Abstract
Methane-producing Archaea can be found in a variety of habitats, including the gastrointestinal tract, where they are linked to various diseases. The majority of current monitoring methods can be slow and laborious. To facilitate gut methanogenic Archaea detection, we investigated flow cytometry for rapid quantification based on the autofluorescent F420 cofactor, an essential coenzyme in methanogenesis. The methanogenic population was distinguishable from the SYBR green (SG) and SYBR green/propidium iodide (SGPI) stained background microbiome based on elevated 452 nm emission in Methanobrevibacter smithii spiked controls. As a proof-of-concept, elevated F420-autofluorescence was used to detect and quantify methanogens in 10 faecal samples and 241 in vitro incubated faecal samples. The methanogenic population in faeces, determined through Archaea-specific 16S rRNA gene amplicon sequencing, consisted of Methanobrevibacter and Methanomassiliicoccus. F420-based methanogen quantification in SG and SGPI-stained faecal samples showed an accuracy of 90 and 100% against Archaea proportions determined with universal primers. When compared to methane and Archaea presence, methanogen categorisation in in vitro incubated faeces exhibited an accuracy of 71 and 75%, with a precision of 42 and 70%, respectively. To conclude, flow cytometry is a reproducible and fast method for the detection and quantification of gut methanogenic Archaea.
1. Introduction
Methanogenic or methane-producing Archaea can be found in a wide variety of habitats, ranging from wetlands to deep-sea vents, but also in the gastrointestinal tracts (GIT) of animals and humans [1,2,3]. In the human GIT, roughly 1.2% of the microbial community is Archaeal [4]. Of all Archaea in the gut microbiome, Methanobrevibacter smithii is the most prevalent methanogen, occurring in 31.3 to 95.7% of human individuals across different ethnic populations [5,6,7,8]. Methanosphaera stadtmanae (prevalence of 1.7 to 29.4% in humans) and members of the recently discovered order Methanomassiliicoccales (0.1 to 80%), which includes Methanomethylophilus (up to 3.3%) and Methanomasiliicoccus (up to 2.9%), are also commonly found [5,7,9,10]. Gut methanogen abundances have been associated with various gastrointestinal diseases. For example, lower methane production and Archaea abundances have been detected in individuals with ulcerative colitis, inflammatory bowel diseases, colonic polyposis and colon cancer compared to healthy individuals, whereas higher abundances have been found in obese individuals [11,12,13]. Methanogen abundances have also been shown to increase with gastrointestinal transit time [8,14].
While extensive research on human gut bacteria has been performed for decades, Archaeal gut microbiome research has only thrived in the past ten years [15]. As a result, research into the exact ecological role of Archaea is still in its infancy. The Archaeal population is hypothesised to benefit from overall microbial metabolism, as bacteria degrade complex dietary compounds and provide the necessary substrates and reducing equivalents for methanogenesis. Most methanogens, such as M. smithii and Methanomasiliicoccus, perform anaerobic respiration via the hydrogenotrophic pathway that utilises H2 to reduce CO2 into CH4 [2,16]. This methanogenesis pathway creates a hydrogen sink, thereby promoting bacterial fermentation and growth since hydrogen accumulation can inhibit bacterial metabolism and the regeneration of the coenzyme NAD+ from NADH [8,14]. Methanogenesis through the conversion of methanol (e.g., in M. stadtmanae) or acetate into methane via the methylotrophic or acetotrophic pathways is less common [17,18,19,20]. Methanol is produced in the gut through pectin degradation, demethylation of endogenous cellular proteins or vitamin B12 synthesis. The non-metabolized methanol enters the liver after intestinal absorption [21,22], and as such, high levels of alcohol contribute to the development of non-alcoholic fatty liver disease. This can be partially prevented by methylotrophic methanogens [22]. At present, acetotrophic methanogenic activity has not been reported in the human gut.
Given the putative role of methanogenic Archaea in gastrointestinal diseases and their important ecological functions, the interest in routinely monitoring and quantifying Archaea within the human gut microbiome is increasing. Archaea detection can be performed indirectly through methane measurements in vivo (breath analysis) or in vitro, or directly with DNA-based methods, such as qPCR, 16S rRNA gene amplicon sequencing and metagenomics, or alternatively through anaerobic culture plating combined with optical microscopy [10,23,24]. These methods have several shortcomings. Breath testing is a qualitative rather than a quantitative method, and readouts can be influenced by oral hygiene [25]. DNA-based methods and anaerobic culture plating can be slow and labour-intensive. To address these issues, we propose flow cytometry based on the autofluorescence properties of cofactor F420 (8-hydroxy-5-deazaflin) as a reliable and fast method to quantify methanogenic Archaea in faecal microbial communities. Cofactor F420 is an auto-fluorescent redox cofactor with absorption and emission peaks at 420 and 472 nm, respectively [26]. It is essential in both hydrogenotrophic and methylotrophic methanogenesis and is present at high concentrations (100 to 400 mg/kg cells) in methanogens [27,28]. The autofluorescence properties of cofactor F420 have already been used to quantify methanogens with fluorescence microscopy after excitation with a violet laser (405 nm) [23]. F420 autofluorescence has also been detected using flow cytometry in methanogenic enrichment cultures and digester samples and in faecal samples [29,30]. While a significant correlation has been established between relative abundances of cofactor F420 auto-fluorescent cell counts and Methanobacteriaceae proportions, absolute quantification of methanogenic Archaea in the human gut microbiome has not been explored yet. We, therefore, validated flow cytometry as a fast, quantitative screening tool to probe methanogenic Archaea in human faecal samples and in vitro reactor samples. In addition, phenotypic fingerprints were computed based on all fluorescent and scatter variables measured through flow cytometry to determine microbial community phenotypic alpha- and beta-diversity [31]. Quantitative Microbiome Profiling based on 16S rRNA gene amplicon sequencing with universal and Archaeal primers and methane measurements was used to benchmark the flow cytometry approach.
2. Materials and Methods
2.1. Validating Flow Cytometry to Identify Methanogenic Archaea in the Human Gut Microbiome Based on F420 Autofluorescence with a Methanobrevibacter Smithii Type Strain
Methanobrevibacter smithii, the most prevalent methanogenic Archaea in the human gut microbiome, was spiked in faecal samples and used as a reference to validate flow cytometry as a quick tool to identify and quantify gut methanogens based on cofactor F420 autofluorescence.
2.1.1. Methanobrevibacter smithii Cultivation and Quantification
Methanobrevibacter smithii type strain DSMZ861 (DSMZ, Braunschweig, Germany) was cultivated in DMSZ1523 medium (Table S1) containing DSMZ320 trace element solution (Table S2) and DSMZ503 seven-vitamins solution (Table S3) according to DSMZ instructions [32,33,34,35]. In short, all components except for bicarbonate, vitamins, cysteine and sulphide were dissolved and boiled. During the subsequent cooldown, the medium was sparged with an 80% H2 and 20% CO2 gas mixture for 45 min. Next, bicarbonate was dispensed in the medium, after which the medium was distributed into anoxic Hungate tubes (Glasgerätebau Ochs, Bovenden, Germany) under a continuous gas flow of 80% H2 and 20% CO2. Next, the tubes were flushed with 80% H2 and 20% CO2 and capped at overpressure. After autoclaving, the filter-sterilised (0.22 μm) vitamin solution, cysteine and sulphide were added to the medium under a continuous gas flow of filter-sterilised (0.22 μm) N2 gas. The pH was adjusted with filter-sterilised (0.22 μm) HCl or NaOH to 6.8–7.0 before inoculation with M. smithii. The inoculated tubes were placed in a stationary 37 °C incubator (Memmert, Schwabach, Germany). Microbial growth started within two days and was visually confirmed through clouding of the medium. M. smithii cultures took 1 week to grow before biomass was harvested and spiked into a faecal slurry.
The M. Smithii culture was quantified after staining with two different stains separately (1% v/v). The used stains were either SYBR® Green I (SG, 100× concentrate, Fisher Scientific, Merelbeke, Belgium) nucleic acid stain dissolved in 0.22 μm-filtered dimethyl sulfoxide (Sigma Aldrich, St. Louis, MO, USA) or SYBR® Green I (10 μL 10,000×, Fisher Scientific, Merelbeke, Belgium) combined with propidium iodide (20 μL of 20 mM propidium iodide, Fisher Scientific, Merelbeke, Belgium) (SGPI) dissolved in 0.22 μm-filtered dimethyl sulfoxide (Sigma Aldrich, St. Louis, MO, USA). The samples were subsequently incubated for 20 min at 37 °C and immediately measured with a CS&T calibrated FACSVerse™ flow cytometer (BD Biosciences, Erembodegem, Belgium), equipped with a violet (405 nm), blue (488 nm) and red (640 nm) laser and blue (Blue1 = 452, Blue2 = 500 nm), green (519 nm) and red (695 nm) emission detectors. The M. smithii pure culture contained 1.48 × 108 ± 2.16 × 106 SG-stained cells mL−1 and 7.09 × 108 ± 4.76 × 108 SGPI-stained cells mL−1.
