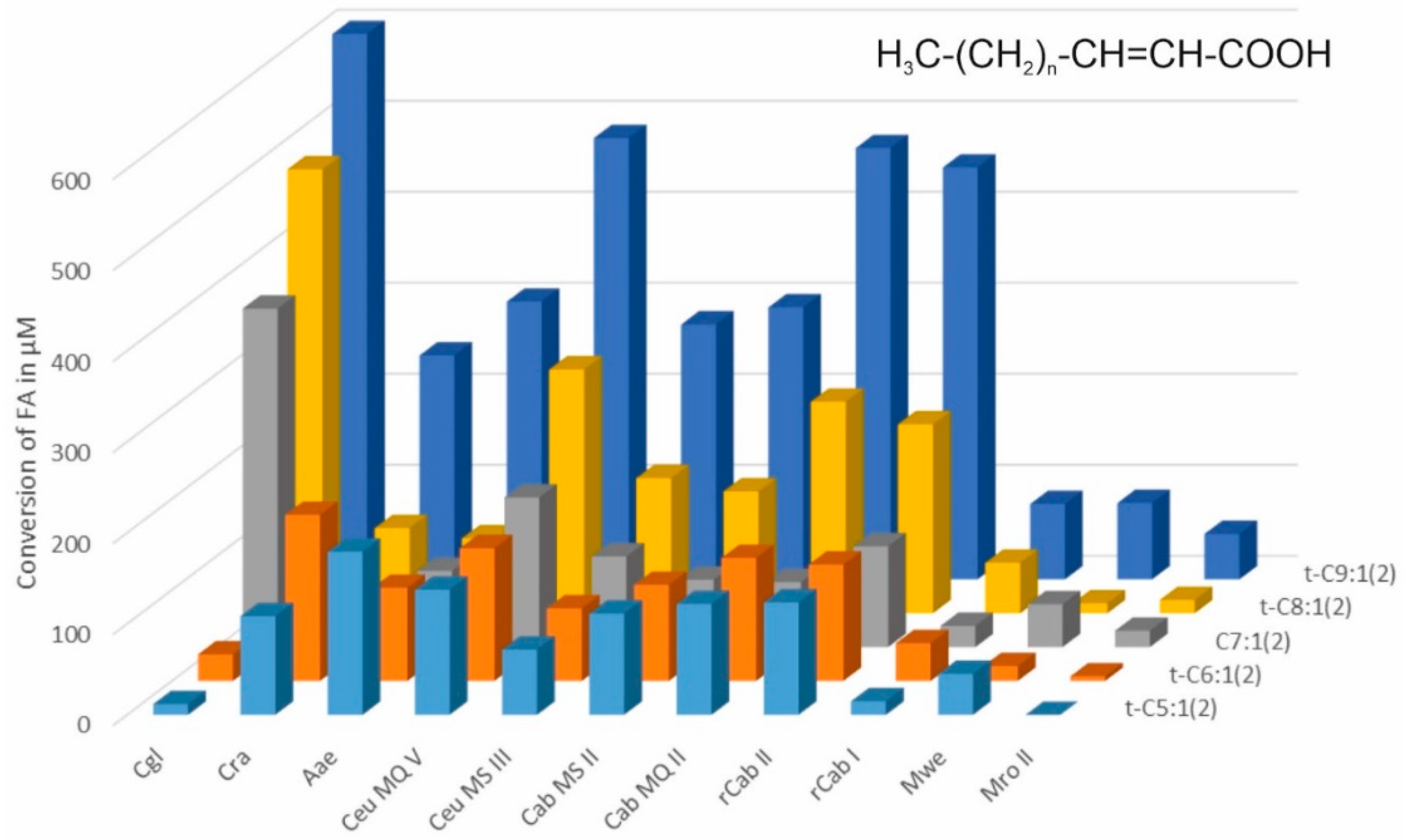
2.2. Conversion of Medium Chained Fatty Acids (MCFAs)
2.3. Preparation of Epoxides via the Prilezhaev Reaction
2.4. Analyses
2.5. Chemicals
3. Results and Discussion