2.1.2. Preparation of Faecal Slurries
Faeces from a non-methanogenic donor, which tested negative for in vitro methane production and consistently lacking endogenous methanogens, as shown by 16S rRNA gene amplicon sequencing in a previous experiment by Minnebo et al. (2021) (Donor 1) [35], were deposited in airtight containers, comprising an AnaeroGen™ sachet (Oxoid, Hampshire, UK) to generate an anaerobic environment. A homogeneous faecal slurry was prepared by homogenising and diluting 20 g of fresh human faeces in 100 mL of 0.1 M anaerobic phosphate buffer pH 6.8 (consisting of 8.8 g L−1 K2HPO4 and 6.8 g L−1 KH2PO4), containing 1 g L−1 sodium thioglycolate as a reducing agent. The homogenate was subsequently centrifuged (3 min, 500× g) to remove particulate matter. The particle-free supernatant or faecal slurry containing the faecal microbes was further analysed with flow cytometry and spiked with M. smithii [36]. Prior to flow cytometry, the faecal samples were further filtered with a 20 μm Filcon syringe filter (BD Biosciences, Erembodegem, Belgium) to remove non-cellular particles.
2.1.3. Flow Cytometry F420 Autofluorescence-Based Detection and Quantification of the Methanogenic Archaea M. smithii in a Spiked Faecal Sample
The spiked faecal slurry consisted of 5 µL of 102 times diluted faecal slurry combined with 5 μL of undiluted M. smithii culture in 485 μL of anaerobic PBS and 5 μL of stain. The 103 times diluted pure M. smithii culture and the 104 times diluted non-methanogenic faecal slurry were analysed as controls (Table S4). All dilutions were made with 0.22 μm-filtered sterile anaerobic PBS. Prior to incubation for 20 min at 37 °C, samples were stained (1% v/v) with either SG or SGPI. The incubated samples were immediately measured with a CS&T calibrated FACSVerse™ flow cytometer (BD Biosciences, Erembodegem, Belgium), equipped with a violet (405 nm), blue (488 nm) and red (640 nm) laser and blue (Blue1 = 452, Blue2 = 500 nm), green (519 nm) and red (695 nm) emission detectors. The emission detectors have bandpass filters and dichroic mirrors with wavelengths of 448/45 and 448/45 nm in Blue1, 528/45 and 500 nm Longpass (reflects light shorter than 500 nm, LP) in Blue2, 527/32 and 507 LP in green, and 700/54 and 665 LP, respectively, in red. Gating of SG- and SGPI-stained samples was performed in Rstudio (v.4.2.0) using the ggCyto package (v.1.24.0) [37,38]. Methanogen cell concentrations were obtained by dividing M. smithii cell counts by the acquisition volume (Figure S1).
2.2. Benchmarking Flow Cytometry to Quantify Endogenous Methanogenic Archaea Based on F420 Autofluorescence against 16S rRNA Gene Amplicon Sequencing
2.2.1. Applying Flow Cytometry to Quantify Endogenous Methanogenic Archaea in Faecal Samples of 10 Individuals Based on F420 Autofluorescence
As a proof-of-concept, endogenous methanogens were quantified in ten different faecal samples derived from ten different healthy individuals (seven women and three men, without diagnosed health concerns and without a history of antibiotic intake six months prior to donation). Faecal slurries were prepared as described above and were diluted 105 times with 0.22 μm-filtered sterile anaerobic PBS (Table S4), except for the sample of donor 5, which required only a 104 times dilution to fall within quantification limits. Dilution, staining, triplicate measurements, methanogen gating and quantification were performed as described above (2.1.3) (Figure S1). To quantify the proportion of methanogens in the microbial community, total microbial cell counts and concentrations were also determined. To this end, gates were defined based on the green and red fluorescence emission channels. All faecal samples were heat-killed (90 °C for 15 min) to distinguish intact and damaged microbial cell populations in SGPI-stained samples. Additionally, 0.22 μm-filtered samples were included as negative controls to identify noise.
2.2.2. Applying Universal and Archaeal-Specific 16S rRNA Gene Amplicon Sequencing to Detect Endogenous Methanogenic Archaea in Faecal Samples of 10 Individuals
Besides flow cytometry, 16S rRNA gene amplicon sequencing was carried out on the faecal samples of the ten donors. DNA extraction of pelletised faecal slurries (10 min, 5000× g) was accomplished through a combination of chemical and mechanical lysis via a bead-beating step, as discussed in [39,40]. The DNA quality was verified on a 1.5% (w/v) agarose gel.
Extracted DNA was sent to LGC Genomics (Teddington, Middlesex, UK) for library preparation and sequencing on an Illumina MiSeq platform, as described by De Paepe et al., 2017 [40]. The V3-V4 region of 16S rRNA gene was amplified through PCR using universal primers (U341F:CCTAYGGGRBGCASCAG and U806R: GGACTACGGGTATCTAAT), as well as Archaeal (340F: CCCTAYGGGGYGCASCAG and 1000R: GGCCATGCACYWCYTCTC) primers nested with the U341F—U806R primer pairs described by Klindworth et al., 2013 and Gantner et al., 2011, respectively [41,42].
2.3. Benchmarking Flow Cytometry to Quantify Methanogenic Archaea in In Vitro Reactor Samples Based on F420 Autofluorescence against Methane Production Measurements and 16S rRNA Gene Amplicon Sequencing
2.3.1. Applying Flow Cytometry to Quantify Endogenous Methanogenic Archaea in SHIME Samples of 5 Individuals Based on F420 Autofluorescence
As a second proof-of-concept, methanogens were also quantified in in vitro gut reactor samples derived from the Simulator of the Human Intestinal Microbial Ecosystem (SHIME). SHIME samples were obtained from a proximal and distal colon environment with total transit times of 21 h (short), 42 h (medium) or 63 h (long) [43]. Since methane production and methanogenic Archaea were absent in the proximal colon, only the distal colon was investigated in this proof-of-concept. In the original study, six donors were included to capture inter-individual variability. Since gas measurements were lacking from donor 6, only five donors were considered for this proof-of-concept [43]. Flow cytometry measurements and data analysis were performed as outlined before (in Section 2.2.1).
2.3.2. Applying Universal 16S rRNA Gene Amplicon Sequencing to Quantify Endogenous Methanogenic Archaea in SHIME Samples of 5 Individuals
Aliquots (1 mL) of the SHIME samples were centrifuged (10 min at 5000× g) and stored at −20 °C for DNA extraction, followed by next-generation 16S rRNA gene amplicon sequencing of the V4 region [44]. Sequencing of the SHIME and control samples (i.e., blanks, negative controls, positive controls with a known composition and a pure Runella slithyformis culture) was performed on an Illumina MiSeq platform (Illumina, Hayward, CA, USA) using Illumina MiSeq v2 chemistry at the VIB Nucleomics core (VIB, Gasthuisberg Campus, Leuven, Belgium). The V4 region of the 16S rRNA gene was amplified through PCR using primers 515F (GTGYCAGCMGCCGCGGTAA and 806R (GGACTACNVGGGTWTCTAAT) according to Vandeputte et al., 2017 [44].
2.3.3. Measuring Methane in SHIME Headspace Samples of 5 Individuals
To establish if methanogen presence is indicative of methanogenic activity, the gas composition was measured in the SHIME. The methane fraction (%) was determined with a Compact GC (Global Analyser Solutions, Breda, The Netherlands), equipped with a Molsieve 5A pre-column and Porabond column (CH4, O2, H2 and N2) and a thermal conductivity detector [45].
2.4. Bioinformatics and Statistical Analysis
All bioinformatics data processing, visualisations and statistical analyses were executed in R version 4.2.0 [38]. All data were visualised with ggplot2_3.3.6 and ggpubr_0.4.0 unless mentioned otherwise [46,47].
The acquired flow cytometry fcs files were submitted to the FlowRepository archive under repository ID FR-FCM-Z5KA and were processed and visualised with the Phenoflow (v.1.1.2) and ggCyto (v.1.24.0) package to determine the methanogenic and total cell populations [31,37].