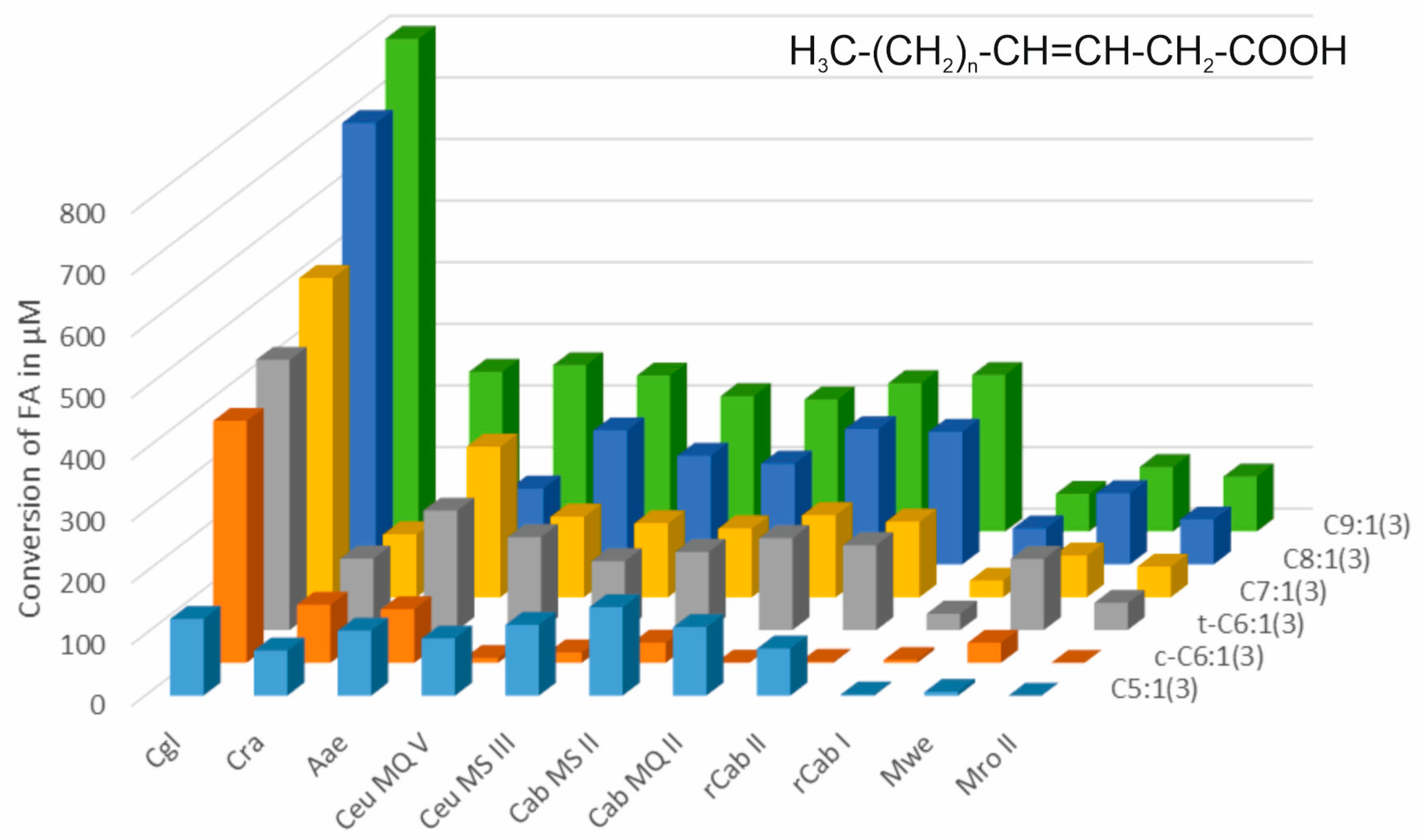
3.1. Terminally Unsaturated FAs
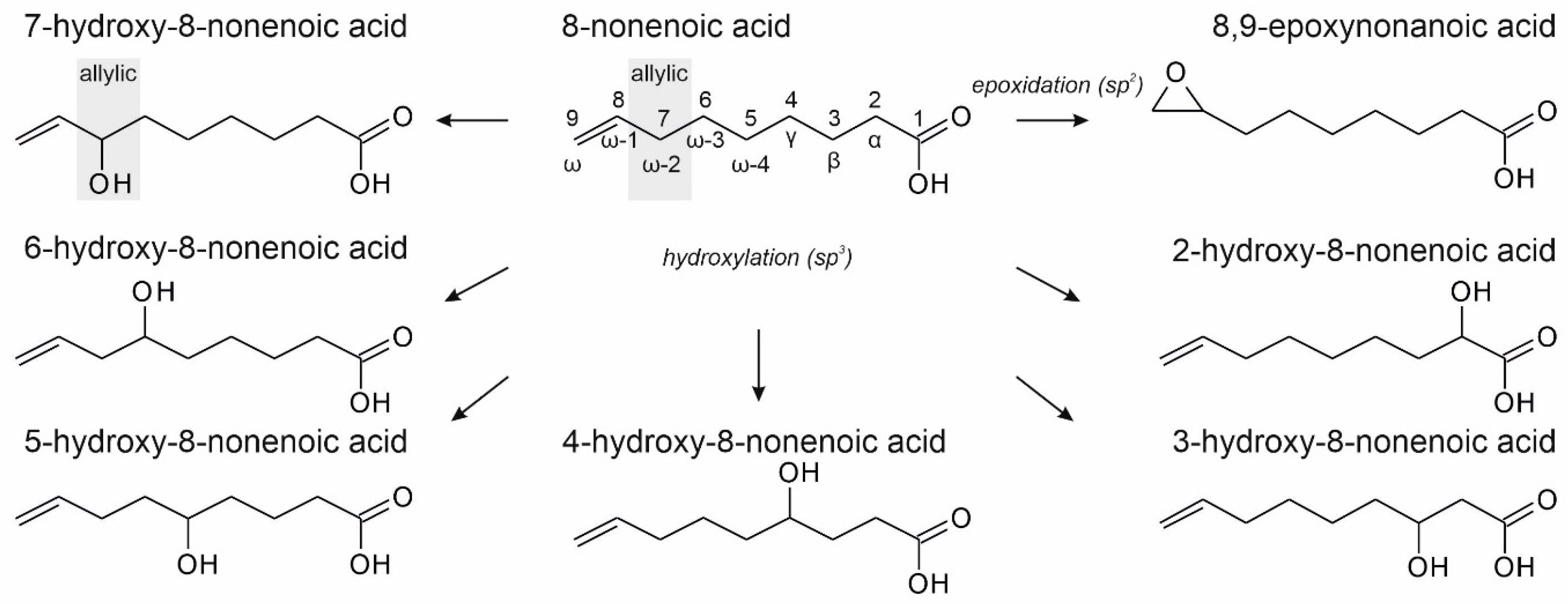
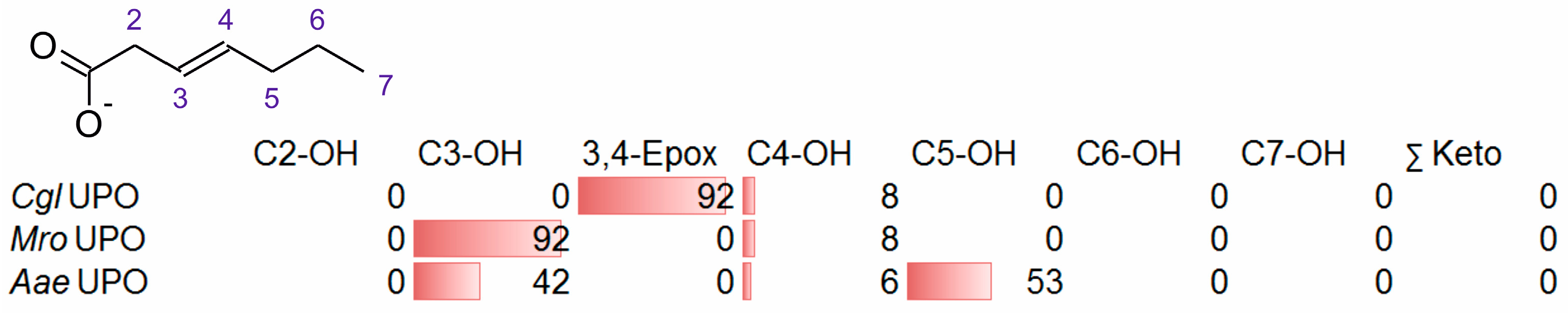
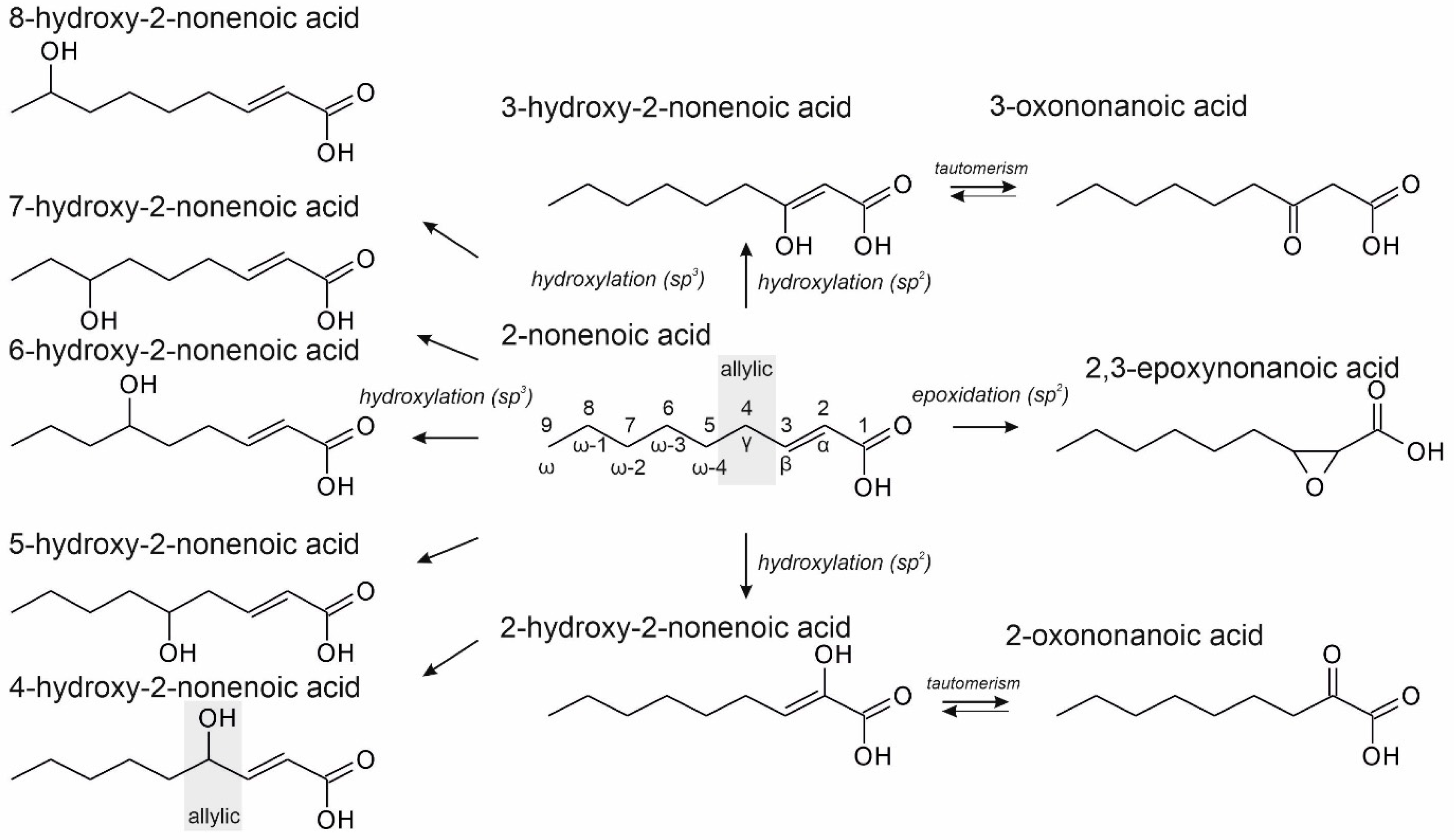
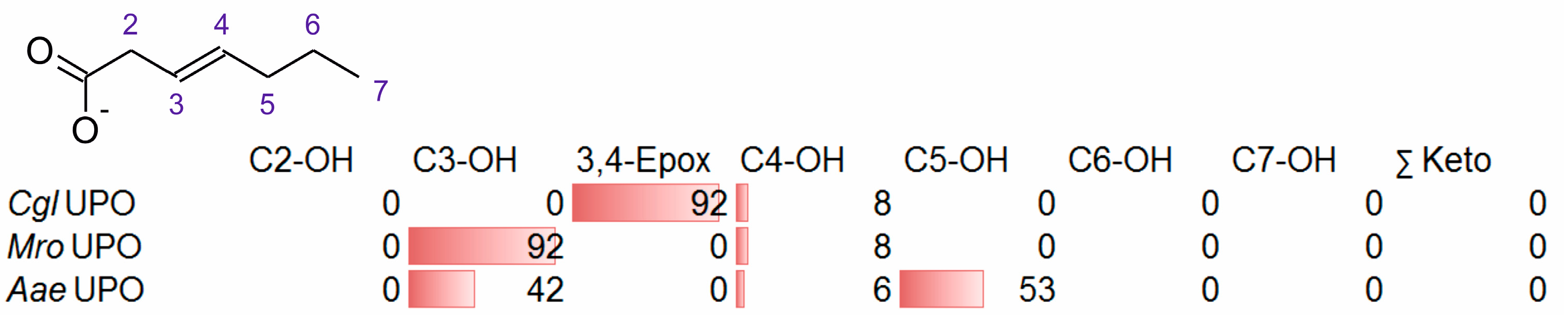