An M. smithii pure culture and a faecal sample lacking methanogens were used as positive and negative controls to set a threshold fluorescence in the Blue1 channel of 2.44 × 103 in SG and 8.38 × 103 in SGPI (lower bound 95% CI) above which cells were considered methanogenic (Figure 1). Since methanogenic populations shifted towards a lower SG fluorescence, polygon gates were drawn on Blue1-Green biplots to maximize the separation and facilitate the identification of the methanogenic populations. The polygon gate contained some noise events, which were quantified in blank samples. Additional sample-specific experimental noise was quantified by measuring 0.22 µm-filtered samples in triplicates. Sample-specific noise events in the methanogenic gate ranged in SG-stained samples between 4.85 × 106 and 1.87 × 109 cells mL−1 with an average of 4.13 × 108 ± 6.63 × 108 cells mL−1 per sample (n = 29) and in SGPI-stained samples, between 3.96 × 107 and 1.55 × 109 cells mL−1 with an average of 4.77 × 108 ± 5.59 × 108 cells mL−1 per sample (n = 29). The definitions of the limit of detection (LOD) and quantification (LOQ) can vary. In this experiment, it is defined as, respectively, 3 and 10 times the average noise level recorded in the entire dataset of 0.22 µm-filtered sterilised samples. To conclude, populations were considered methanogenic if the F420 auto-fluorescent Blue1 signal exceeded 2.44 × 103 in SG and 8.38 × 103 in SGPI, and if the cell counts in the methanogen gate exceeded the LOD (1.23 × 109 cells mL−1 in SG and 1.43 × 109 cells mL−1 in SGPI). Furthermore, populations were considered quantifiable if the cell counts in the methanogen gate surpassed the LOQ (4.13 × 109 cells mL−1 in SG and 4.77 × 109 cells mL−1 in SGPI).
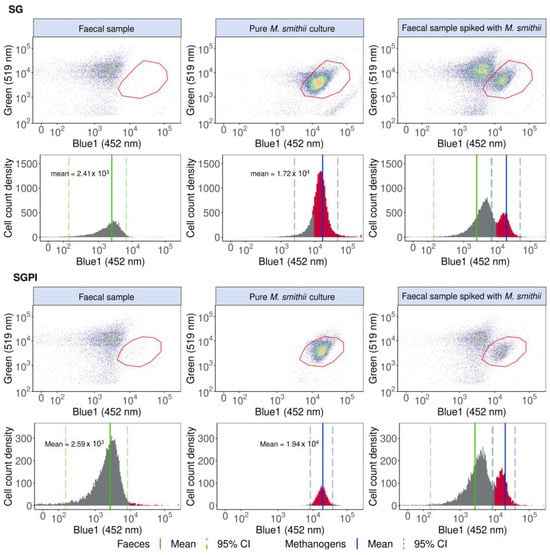
Figure 1.
M. smithii cells can be identified, singled out and quantified in a pure M. smithii culture and faecal sample spiked with M. smithii based on elevated 452 nm (Blue1) fluorescence derived from F420 cofactor autofluorescence in SYBR® Green—propidium iodide (SGPI) or SYBR® Green (SG) stained samples. The one-dimensional histograms of the 452 nm emissions are depicted with the F420 autofluorescence peaks in red. The mean Blue1 fluorescence intensities of the faecal samples and pure methanogenic cultures are shown with a green and blue line, respectively, and the 95% CI is demarcated with dotted lines. Cofactor F420 autofluorescence in the Blue2 (500 nm) and forward and sideward scatter are visualised in Figure S4.
The phenotypic β diversity of the faecal microbiome fingerprint was assessed based on the flow cytometry data collected in the fluorescent Green (519 nm), Red (695 nm), Blue1 (452 nm) and Blue2 (500 nm), and forward and sideward scatter channels with Phenoflow (v1.1.2) according to Props et al., 2016. The data were first arcsine-transformed and subsequently normalised by dividing all values by the maximum Green fluorescence intensity.
The amplicon sequencing data was submitted to the European Bioinformatics Institute’s (EBI) European Nucleotide Archive (ENA) under accession number ERP138722 and was processed with the mothur software package (v.1.44.3) as discussed by De Paepe et al., 2017 and classified with Silva.nr_v138 [48]. The mothur processed amplicon sequencing data, consisting of a read count table (containing the number of reads observed for each OTU in each sample) and the taxonomic annotations, were subjected to quality control and further processed in R version 4.2.0 [38]. Singletons and OTUs were present in less than 5% of the samples or with read counts not exceeding 0.5 times the number of samples that were removed [49]. The read counts and reproducibility of the samples were deemed satisfactory (Figure S2). Rarefaction curves of all samples were additionally constructed to ensure sufficient sequencing depths (Figure S3) (vegan_2.5-7) [50].
The proportional microbial community compositions of the samples were inspected with phyloseq (v.1.36.0) at the genus level [51]. Absolute microbial abundances were obtained by multiplying the total cell counts with the rarefied copy-number corrected (using the RDP classifier tool, RDP 16S rRNA training set 16) relative microbial community profiles according to the Quantitative Microbiome Profiling (QMP) method [44,45]. QMP was only applied to the data obtained with the universal primer set to compare absolute Archaea abundances with methanogen cell counts. The comparison was performed with Spearman’s rank correlation coefficients, which were determined using the Stats package (v.4.2.0) [38]. Likewise, relative microbial community compositions obtained with the universal primer set were compared with Spearman’s rank correlation coefficients with the methanogen proportions calculated based on the flow cytometry data also using the Stats package (v.4.2.0) [38].
In addition, methanogen classification (i.e., F420 Blue1 > 2.44 × 103 in SG and >8.38 × 103 in SGPI-stained samples and cell counts > LOD, see definition before) was benchmarked against the 16S rRNA gene amplicon sequencing results. The 16S samples were marked as Archaea positive when more than 1 Archaea read was present after singletons and OTUs present in less than 5% of the samples or with read counts not exceeding 0.5 times the number of samples removed. In in vitro samples, the predictive value of methane production as a marker for methanogen presence was determined.
The F1-score, accuracy ([true positive + true negative]/[total]), sensitivity ([true positive ]/[true positive + false negative]), specificity ([true negative]/[true negative + false positive]), and negative predictive values ([true negative]/[true negative + false negative]) of the methanogen classification were examined using the confusion matrix function of the Caret package (v.6.0-92) [52]. The F1-score was calculated as a harmonic mean of precision ([true positive]/[true positive + false positive) and sensitivity.
2.5. Ethical Approval
Research with human faecal material was conducted in accordance with the ethical approval obtained from the Ethical Committee of the Ghent University Hospital (B670201836318).
3. Results
Flow cytometry was validated as a rapid tool to identify and accurately (% error~36–41%) and precisely (CV~10–28%) quantify methanogenic Archaea in faecal samples based on significantly elevated 452 nm emission derived from F420 cofactor autofluorescence (p < 1 × 10−22). F420 autofluorescence (with a peak emission at 472 nm [26]) was most clearly detected at 452 nm and, to a lesser extent, at 500 nm (Figure S4). F420 detection was, moreover, compatible with commonly used SYBR® Green—propidium iodide (SGPI) and SYBR® Green (SG) staining in gut microbiome research [44,45].
Pure M. smithii cultures stained with SG and SGPI showed a seven-fold higher fluorescence intensity at 452 nm compared to a faecal sample without methanogenic Archaea. In our specific flow cytometer, the mean 452 nm fluorescence intensity was 1.72 × 104 (95% CI: 2.44 × 103 to 4.97 × 104) for SG and 1.94 × 104 (95% CI: 8.38 × 103 to 3.78 × 104) for SGPI, while the mean fluorescence intensity for the faecal sample was 2.41 × 103 (95% CI: 1.50 × 102 to 6.51 × 103)for SG and 2.59 × 103 (95% CI: 1.62 × 102 to 8.15 × 103) for SGPI. Besides an increased 452 nm fluorescence, M. smithii cells shifted towards a lower SG (green, 519 nm) fluorescence, which can aid in the identification and distinction of methanogenic Archaea in a more complex background. M. smithii SG and SGPI-stained populations were therefore also gated in blue1 (452 nm) vs. green (519 nm) emission biplots (Figure 1).
The methanogen population, delineated based on pure M. smithii cultures, could be singled out and distinguished from the background faecal microbes in M. smithii-spiked faecal slurries (Figure 1).
Methanogenic Archaea quantification was precise, accurate and fast (max. 2 min per sample). In SG-stained spiked faecal samples, 9.44 × 105 ± 9.68 × 104 M. smithii cells mL−1 were detected, whereas, after spike-in, 1.48 × 106 ± 2.16 × 104 M. smithii cells mL−1 were expected, resulting in a percent error of 36.21% and a coefficient of variation of 10.2%. SGPI-stained spiked faecal samples exhibited a higher variation (coefficient of variation of 27.8%). The difference between the expected (7.09 × 106 ± 4.76 × 106 M. smithii cells mL−1) and detected (4.17 × 106 ± 1.16 × 106 M. smithii cells mL−1) M. smithii concentration also increased compared to the SG-stained samples, resulting in a larger percent error of 41%.
The detected 452 nm emission was specific to methanogenic Archaea, as the polygon gate in biplots displaying the faecal sample lacking Archaea was empty apart from a few noise events of 29.37 ± 7.13 cells mL−1 in SGPI and 2.71 ± 0.24 cells mL−1 in SG.
Benchmarking against 16S rRNA gene amplicon sequencing and methane measurements further confirmed the application potential of F420-based flow cytometric detection of methanogens in faecal samples and incubated in vitro gut microbiota samples.