3.2. Unsaturated FAs with the Double Bond at the β,γ-Position
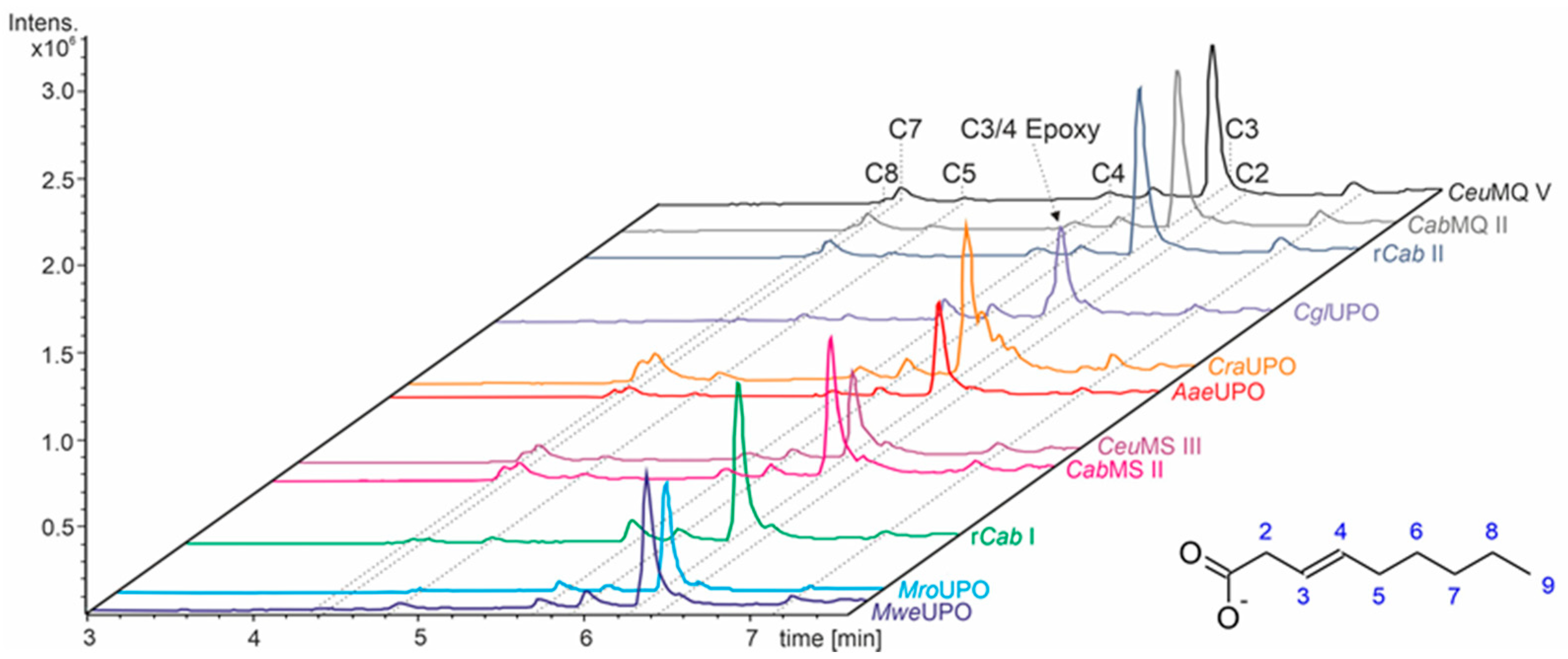
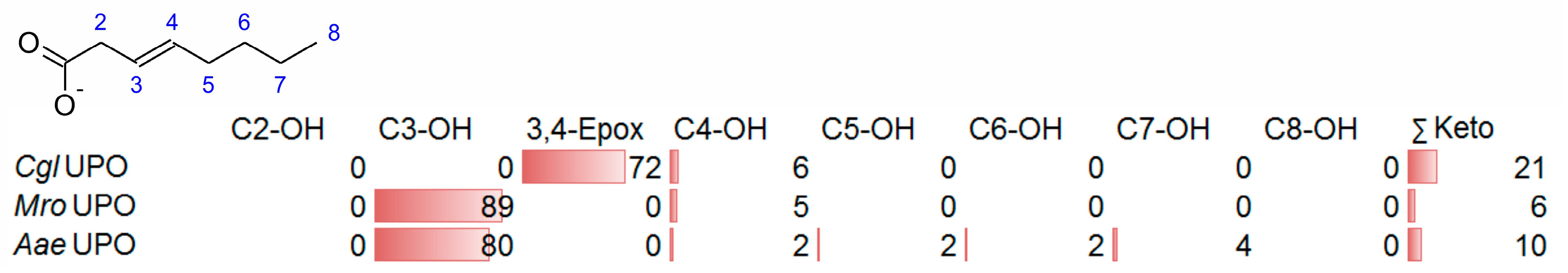
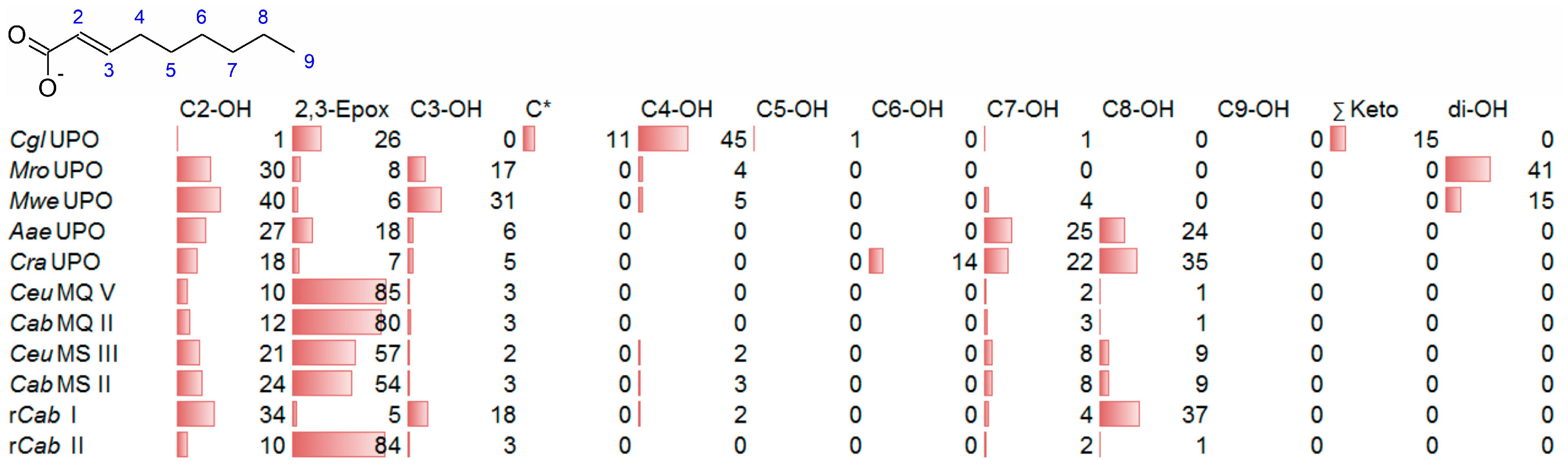
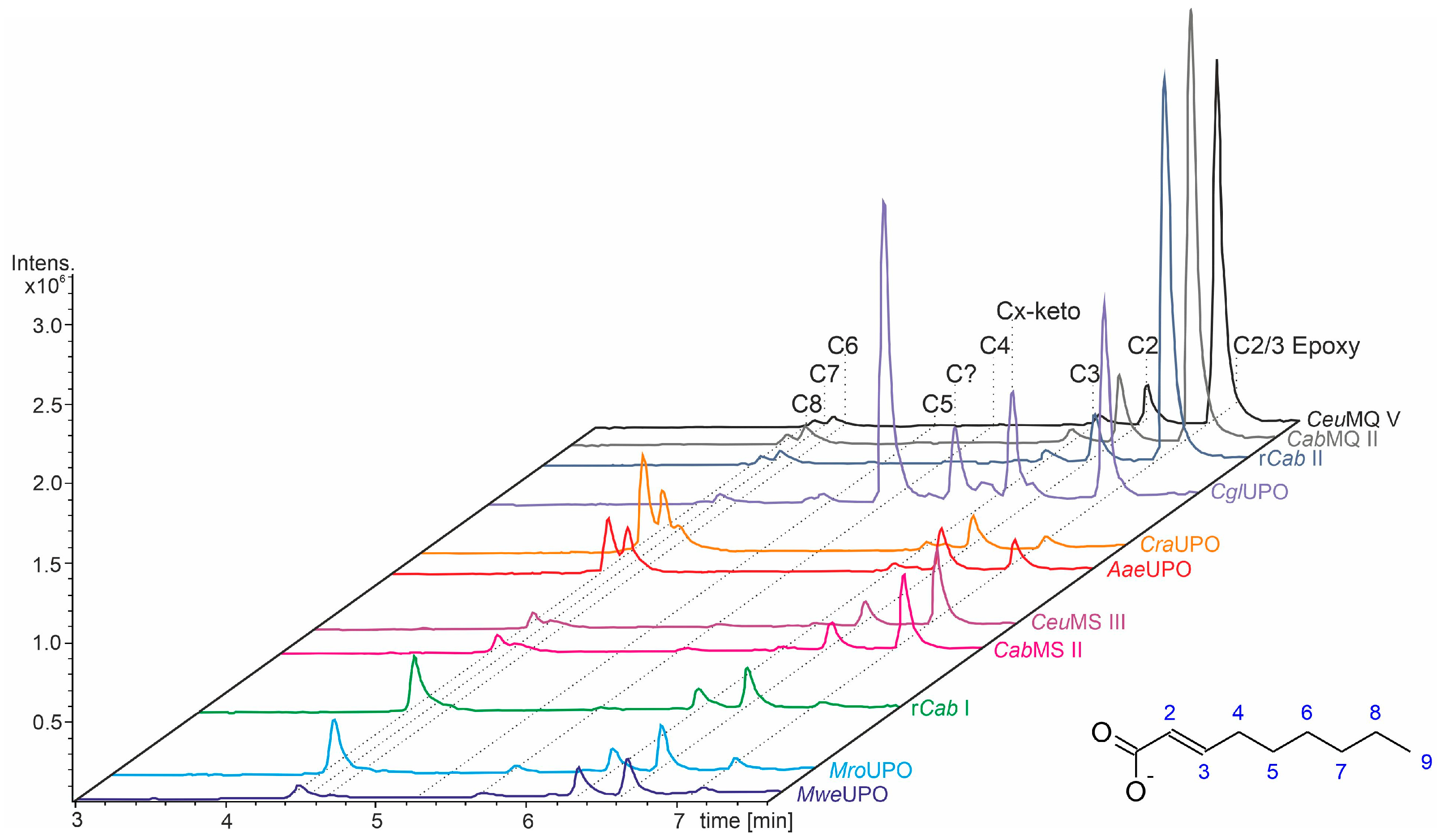
3.3. Unsaturated FAs with the Double Bond at α,β-Position
- UPOs mainly forming the epoxide between C2 and C3 (α,β-position) with yields of about 80% and producing minor hydroxylation products at different positions (C2, C3, C7 and C8). This group included wild-type and recombinant UPOs deriving from dark-spored agaric Basidiomycota (genera Candolleomyces, Psathyrellaceae), i.e., CeuMQ V, CabMQ II and rCab II.