3.1. Detection and Quantification of Endogenous Methanogens Is Consistent between Flow Cytometry and 16S rRNA Gene Amplicon Sequencing in Faecal Samples and In Vitro Incubated Faecal Samples
Flow cytometry identified endogenous methanogens in the faecal slurries of three donors (1, 2 and 3) stained with SG and two donors (1 and 3) stained with SGPI (Figure 2A). The measured methanogen cell counts were higher than the LOD (three times the average noise level of all 0.22 µm-filtered sterilised samples). The presence of methanogens was confirmed with 16S rRNA gene amplicon sequencing in the faecal samples of two donors (1 and 3) with universal primers and four donors (1, 2, 3 and 5) with Archaeal primers (Figure 2C). Flow cytometry, thus, correctly predicted the presence of methanogens in three out of four donors with SG staining and thereby outperformed 16S rRNA gene amplicon sequencing with universal primers, which did not detect lower abundances of Archaea in donors 2 and 5. This is reflected in high accuracy (90%) and F1 score (0.86) in a classification model comparing flow cytometry in combination with SG staining to Archaea-specific sequencing as a reference method (Figure 2B). The Accuracy (80%) and F1 scores (0.67) dropped with classification based on SGPI staining combined with flow cytometry and 16S rRNA gene amplicon sequencing using Archaeal primers.
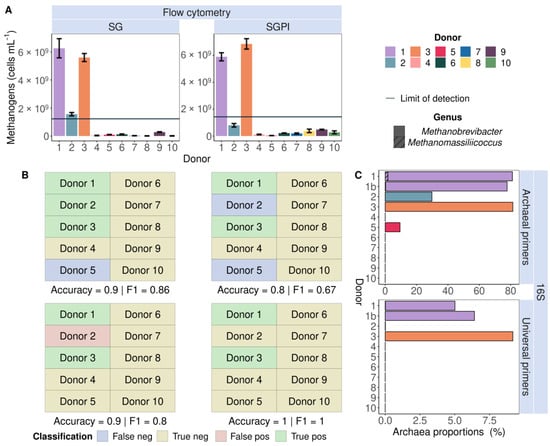
Figure 2.
Flow cytometric detection of cofactor F420 auto-fluorescence (A) corresponded with 16S rRNA gene amplicon (16S) Archaea presence (C) in nine out of ten SYBR® Green (SG) and eight out of ten SYBR® Green—propidium iodide (SGPI)-stained faecal slurries derived from ten different healthy donors. Universal primers failed to identify Archaea in donors 2 and 5, who carried lower abundances of Methanobrevibacter (no pattern fill) and Methanomassiliicoccus (hatched bars) according to Archaea-specific sequencing, which served as a benchmark. The flow cytometry-based methanogenic Archaea detection yielded the highest accuracy and F1-score due to the highest number of true positive (green) and true negative (yellow) and the smallest number of false positive (red) and false negative (purple) classifications. (B) Donors were classified as ‘methanogen positive’ when methanogenic Archaea cell counts were higher than the limit of detection (LOD, full line).
Since the more sensitive and Archaea-specific nested sequencing approach does not allow quantification of the Archaea:Bacteria ratio, methanogen proportions obtained with flow cytometry were only compared to Archaea proportions derived from sequencing with universal primers. When stained with SG and SGPI, only donors 1 and 3 showed values above the LOQ (10 times the average filtered background noise) (Figure 3). As a result, the dataset was too small to calculate correlations. However, when not taking the LOQ into account, correlations were discovered. Flow cytometry-based methanogen proportions were consistently lower but strongly and significantly positively correlated with sequencing-based Archaea proportions. The SG-stained samples displayed a higher Spearman’s rank correlation coefficient (0.91, p = 8.23 × 10−5) compared to the SGPI-stained samples (0.7, p = 0.017) (Figure 3A).
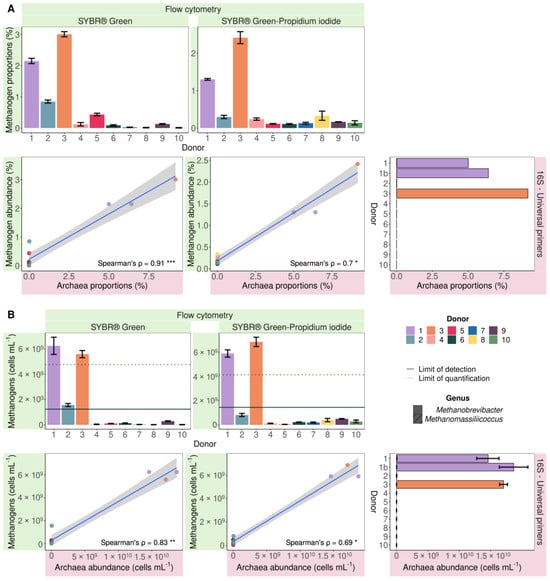
Figure 3.
Proportional (A) and absolute (B) flow cytometric methanogenic Archaea abundance based on cofactor F420 autofluorescence of SYBR® Green and SYBR® Green—propidium iodide-stained samples significantly positively correlated (Spearman’s rank correlation) with 16S rRNA gene amplicon Archaea abundances in 10 faecal slurries derived from 10 different healthy donors. The Archaea community consisted of Methanobrevibacter (no fill pattern) and Methanomassiliicoccus (hatched bars). Spearman’s rank correlation coefficients (ρ) are depicted in the bottom right corner of every plot. *** = p < 0.001, ** = p < 0.01 and * p < 0.05. The limit of detection and the limit of quantification are depicted with a full and dashed line, respectively.
Archaea made up 9.18% of the sequenced microbial community in donor 3, whereas methanogens accounted for 3.01 ± 0.08% of SG-stained cells versus 2.20 ± 0.19% of SGPI-stained cells. In donor 1, a similar difference was observed. Sequencing indicated a proportional Archaea abundance of 5.72 ± 0.70%, which is larger than the methanogen percentages in the SG (2.15 ± 0.08%) and SGPI (1.43 ± 0.02%) stained microbial community. While sequencing of 16S rRNA gene amplicons generated with universal primers did not detect Archaea in donors 2 and 5, flow cytometry revealed methanogen proportions of 0.85 ± 0.05 and 0.43 ± 0.04%, respectively, following SG staining. Even though these methanogen counts were below the LOQ, they stood out from the remaining samples. Archaea absence in the other samples coincided with below-noise event counts in the methanogen gate, which corresponded to 0.06 ± 0.06% of total events in SG-stained and 0.18 ± 0.09% in SGPI-stained samples.
While SG staining is more accurate in methanogen classification, SGPI staining provides additional information about the physiological state of cells, more precisely about membrane integrity. The percentage of total intact cells ranged between 60.87 ± 0.46% in donor 1 and 92.13 ± 0.87% in donor 10, with an average of 80.46 ± 9.35%. Of the samples marked as methanogen positive, besides donor 1, donors 2, 3 and 5 had percentages of 71.63 ± 1.67, 76.49 ± 2.68 and 83.61 ± 0.72%, respectively (Figure S5A). The methanogenic subpopulation was mostly intact in donors 1 (~98%) and 3 (~97%) (Figure S5B). This was much lower for the remaining donors, as only 28.2 ± 6.0% of the counted methanogens were intact (Figure S5B). Besides the additional information about cell membrane integrity and the lower detection limits compared to standard universal 16S rRNA gene amplicon sequencing, flow cytometry offers absolute instead of relative quantification.
The faecal microbiomes of donors 1 and 3, which were proportionally enriched in methanogens compared to the other donors, contained the highest absolute methanogen cell counts. In SG-stained samples, similar methanogen counts were recorded in donor 1 (6.26 × 109 ± 6.83 × 108 cells mL−1) and donor 3 (5.60 × 109 ± 2.86 × 108 cells mL−1). SGPI-stained faecal samples contained similar concentrations of 5.89 × 109 ± 3.02 × 108 cells mL−1 in donor 1 and slightly higher 6.85 × 109 ± 3.87 × 108 cells mL−1 in donor 3 (Figure 3B). The difference between SG-stained (1.56 × 109 ± 1.20 × 108 cells mL−1) and SGPI-stained (8.01 × 108 ± 1.31 × 108 cells mL−1) methanogen counts was equally small in donor 2. However, the SGPI-stained methanogen counts remained below the LOD and were therefore not counted as ‘methanogen positive’ in the methanogen classification (Figure 2A). While donor 5 showed a higher percentage of methanogenic Archaea after staining with SG relative to the remaining donors, the methanogen load was not different. The average flow cytometry methanogen counts of the remaining donors showed more variation within and between staining protocols 8.32 × 107 ± 1.02 × 108 (SG) and 2.8 × 108 ± 1.41 × 108 cells mL−1 (SGPI) and was 1–2 log units smaller compared to donor 1–3. This indicates that the precision of the flow cytometry-based methanogen quantification is reduced at lower methanogen counts.
The composition of the methanogenic population cannot be inferred from flow cytometry data, but sequencing showed that the genus Methanobrevibacter dominated the Archaeal community when present. The genus Methanomassiliicoccus was also detected in donor 1 with both primers but at a more than 45-fold lower abundance compared to Methanobrevibacter (Figure 3).
While flow cytometry cannot be used to directly identify the taxonomic composition of microbial communities, shifts in composition can result in shifts in flow cytometry fingerprints. This is due to, amongst others, differences in genome size and thus the amount of DNA that can be stained, the cell size and cell granularity. It is thus informative to assess the population spread in cytograms through the calculation of phenotypic β diversity. A phenotypic β-diversity analysis of the entire microbial population based on six parameters (Blue1 (452 nm), Blue2 (500 nm), Green (519 nm) and Red (695 nm)) separated the methanogen-positive donors 1, 2, 3 and 5 from the other donors (Figure S6). A classic PCoA ordination of the 16S rRNA gene amplicon community composition determined with Archaeal primers demonstrated no distinct clustering of Archaea-positive donors. This is in line with our results since donors 2 and 5 could only be identified as Archaea-positive after a nested PCR approach to enrich Archaeal species prior to amplification with universal primers (Figure 4B). On the other hand, in the PCoA of the Archaea-enriched community sequenced with Archaea-specific primers, donors 2 and 5, just like donors 1 and 3, clustered separately from the other donors with no clear differences between the quantitative and proportional microbial profiles (Figure 4B).
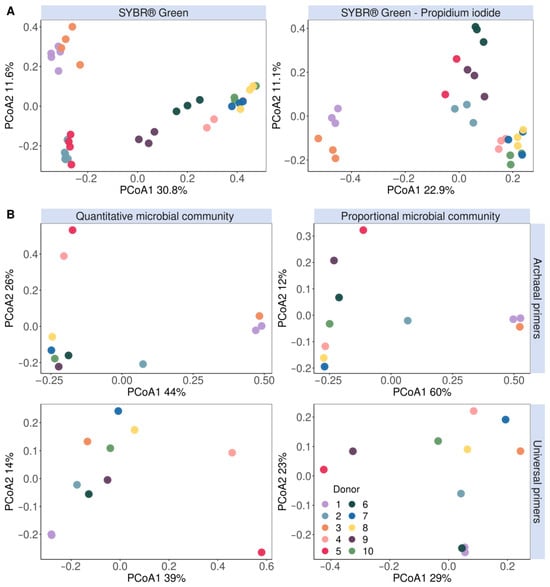
Figure 4.
Microbial community β-diversity assessment clustered methanogens according to their (A) flow cytometric fingerprint data stained with either SYBR® Green (left) or a SYBR® Green—propidium iodide staining mix (right) and (B) principal coordinates analysis (based on Bray–Curtis dissimilarity) of the genus-level quantitative microbial community compositions (left) and proportional microbial community composition (right). The fingerprint was based on the isolated methanogen information of six phenotypic parameters (four fluorescence and two scatter signals). The V3–V4 region of the 16S rRNA gene was amplified with either Archaeal (top) or universal (bottom) primers.
A similar clustering pattern was observed in the phenotypic β-diversity analysis of the methanogenic population based on six parameters (Blue1 (452 nm), Blue2 (500 nm), Green (519 nm) and Red (695 nm) fluorescence and the forward and sideward scatter signals) (Figure 4A). Interestingly, donors 2 and 5 clustered together and apart from the other donors in the SG-stained phenotypic β-diversity and quantitative microbial community amplified with Archaeal primers (Figure 4).
The flow cytometry methanogen detection tool was also demonstrated on 241 SGPI-stained samples obtained from the dataset of Minnebo et al., 2023, in which the SHIME transit times were adjusted to short, medium and long [43]. Comparing flow cytometric analysis with 16S rRNA gene amplicon sequencing (universal primers on V4 region) on methanogenic Archaea abundances as a reference method yielded an accuracy of 75% (p = 7.90 × 10−5). Furthermore, the detection tool had a precision of 70%, positive and negative predictive values of 0.65 and 0.82, and an F1-score of 0.67 (Table S7). Two methane-producing Archaea were detected in the dataset, namely Methanobrevibacter in donors 2 to 5 and Methanosphaera in donor 2.
3.2. Methanogens Clustered according to In Vitro Methane Production and Methanogenic Archaea Presence in In Vitro Incubated Faecal Samples
To confirm not only the presence but also the activity of methanogens in the human gut microbiome, methane production is commonly measured in in vitro incubated faecal samples. Therefore, besides sequencing and flow cytometry, the methane production in the SHIME headspace was also measured [43]. Since no methane production or Archaea abundances were observed in the proximal colon, only the distal colon was investigated in this proof-of-concept.
Of the five donors examined in this experiment (donor 6 was excluded as no gas was measured), donor 1 showed no methanogenic activity nor Archaea presence based on gas analyses and 16S rRNA gene amplicon sequencing (universal primers on V4 region), respectively. In donor 2, only Archaea were observed, but no methanogenic activity. Donor 3, 4 and 5 hosted methanogenic Archaea and demonstrated methane production, of which in donor 3 only in the long distal transit time. With methane production as a reference method, compared to sequencing, a lower accuracy of 71% (p = 0.013), a precision of 42%, a sensitivity of 65%, negative predictive values of 0.87 and an F1-score of 0.51 were achieved (n = 241, Table S7).
A phenotypic β-diversity analysis was performed on SHIME samples obtained throughout the 10-day stabilisation phase (n = 74), during which the microbial community adapts to an in vitro environment. This ordination revealed a separate clustering of methane and non-methane producers in the distal colon. In the short and medium SHIME transit times, donors 1, 2 and 3 did not produce methane and were consistently separated from the methane-producing donors 4 and 5 based on flow cytometry fingerprints. A long SHIME transit time induced methane production in donor 3, which was also evident by a regrouping of donor 3 in the methane-producing cluster together with donors 4 and 5 (Figure 5). Donor 2’s samples clustered separately, in which Archaea was observed, but no methane production was observed in the distal colon of the medium and long transit.
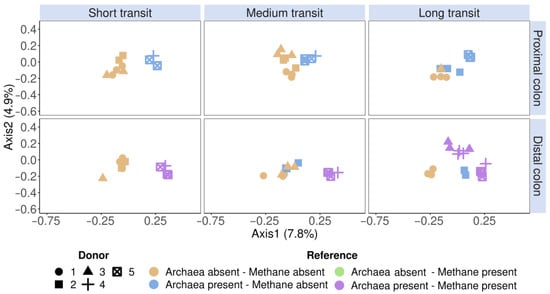
Figure 5.
Community β-diversity assessment of the flow cytometric SHIME fingerprint data clustered according to the reference methods (methanogenic Archaea presence and SHIME methane presence). The fingerprint was based on the isolated methanogen information of six phenotypic parameters (four fluorescence and two scatter signals), stained with an SYBR® Green—propidium iodide staining mix. Archaea presence was verified with 16S rRNA gene amplicon sequencing of the V4 region, on which methanogenic Archaea and methane presence were coloured according to the presence or absence. Methane presence was verified through methane production. The total SHIME transit times were adapted into three categories, namely short (21 h), medium (42 h) and long (63 h). Ordination was performed with principal coordinates analysis (PCoA) based on Bray–Curtis dissimilarity.
In addition to a snapshot moment during stabilisation, the methanogen detection tool was also evaluated in the SHIME start-up phase (i.e., before pumps are initiated), during stabilisation, and when a stabilised microbial community was achieved of donors 1 and 4 in the distal colon. These donors were deliberately chosen because they contrasted in Figure 5. Donor 1 had no methanogenesis nor Archaea, while donor 4 had. The SHIME model enables longitudinal monitoring of the methanogenic Archaea fraction and microbial community through extension, which is critical in tracking the formation of the microbiome. Donor 1 showed no methane production throughout the entire experiment. However, in the long SHIME transit time, a proportional Archaea abundance of 0.5% was detected, which incidentally corresponded with a methanogen-positive classification of the flow cytometry tool (Figure 6). Donor 4 portrayed methane production and carried Archaea as determined through sequencing and flow cytometry (Figure 6A,B). Furthermore, the different methanogenic cell counts and SHIME phases clustered differently according to the absence or presence of methane and methanogenic Archaea. For example, in donor 1 in the long SHIME transit, the start-up phase was clustered differently from the other two phases. In addition, in the long transit, each SHIME phase was clustered differently in donor 4. This may indicate a shift in the microbial community (Figure 6C).