- One UPO forming the epoxide with yields around 25% but favoring hydroxylation at the C4 position (about 45%); furthermore, the enzyme formed a so far unidentified oxygenation product (C?) and a keto-product (4-oxo-trans-2-nonenoic acid, which was probably deriving from a second hydroxylation at the C4 position (second hydroxylation at the C4 position gives a gem-diol that is in equilibrium with the chemically favored oxo-FA, 14%). CglUPO was the only enzyme in this group.
- UPOs favoring subterminal hydroxylation at C8 (26–36%), C7 (25–29%), and C6 (14%) and additionally, forming the epoxide (5–18%) and other hydroxylation products at C2/3 (about 20% and 3%, respectively). This group included UPOs from dark-spored agaric Basidiomycota, AaeUPO and CraUPO.
- Enzymes yielding the epoxide (about 55%) to some extent as well as hydroxylating subterminally and at the C2 position (about 20%). Two wild-type UPOs, CeuMS III and CabMS II (both from the genus Candolleomyces).
- A UPO hydroxylating at C2 (34%), C3 (18%) and favorably at the C8 (ω-1; 37%) position. One recombinant enzyme, rCab I (from the genus Candolleomyces).
- Two enzymes favorably hydroxylating close to the carboxylic group at C2 (28–35%), C3 (25–28%), and C4 (4–5%) positions, and yielding the epoxide to minor extent; in addition, they catalyze a second hydroxylation yielding dihydroxylated products. This group includes UPOs from bright-spored agaric Basidiomycota (genus Marasmius), MroUPO and MweUPO.
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hofrichter, M.; Kellner, H.; Herzog, R.; Karich, A.; Kiebist, J.; Scheibner, K.; Ullrich, R. Peroxide-mediated oxygenation of organic compounds by fungal peroxygenases. Antioxidants 2022, 11, 163. [Google Scholar] [CrossRef]
- Hofrichter, M.; Kellner, H.; Herzog, R.; Karich, A.; Liers, C.; Scheibner, K.; Kimani, V.W.; Ullrich, R. Fungal peroxygenases: A phylogenetically old superfamily of heme enzymes with promiscuity for oxygen transfer reactions. In Grand Challenges in Fungal Biotechnology; Nevalainen, H., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 369–403. [Google Scholar]
- Ullrich, R.; Karich, A.; Hofrichter, M. Fungal peroxygenases—A versatile tool for biocatalysis. In Encyclopedia of Mycology; Zaragoza, Ó., Casadevall, A., Eds.; Elsevier: Oxford, UK, 2021; pp. 260–280. [Google Scholar]
- Munro, A.W.; Girvan, H.M.; Mason, A.E.; Dunford, A.J.; McLean, K.J. What makes a P450 tick? Trends Biochem. Sci. 2013, 38, 140–150. [Google Scholar] [CrossRef]
- Ullrich, R.; Nüske, J.; Scheibner, K.; Spantzel, J.; Hofrichter, M. Novel haloperoxidase from the agaric basidiomycete Agrocybe aegerita oxidizes aryl alcohols and aldehydes. Appl. Environ. Microbiol. 2004, 70, 4575–4581. [Google Scholar] [CrossRef] [Green Version]
- Karich, A.; Ullrich, R.; Scheibner, K.; Hofrichter, M. Fungal unspecific peroxygenases oxidize the majority of organic EPA Priority Pollutants. Front. Microbiol. 2017, 8, 1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karich, A.; Scheibner, K.; Ullrich, R.; Hofrichter, M. Exploring the catalase activity of unspecific peroxygenases and the mechanism of peroxide-dependent heme destruction. J. Mol. Catal. B Enzym. 2016, 134, 238–246. [Google Scholar] [CrossRef]
- Gutierrez, A.; Babot, E.D.; Ullrich, R.; Hofrichter, M.; Martinez, A.T.; del Rio, J.C. Regioselective oxygenation of fatty acids, fatty alcohols and other aliphatic compounds by a basidiomycete heme-thiolate peroxidase. Arch. Biochem. Biophys. 2011, 514, 33–43. [Google Scholar] [CrossRef] [Green Version]
- Olmedo, A.; Aranda, C.; del Río, J.C.; Kiebist, J.; Scheibner, K.; Martínez, A.T.; Gutiérrez, A. From alkanes to carboxylic acids: Terminal oxygenation by a fungal peroxygenase. Angew. Chem. Int. Ed. 2016, 55, 12248–12251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carro, J.; González-Benjumea, A.; Fernández-Fueyo, E.; Aranda, C.; Guallar, V.; Gutiérrez, A.; Martínez, A.T. Modulating fatty acid epoxidation vs hydroxylation in a fungal peroxygenase. ACS Catal. 2019, 9, 6234–6242. [Google Scholar] [CrossRef] [Green Version]
- Aranda, C.; Olmedo, A.; Kiebist, J.; Scheibner, K.; del Río, J.C.; Martínez, A.T.; Gutiérrez, A. Selective epoxidation of fatty acids and fatty acid methyl esters by fungal peroxygenases. ChemCatChem 2018, 10, 3964–3968. [Google Scholar] [CrossRef] [Green Version]
- González-Benjumea, A.; Carro, J.; Renau-Mínguez, C.; Linde, D.; Fernández-Fueyo, E.; Gutiérrez, A.; Martínez, A.T. Fatty acid epoxidation by Collariella virescens peroxygenase and heme-channel variants. Catal. Sci. Technol. 2020, 10, 717–725. [Google Scholar] [CrossRef] [Green Version]
- Municoy, M.; González-Benjumea, A.; Carro, J.; Aranda, C.; Linde, D.; Renau-Mínguez, C.; Ullrich, R.; Hofrichter, M.; Guallar, V.; Gutiérrez, A.; et al. Fatty-acid oxygenation by fungal peroxygenases: From computational simulations to preparative regio- and stereoselective epoxidation. ACS Catal. 2020, 10, 13584–13595. [Google Scholar] [CrossRef]
- Olmedo, A.; Rio, J.C.D.; Kiebist, J.; Ullrich, R.; Hofrichter, M.; Scheibner, K.; Martinez, A.T.; Gutierrez, A. Fatty acid chain shortening by a fungal peroxygenase. Chemistry 2017, 23, 16985–16989. [Google Scholar] [CrossRef] [Green Version]
- Olmedo, A.; Ullrich, R.; Hofrichter, M.; del Río, J.C.; Martínez, Á.T.; Gutiérrez, A. Novel fatty acid chain-shortening by fungal peroxygenases yielding 2C-shorter dicarboxylic acids. Antioxidants 2022, 11, 744. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Peng, W.; Li, Z.; You, C.; Zhao, Y.; Tang, D.; Wang, B.; Li, S. Unexpected reactions of α,β-unsaturated fatty acids provide insight into the mechanisms of CYP152 peroxygenases. Angew. Chem. Int. Ed. 2021, 60, 24694–24701. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Lee, J.-H.; Younes, S.H.H.; Tonin, F.; Hagedoorn, P.-L.; Pichler, H.; Baeg, Y.; Park, J.-B.; Kourist, R.; Hollmann, F. Photobiocatalytic synthesis of chiral secondary fatty alcohols from renewable unsaturated fatty acids. Nat. Commun. 2020, 11, 2258. [Google Scholar] [CrossRef]
- Schönfeld, P.; Wojtczak, L. Short- and medium-chain fatty acids in energy metabolism: The cellular perspective. J. Lipid Res. 2016, 57, 943–954. [Google Scholar] [CrossRef] [Green Version]
- Frampton, J.; Murphy, K.G.; Frost, G.; Chambers, E.S. Short-chain fatty acids as potential regulators of skeletal muscle metabolism and function. Nat. Metab. 2020, 2, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Paul, C.E.; Hilberath, T.; Jongkind, E.P.J.; Zhang, W.; Alcalde, M.; Hollmann, F. Peroxygenase-Promoted Enzymatic Cascades for the Valorisation of Fatty Acids. ChemCatChem 2023, 15, e202300411. [Google Scholar] [CrossRef]
- Kiebist, J.; Schmidtke, K.U.; Zimmermann, J.; Kellner, H.; Jehmlich, N.; Ullrich, R.; Zander, D.; Hofrichter, M.; Scheibner, K. A peroxygenase from Chaetomium globosum catalyzes the selective oxygenation of testosterone. Chembiochem 2017, 18, 563–569. [Google Scholar] [CrossRef] [Green Version]
- Anh, D.H.; Ullrich, R.; Benndorf, D.; Svatos, A.; Muck, A.; Hofrichter, M. The coprophilous mushroom Coprinus radians secretes a haloperoxidase that catalyzes aromatic peroxygenation. Appl. Environ. Microbiol. 2007, 73, 5477–5485. [Google Scholar] [CrossRef] [Green Version]
- Kimani, V.W. New Secretory Peroxidases and Peroxygenases from Saprotrophic Fungi of Kenyan Forests; TU Dresden: Dresden, Germany, 2020. [Google Scholar]
- Santos, P.G.D.; Hoang, M.D.; Kiebist, J.; Kellner, H.; Ullrich, R.; Scheibner, K.; Hofrichter, M.; Liers, C.; Alcalde, M.; Master, E.R. Functional expression of two unusual acidic peroxygenases from Candolleomyces aberdarensis in yeasts by adopting evolved secretion mutations. Appl. Environ. Microbiol. 2021, 87, e00878-00821. [Google Scholar] [CrossRef]
- Ullrich, R.; Poraj-Kobielska, M.; Scholze, S.; Halbout, C.; Sandvoss, M.; Pecyna, M.J.; Scheibner, K.; Hofrichter, M. Side chain removal from corticosteroids by unspecific peroxygenase. J. Inorg. Biochem. 2018, 183, 84–93. [Google Scholar] [CrossRef]