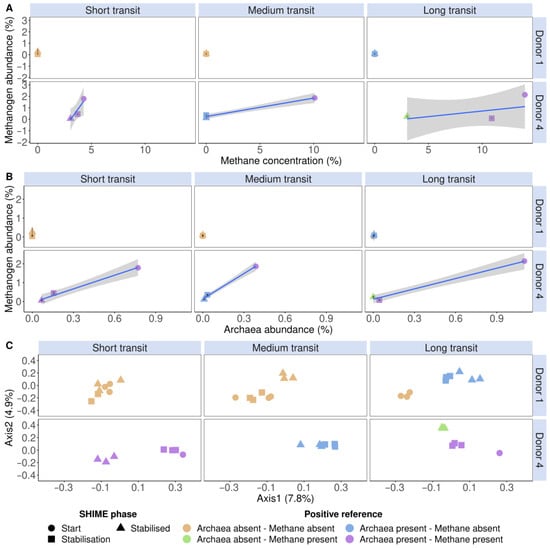
Figure 6.
The detected proportional methanogen counts (%) of donors 1 and 4 in the distal colon, stained with a SYBR® Green—propidium iodide staining mix, increased with methane concentration (%) (A) and proportional Archaea abundance (%) (B). (C) Community β-diversity assessment of the flow cytometric SHIME fingerprint data, stained with a SYBR® Green—propidium iodide staining mix, clustered according to methanogenic Archaea presence and methane presence. The fingerprint was based on the isolated methanogen information of six phenotypic parameters (four fluorescence and two scatter signals). Archaea presence was verified with 16S rRNA gene amplicon sequencing of the V4 region, on which methanogenic Archaea and methane presence were coloured according to the presence or absence. Methane presence was verified using methane production. The total SHIME transit times were adapted into three categories, namely short (21 h), medium (42 h) and long (63 h). Ordination was performed with principal coordinates analysis (PCoA) based on Bray–Curtis dissimilarity.
4. Discussion
Flow cytometry-based detection of methanogen-specific F420 autofluorescence was highly reproducible, accurate (71–100%) and sensitive (65–100%) in proof-of-concept studies with faecal slurries and in vitro incubated faecal slurries from different healthy individuals. F420-based methanogen detection was moreover compatible with widely used staining methods to measure total (SG) or viable and permeabilised (SGPI) cell counts in faecal and in vitro incubated faecal samples [43,44,45,53,54,55]. Despite a limited dataset of 10 donors, as a proof-of-concept, this technique demonstrates potential as a methanogen detection method.
A slight cell loss was observed, which may be due to cell death and disintegration during the flow cytometry measurement delay after spike-in. The correlations between spiked-in M. smithii cell counts and detected methanogenic Archaea abundances were higher with SG compared to SGPI staining. This was also reflected in higher accuracies and overall model performance scores in SG-stained samples. The presence of viability in SGPI-stained samples could explain this reduced resolution. A possible explanation is that PI also stains extracellular nucleic acids, which can cause more “noise” to be perceived [56].
Despite the strong correlations, the methanogen counts from flow cytometry were always lower than the copy-number corrected counts from 16S rRNA gene sequencing. For example, in the donors with the highest methanogenic populations, donors 1 and 3 counted more than 3.8-fold fewer methanogens than those detected via universal primers. This discrepancy is probably because cells are less stable than DNA, and flow cytometry-based single-cell detection is likely affected by cell disintegration or deterioration of cofactor F420 in damaged cells. Roughly 61 and 76% of the microbial cells in donors 1 and 3, respectively, were intact. Yet, in the same donors, 97 and 98% of the counted methanogens were intact, which supports the hypothesis. Assuming that the amount of methanogens is equally spread over the intact and damaged fractions, 39 and 24% of methanogens were not counted in donors 1 and 3, respectively. Taking into account this factor, the actual methanogen counts would be 1.03 × 1010 ± 1.12 × 109 and 7.37 × 109 ± 3.77 × 108 cells mL−1, which is closer to the 1.66 × 1010 ± 2.03 × 109 and 1.71 × 1010 ± 6.37 × 108 cells per mL measured with copy-number corrected 16S rRNA gene amplicon sequencing.
Despite the increased DNA stability, which allows for the quantification of the damaged/dead as well as viable Archaea population, standard sequencing with universal primers is less appropriate for the identification of methanogens occurring at lower abundances. This is due to the detection limit of standard amplicon sequencing. The term “universal primers” can be misleading, as no primer or binding site can truly target all taxa. A nested primer design enriching Archaea prior to 16S rRNA gene amplicon sequencing identified two additional Archaea-carrying donors, namely 2 and 5. In a study by Pausan et al., 2019, who performed an extensive primer evaluation for the characterisation of the gastrointestinal archaeome, they uncovered that universal primers were mostly unsuitable [57]. While their best Archaeal primer pairs detected 81 Archaeal ribosomal sequence variants, and the average Archaeal primer pairs (n = 32) identified more than 17, their best universal primer pair (n = 4) only detected 2 variants. The underestimation of Archaea presence with universal primers may also be the reason why SHIME samples (515F—806R on the V4 region instead of U341F—U806R on the V3-V4 region in faecal samples) portrayed a lower accuracy of 75%. As a result, the SG-stained samples detect more methanogens than the SGPI-stained samples, but the latter is still equal to the presence of methanogens detected through universal primers.
Interestingly, the methanogenic cell counts and SHIME phases clustered differently according to methane production and Archaea presence. The highest observed flow cytometry methanogen counts always corresponded to both archaeal and methanogen presence, while lower counts corresponded to lower methane production or the absence of methane altogether. However, the lack of methane does not equate to the lack of methanogens but could rather be the lack of methanogenic activity, which was confirmed in our experiment.
Methane was detected in the stabilised community in the long SHIME transit of donor 4, but no Archaea were detected by sequencing in this sample. Additionally, this particular sample exhibited a distinct clustering pattern compared to the other samples. This was also associated with a small increase in detected methanogen counts. A recent discovery revealed how methane production can also occur in microorganisms other than Archaea, specifically in the presence of reactive oxygen species [58]. The researchers also noted how the population growth and methane formation ceased with oxygen depletion. As oxygen concentrations are extremely low, methane production via these pathways is highly unlikely in the colon environment. We can, therefore, assume that methane production in the SHIME and the gut is predominantly accomplished through methanogenic Archaea. A potential explanation as to why methane was detected but not Archaea is the washout of methanogens in the SHIME suspension while methane remained in the gas fraction. This is also confirmed by the higher Archaeal abundances during the start and stabilisation phase.
5. Conclusions
Methanogens could be detected and quantified with flow cytometry by measuring F420-autofluorescence. These results were confirmed using archaeal amplicon sequencing and methane measurements. With a Spearman’s rank correlation coefficient of 0.83 and model accuracy of 90%, staining with SG outperformed SGPI (ρ = 0.69, accuracy = 80%). Nevertheless, methanogen classification and quantification with SGPI were still equivalent to the detection with universal primers. Methanogen classification in in vitro incubated faecal samples portrayed an accuracy of 71 and 75% when benchmarked against methane and Archaea presence as reference methods, respectively. In addition, phenotypic fingerprinting clustered the Archaeal positive samples consistently together over the Archaeal negative samples. These results indicate that employing flow cytometry to detect and quantify methanogens is promising and recommended in gut microbial research.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/applmicrobiol4010012/s1, Figure S1: Visualisation of cofactor F420 auto fluorescence (gated in red) in Blue1 (452 nm) and Blue2 (500 nm) fluorescence channels as a function of the Green (519 nm) fluorescence channels, Blue2 fluorescence as a function of the Blue1 fluorescence channels and the microbial community visualisation in the forward scatter as a function of the sidewards scatter channels of technical replicates of a faecal sample derived from donor 1 to 10. The replicates were stained with either SYBR® Green (left) or a SYBR® Green–propidium iodide staining mix (right). The proportional (left y-axis) and absolute (right y-axis) cell counts are depicted as a bar chart on the right, with the detected methanogen population in the filtered sample depicted as a horizontal line. The absolute methanogen population below and above the filtered sample population is depicted in blue and red, respectively; Figure S2: (A) Number of unique taxa in the 16S rRNA gene amplicon sequencing samples. (B) Proportional abundance (%) of the 15 most abundant genera in all samples. Less abundant genera are pooled into “Other”. The V3-V4 region of 16S rRNA gene was amplified with either universal (Univ, U341F–U806) or Archaeal (Arch, 340F–1000R, nested with U341F–U806R) primers. Higher level taxa are to be interpreted as unclassified genus belonging to the respective taxon; Figure S3: Rarefaction curves of 16S rRNA gene sequence count data. The V3-V4 region of 16S rRNA gene was amplified with either universal (Univ, U341F–U806) or Archaeal (Arch, 340F–1000R, nested with U341F–U806R) primers. The raw data corresponding to the sample identifiers can be found in the EBI ENA submission (ERP138722); Figure S4: Flow cytometric visualisation and gating of the cofactor F420 auto fluorescence (gated in red) in the Blue2 (500 nm) fluorescence channel as a function of the Blue1 (452 nm) fluorescence channel and community visualisation in the forward scatter channel in function of the sideward scatter channel. The one-dimensional histograms of the 452 and 500 nm emissions are depicted below the density plots with the F420 auto fluorescence peaks in red. The samples were stained with either SYBR® Green–propidium iodide (A) or SYBR® Green (B), of a faecal sample, pure M. smithii culture and faecal sample spiked with M. smithii; Figure S5: (A) The total percentage of intact cells. Methanogen cell counts (B) of all samples stained with Sybr Green–Propidium iodide (purple), and its subsequent intact fraction (green) and damaged fraction (orange); Figure S6: Community β diversity assessment of the flow cytometric fingerprint data of all cellular information based on six phenotypic parameters (4 fluorescence and 2 scatter signals), stained with SYBR® Green. Ordination was performed with principal coordinates analysis (PCoA) based on Bray-Curtis dissimilarity; Table S1: Components of DMSZ medium 1523. Modified Methanobacterium medium (DSMZ, 2015); Table S2: Components of trace element solution SL-10, derived from DSMZ medium 320 [2]; Table S3: Components in the seven vitamins solution, derived from DSMZ medium 503 [3]; Table S4: Reducing anaerobic phosphate buffer (pH 6.8) was sparged and flushed with N2-gas prior to autoclaving. Table S5: Classification measures (accuracy, p-value, sensitivity, specificity, precision, F1-score and positive and negative predictive values) of the methanogen classification method on ten faecal samples, stained with either SGPI or SG, as verified with 16S rRNA gene amplicon (16S) Archaea abundances, sequenced with either universal or Archaeal primers. Methanogen classifications were marked as positive when methanogen cell counts were higher than both the sum of the average, sample wide, filtered background noise and standard deviation, and the sample specific filtered background noise; Table S6: Confusion matrices of the predicted methanogen classification method (FCM) on ten faecal samples, stained with either SGPI or SG, as verified with 16S rRNA gene amplicon (16S) Archaea abundances, sequenced with either universal or Archaeal primers. Methanogen classifications were marked as positive when methanogen cell counts were higher than both the sum of the average, sample wide, filtered background noise and standard deviation, and the sample specific filtered background noise; Table S7: Classification measures (A) (accuracy, p-value, sensitivity, specificity, precision, F1-score and positive and negative predictive values) and confusion matrix (B) of the predicted methanogen classification method (FCM) on 241 SHIME samples, stained with SGPI and verified with the presence of methane (CH4) in SHIME headspace and the presence of methanogenic Archaea. Methanogen classifications were marked as positive when methanogen cell counts were higher than the average, sample wide, filtered background noise and standard deviation.
Author Contributions
Y.M., T.V.d.W. and N.B. conceived the experiments. Y.M. recruited the donors, planned and performed the experiments, analysed its results and wrote the manuscript. K.D.P. and R.P. contributed to data interpretation. T.L. extracted the DNA samples and provided the 16S rRNA gene amplicon sequencing platform. K.D.P., T.V.d.W., T.L. and N.B. reviewed and edited the manuscript. The experiments were supervised by T.V.d.W. and N.B. All authors have read and agreed to the published version of the manuscript.
Funding
Y.M. is a recipient of a grant from the FWO (EOS program no. 30770923).
Institutional Review Board Statement
Research with human faecal material was conducted in accordance with the ethical approval obtained from the Ethical Committee of the Ghent University hospital Hospital (B670201836318).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The amplicon sequencing data were submitted to the European Bioinformatics Institute’s (EBI) European Nucleotide Archive (ENA) under accession number PRJEB53907. The acquired flow cytometry fcs files were submitted to the FlowRepository archive under repository ID FR-FCM-Z5KA.
Acknowledgments
We express our gratitude to Ghent University’s Special Research Fund for their support in acquiring the flow cytometer (BOF.BAS.2022.0014.01).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ver Eecke, H.C.; Butterfield, D.A.; Huber, J.A.; Lilley, M.D.; Olson, E.J.; Roe, K.K.; Evans, L.J.; Merkel, A.Y.; Cantin, H.V.; Holden, J.F. Hydrogen-limited growth of hyperthermophilic methanogens at deep-sea hydrothermal vents. Proc. Natl. Acad. Sci. USA 2012, 109, 13674–13679. [Google Scholar] [CrossRef] [PubMed]
- Gaci, N.; Borrel, G.; Tottey, W.; O’Toole, P.W.; Brugère, J.-F. Archaea and the human gut: New beginning of an old story. World J. Gastroenterol. WJG 2014, 20, 16062. [Google Scholar] [CrossRef] [PubMed]
- Angle, J.C.; Morin, T.H.; Solden, L.M.; Narrowe, A.B.; Smith, G.J.; Borton, M.A.; Rey-Sanchez, C.; Daly, R.A.; Mirfenderesgi, G.; Hoyt, D.W.; et al. Methanogenesis in oxygenated soils is a substantial fraction of wetland methane emissions. Nat. Commun. 2017, 8, 1567. [Google Scholar] [CrossRef] [PubMed]
- Kumpitsch, C.; Fischmeister, F.P.S.; Mahnert, A.; Lackner, S.; Wilding, M.; Sturm, C.; Springer, A.; Madl, T.; Holasek, S.; Högenauer, C.; et al. Reduced B12 uptake and increased gastrointestinal formate are associated with archaeome-mediated breath methane emission in humans. Microbiome 2021, 9, 193. [Google Scholar] [CrossRef] [PubMed]
- Dridi, B.; Henry, M.; El Khéchine, A.; Raoult, D.; Drancourt, M. High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS ONE 2009, 4, e7063. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and Fungi of the Human Gut Microbiome: Correlations with Diet and Bacterial Residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef] [PubMed]
- Chibani, C.M.; Mahnert, A.; Borrel, G.; Almeida, A.; Werner, A.; Brugère, J.-F.; Gribaldo, S.; Finn, R.D.; Schmitz, R.A. A catalogue of 1167 genomes from the human gut archaeome. Nat. Microbiol. 2021, 7, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Hoegenauer, C.; Hammer, H.F.; Mahnert, A.; Moissl-Eichinger, C. Methanogenic archaea in the human gastrointestinal tract. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 805–813. [Google Scholar] [CrossRef]
- Vanderhaeghen, S.; Lacroix, C.; Schwab, C. Methanogen communities in stools of humans of different age and health status and co-occurrence with bacteria. FEMS Microbiol. Lett. 2015, 362, 92. [Google Scholar] [CrossRef]
- Borrel, G.; McCann, A.; Deane, J.; Neto, M.C.; Lynch, D.B.; Brugère, J.F.; O’Toole, P.W. Genomics and metagenomics of trimethylamine-utilizing Archaea in the human gut microbiome. ISME J. 2017, 11, 2059–2074. [Google Scholar] [CrossRef]
- Basseri, R.J.; Basseri, B.; Pimentel, M.; Chong, K.; Youdim, A.; Low, K.; Hwang, L.; Soffer, E.; Chang, C.; Mathur, R. Intestinal Methane Production in Obese Individuals Is Associated with a Higher Body Mass Index. Gastroenterol. Hepatol. 2012, 8, 22–28. [Google Scholar]
- Ghavami, S.B.; Rostami, E.; Sephay, A.A.; Shahrokh, S.; Balaii, H.; Aghdaei, H.A.; Zali, M.R. Alterations of the human gut Methanobrevibacter smithii as a biomarker for inflammatory bowel diseases. Microb. Pathog. 2018, 117, 285–289. [Google Scholar] [CrossRef]
- Coker, O.O.; Wu, W.K.K.; Wong, S.H.; Sung, J.J.; Yu, J. Altered Gut Archaea Composition and Interaction With Bacteria Are Associated With Colorectal Cancer. Gastroenterology 2020, 159, 1459–1470.e5. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.W.; Shorten, P.R.; Altermann, E.H.; Roy, N.C.; McNabb, W.C. Hydrogen cross-feeders of the human gastrointestinal tract. Gut Microbes 2019, 10, 270. [Google Scholar] [CrossRef] [PubMed]
- Bang, C.; Schmitz, R.A. Archaea: Forgotten players in the microbiome. Emerg. Top. Life Sci. 2018, 2, 459–468. [Google Scholar]
- Dridi, B.; Fardeau, M.-L.; Ollivier, B.; Raoult, D.; Drancourt, M. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2012, 62, 1902–1907. [Google Scholar] [CrossRef]
- Miller, T.L.; Wolin, M.J. Methanosphaera stadtmaniae gen. nov., sp. nov.: A species that forms methane by reducing methanol with hydrogen. Arch. Microbiol. 1985, 141, 116–122. [Google Scholar] [CrossRef]
- Karakashev, D.; Batstone, D.J.; Trably, E.; Angelidaki, I. Acetate oxidation is the dominant methanogenic pathway from acetate in the absence of Methanosaetaceae. Appl. Environ. Microbiol. 2006, 72, 5138–5141. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.P.; Conway, P.L.; Schlundt, J. Methanogens in humans: Potentially beneficial or harmful for health. Appl. Microbiol. Biotechnol. 2018, 102, 3095–3104. [Google Scholar] [CrossRef]
- Söllinger, A.; Urich, T. Methylotrophic methanogens everywhere—Physiology and ecology of novel players in global methane cycling. Biochem. Soc. Trans. 2019, 47, 1895–1907. [Google Scholar] [CrossRef]
- Dorokhov, Y.L.; Shindyapina, A.V.; Sheshukova, E.V.; Komarova, T.V. Metabolic methanol: Molecular pathways and physiological roles. Physiol. Rev. 2015, 95, 603–644. [Google Scholar] [CrossRef]
- Oliphant, K.; Allen-Vercoe, E. Macronutrient metabolism by the human gut microbiome: Major fermentation by-products and their impact on host health. Microbiome 2019, 7, 91. [Google Scholar] [CrossRef]
- Dridi, B. Laboratory tools for detection of archaea in humans. Clin. Microbiol. Infect. 2012, 18, 825–833. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vlasova, A.V.; Isakov, V.A.; Pilipenko, V.I.; Sheveleva, S.A.; Markova, Y.M.; Polyanina, A.S.; Maev, I.V. Methanobrevibacter smithii in irritable bowel syndrome: A clinical and molecular study. Ter. Arkhiv 2019, 91, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Erdrich, S.; Tan, E.C.K.; Hawrelak, J.A.; Myers, S.P.; Harnett, J.E. Hydrogen–methane breath testing results influenced by oral hygiene. Sci. Rep. 2021, 11, 26. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, P.; Toms-Wood, A.; Wolfe, R.S. Isolation and Properties of a Fluorescent Compound, Factor420, from Methanobacterium Strain M.o.H. J. Bacteriol. 1972, 112, 527. [Google Scholar] [CrossRef]