- Gröbe, G.; Ullrich, R.; Pecyna, M.J.; Kapturska, D.; Friedrich, S.; Hofrichter, M.; Scheibner, K. High-yield production of aromatic peroxygenase by the agaric fungus Marasmius rotula. AMB Express 2011, 1, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiebist, J. Oxyfunktionalisierung Pharmazeutisch Relevanter Strukturen Durch Unspezifische Peroxygenasen der Familie I aus Dikarya-Pilzen; TU Dresden: Dresden, Germany, 2018. [Google Scholar]
- Barnes, R.H.; Rusoff, I.I.; Miller, E.S.; Burr, G.O. Relationship between unsaturation and ultraviolet absorption spectra of various fats and fatty acids. Ind. Eng. Chem. Anal. Ed. 1944, 16, 385–386. [Google Scholar] [CrossRef]
- Burr, G.O.; Miller, E.S. Ultraviolet absorption spectra of fatty acids and their application to chemical problems. Chem. Rev. 1941, 29, 419–438. [Google Scholar] [CrossRef]
- Grechkin, A.N.; Kuramshin, R.A.; Safonova, E.Y.; Mukhtarova, L.S.; Latypov, S.K.; Ilyasov, A.V. The keto-enol tautomerism and the redox conversions of α-ketol fatty acids. Chem. Phys. Lipids 1993, 66, 199–208. [Google Scholar] [CrossRef]




| Enzyme (UPO Abbreviation) | Fungal Species | Specific Activity (UVA/mg) * | Isoelectric Point (pI) | Reference |
|---|---|---|---|---|
| CglUPO | Chaetomium globosum | 3.9 | 4.3–4.7 (5.59 *) | [21] |
| CraUPO | Coprinellus radians | 72.0 | 3.8–4.2 | [22] |
| AaeUPO | Cyclocybe (Agrocybe) aegerita | 82.0 | 4.9–6.1 | [5] |
| CeuMQ V | Candolleomyces eurysporus | 12.5 | 4.25 | [23] |
| CeuMS III | C. eurysporus | 11.0 | 7.6 | herein |
| CabMS II | C. aberdarensis | 72.0 | 5.1 | herein |
| CabMQ II | C. aberdarensis | 172.0 | 4.1 | [23] |
| rCab II | C. aberdarensis (heterologously expressed in Pichia pastoris) | 21.0 | 5.1 | [24] |
| rCab I | C. aberdarensis (heterologously expressed in in Pichia pastoris) | 21.0 | n.d. | [24] |
| MweUPO | Marasmius wettsteinii | 50.0 | 5.1 ** | [2,25] |
| MroUPO | M. rotula | 25.4 | 5.0–5.3 | [26] |
| Number of Carbon Atoms | Substrates (Unsaturated Short- and Medium-Chain Fatty Acids) | Abbreviation and Double Bond Position (according to IUPAC Nomenclature) | Acetonitrile Concentration |
|---|---|---|---|
| 9 | trans-2-nonenoic acid | t-C9:1(2) | 55% |
| 3-nonenoic acid | m-C9:1(3) | ||
| 8-nonenoic acid | C9:1(8) | ||
| 8 | trans-2-octenoic acid | t-C8:1(2) | 50% |
| 3-octenoic acid | m-C8:1(3) | ||
| 7-octenoic acid | C8:1(7) | ||
| 7 | 2-heptenoic acid | m-C7:1(2) | 40% |
| 3-heptenoic acid | m-C7:1(3) | ||
| 6-heptenoic acid | C7:1(6) | ||
| 6 | cis-2-hexenoic acid | c-C6:1(2) | 30% |
| trans-2-hexenoic acid | t-C6:1(2) | ||
| trans-3-hexenoic acid | t-C6:1(3) | ||
| 5-hexenoic acid | C6:1(5) | ||
| 5 | trans-2-pentenoic acid | t-C5:1(2) | 15% |
| 3-pentenoic acid | m-C5:1(3) | ||
| 4-pentenoic acid | C5:1(4) | ||
| 4 | 3-butenoic acid | C4:1(3) | 7% |
| (vinylacetic acid) | |||
| 3 | 2-propenoic acid | C3:1(2) | 5% |
| (acrylic acid) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karich, A.; Salzsieder, F.; Kluge, M.; Alcalde, M.; Ullrich, R.; Hofrichter, M. Conversion of Unsaturated Short- to Medium-Chain Fatty Acids by Unspecific Peroxygenases (UPOs). Appl. Microbiol. 2023, 3, 826-840. https://doi.org/10.3390/applmicrobiol3030057
Karich A, Salzsieder F, Kluge M, Alcalde M, Ullrich R, Hofrichter M. Conversion of Unsaturated Short- to Medium-Chain Fatty Acids by Unspecific Peroxygenases (UPOs). Applied Microbiology. 2023; 3(3):826-840. https://doi.org/10.3390/applmicrobiol3030057
Chicago/Turabian StyleKarich, Alexander, Fabian Salzsieder, Martin Kluge, Miguel Alcalde, René Ullrich, and Martin Hofrichter. 2023. "Conversion of Unsaturated Short- to Medium-Chain Fatty Acids by Unspecific Peroxygenases (UPOs)" Applied Microbiology 3, no. 3: 826-840. https://doi.org/10.3390/applmicrobiol3030057
APA StyleKarich, A., Salzsieder, F., Kluge, M., Alcalde, M., Ullrich, R., & Hofrichter, M. (2023). Conversion of Unsaturated Short- to Medium-Chain Fatty Acids by Unspecific Peroxygenases (UPOs). Applied Microbiology, 3(3), 826-840. https://doi.org/10.3390/applmicrobiol3030057