- Eirich, L.D.; Vogels, G.D.; Wolfe, R.S. Distribution of coenzyme F420 and properties of its hydrolytic fragments. J. Bacteriol. 1979, 140, 20. [Google Scholar] [CrossRef] [PubMed]
- Grinter, R.; Greening, C. Cofactor F420: An expanded view of its distribution, biosynthesis and roles in bacteria and archaea. FEMS Microbiol. Rev. 2021, 45, fuab021. [Google Scholar] [CrossRef]
- Lambrecht, J.; Cichocki, N.; Hübschmann, T.; Koch, C.; Harms, H.; Müller, S. Flow cytometric quantification, sorting and sequencing of methanogenic archaea based on F420 autofluorescence. Microb. Cell Fact. 2017, 16, 180. [Google Scholar] [CrossRef]
- Bellais, S.; Nehlich, M.; Ania, M.; Duquenoy, A.; Mazier, W.; Engh, G.v.D.; Baijer, J.; Treichel, N.S.; Clavel, T.; Belotserkovsky, I.; et al. Species-targeted sorting and cultivation of commensal bacteria from the gut microbiome using flow cytometry under anaerobic conditions. Microbiome 2022, 10, 24. [Google Scholar] [CrossRef]
- Props, R.; Monsieurs, P.; Mysara, M.; Clement, L.; Boon, N. Measuring the biodiversity of microbial communities by flow cytometry. Methods Ecol. Evol. 2016, 7, 1376–1385. [Google Scholar] [CrossRef]
- Balch, W.E.; Fox, G.E.; Magrum, L.J.; Woese, C.R.; Wolfe, R.S. Methanogens: Reevaluation of a unique biological group. Microbiol. Rev. 1979, 43, 260–296. [Google Scholar] [CrossRef]
- DSMZ. Medium Number 1523. Modified Methanobacterium Medium. Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH. List of Recommended Media for Microorganisms. 2015. Available online: https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium1523.pdf (accessed on 16 June 2022).
- DSMZ. Medium Number 320. Clostridium cellulovorans Medium. Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH. List of Recommended Media for Microorganisms. 2020. Available online: https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium320.pdf (accessed on 16 June 2022).
- DSMZ. Medium Number 503. Anaerobic Freshwater (fwm) Medium. Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH. List of Recommended Media for Microorganisms. 2022. Available online: https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium503.pdf (accessed on 16 June 2022).
- De Boever, P.; Deplancke, B.; Verstraete, W. Fermentation by Gut Microbiota Cultured in a Simulator of the Human Intestinal Microbial Ecosystem Is Improved by Supplementing a Soygerm Powder. J. Nutr. 2000, 130, 2599–2606. [Google Scholar] [CrossRef] [PubMed]
- Van, P.; Jiang, W.; Gottardo, R.; Finak, G. ggCyto: Next generation open-source visualization software for cytometry. Bioinformatics 2018, 34, 3951–3953. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 8 January 2024).
- Geirnaert, A.; Wang, J.; Tinck, M.; Steyaert, A.; Abbeele, P.V.D.; Eeckhaut, V.; Vilchez-Vargas, R.; Falony, G.; Laukens, D.; De Vos, M.; et al. Interindividual differences in response to treatment with butyrate-producing Butyricicoccus pullicaecorum 25-3T studied in an in vitro gut model. FEMS Microbiol. Ecol. 2015, 91, fiv054. [Google Scholar] [CrossRef] [PubMed]
- De Paepe, K.; Kerckhof, F.; Verspreet, J.; Courtin, C.M.; Van de Wiele, T. Inter-individual differences determine the outcome of wheat bran colonization by the human gut microbiome. Environ. Microbiol. 2017, 19, 3251–3267. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic. Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Gantner, S.; Andersson, A.F.; Alonso-Sáez, L.; Bertilsson, S. Novel primers for 16S rRNA-based archaeal community analyses in environmental samples. J. Microbiol. Methods 2011, 84, 12–18. [Google Scholar] [CrossRef]
- Minnebo, Y.; Delbaere, K.; Goethals, V.; Raes, J.; Van de Wiele, T.; De Paepe, K. Gut microbiota response to in vitro transit time variation is mediated by microbial growth rates, nutrient use efficiency and adaptation to in vivo transit time. Microbiome 2023, 11, 240. [Google Scholar] [CrossRef]
- Vandeputte, D.; Kathagen, G.; D’Hoe, K.; Vieira-Silva, S.; Valles-Colomer, M.; Sabino, J.; Wang, J.; Tito, R.Y.; De Commer, L.; Darzi, Y.; et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 2017, 551, 507–511. [Google Scholar] [CrossRef]
- Minnebo, Y.; De Paepe, K.; Raes, J.; Van de Wiele, T. Nutrient load acts as a driver of gut microbiota load, community composition and metabolic functionality in the simulator of the human intestinal microbial ecosystem. FEMS Microbiol. Ecol. 2021, 97, 111. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis. 2016. Available online: https://ggplot2.tidyverse.org/ (accessed on 8 January 2024).
- Kassambara, A. ggpubr: ‘ggplot2′ Based Publication Ready Plots. R Package Version 0.2.4.999. 2019. Available online: https://rpkgs.datanovia.com/ggpubr/ (accessed on 8 January 2024).
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic. Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.5-6. 2019. Available online: https://cran.r-project.org/package=vegan (accessed on 8 January 2024).
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Geirnaert, A.; Calatayud, M.; Grootaert, C.; Laukens, D.; Devriese, S.; Smagghe, G.; De Vos, M.; Boon, N.; Van De Wiele, T. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep. 2017, 7, 11450. [Google Scholar] [CrossRef]
- Van Herreweghen, F.; De Paepe, K.; Roume, H.; Kerckhof, F.-M.; Van de Wiele, T. Mucin degradation niche as a driver of microbiome composition and Akkermansia muciniphila abundance in a dynamic gut model is donor independent. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, D.; De Commer, L.; Tito, R.Y.; Kathagen, G.; Sabino, J.; Vermeire, S.; Faust, K.; Raes, J. Temporal variability in quantitative human gut microbiome profiles and implications for clinical research. Nat. Commun. 2021, 12, 6740. [Google Scholar] [CrossRef]
- Rosenberg, M.; Azevedo, N.F.; Ivask, A. Propidium iodide staining underestimates viability of adherent bacterial cells. Sci. Rep. 2019, 9, 6483. [Google Scholar] [CrossRef]
- Pausan, M.R.; Csorba, C.; Singer, G.; Till, H.; Schöpf, V.; Santigli, E.; Klug, B.; Högenauer, C.; Blohs, M.; Moissl-Eichinger, C. Exploring the Archaeome: Detection of Archaeal Signatures in the Human Body. Front. Microbiol. 2019, 10, 2796. [Google Scholar] [CrossRef]
- Ernst, L.; Steinfeld, B.; Barayeu, U.; Klintzsch, T.; Kurth, M.; Grimm, D.; Dick, T.P.; Rebelein, J.G.; Bischofs, I.B.; Keppler, F. Methane formation driven by reactive oxygen species across all living organisms. Nature 2022, 603, 482–487. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).