Comparative Analysis of Ligninolytic Potential among Pleurotus ostreatus and Fusarium sp. with a Special Focus on Versatile Peroxidase
Abstract
Highlights
- The lignin in paddy and wheat straws is highly resistant to degradation, and burning the same is a global warming issue.
- Dye degrading ability and expression of three vp genes were studied in Pleurotus ostreatus and Fusarium sp. cultures.
- High enzymatic activity and the vp2 gene were observed in the Fusarium sp. culture, which makes it an alternate fungal species for lignin degradation.
Abstract
1. Introduction
2. Method and Materials
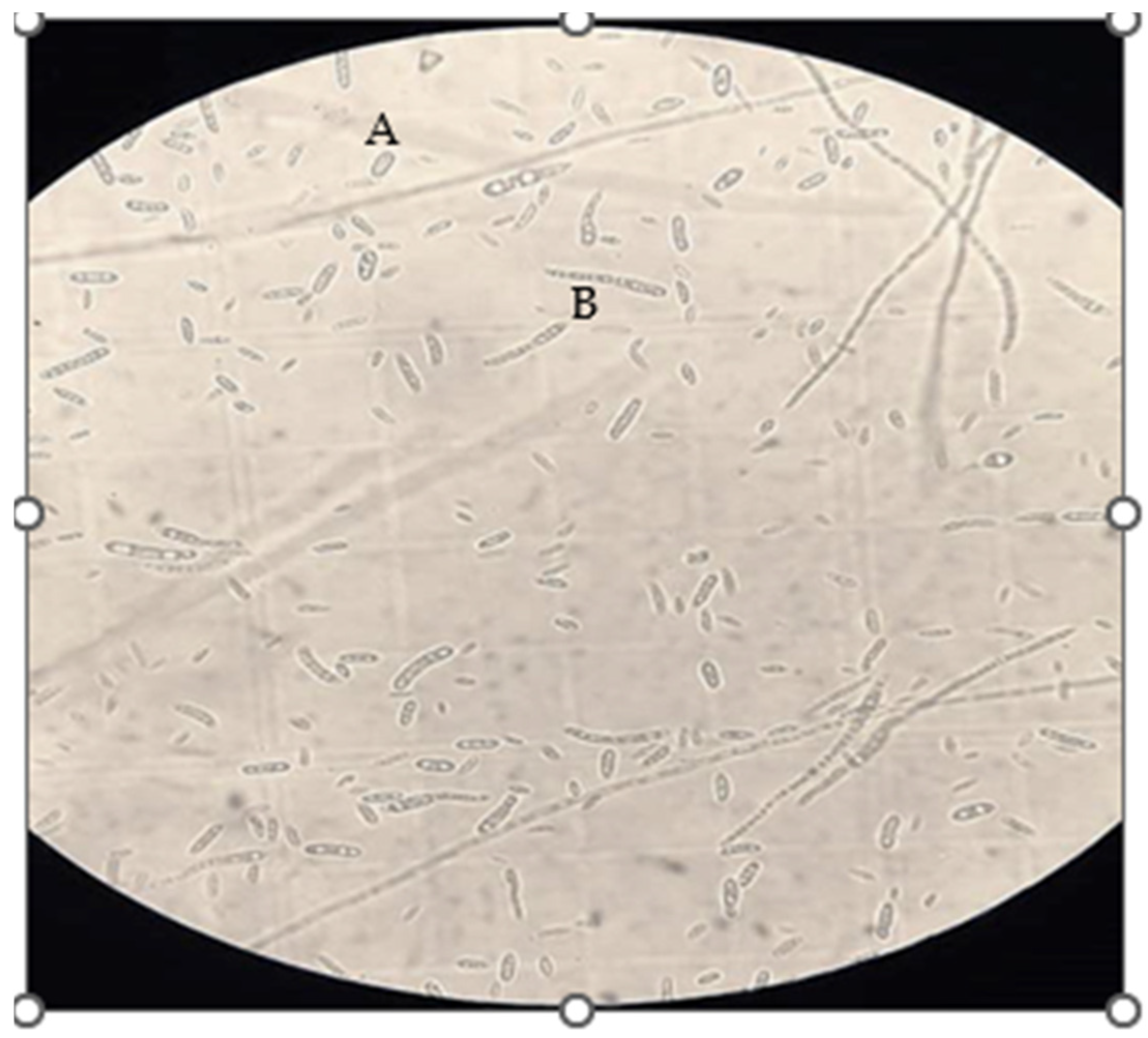
2.1. Fungal Isolation
2.2. Qualitative Screening
2.3. Quantitative Estimation of Enzyme Activity
2.4. Analytical Procedure
- Z = Enzyme activity (UL−1); ∆A: is change in OD per unit of time;
- V = assay volume in mL;
- ∆t = change in time (minutes); ε = Extinction Coefficient (M−1cm−1); and
- d = path length (cm).
2.5. DNA Isolation and Molecular Identification
3. Results and Discussion
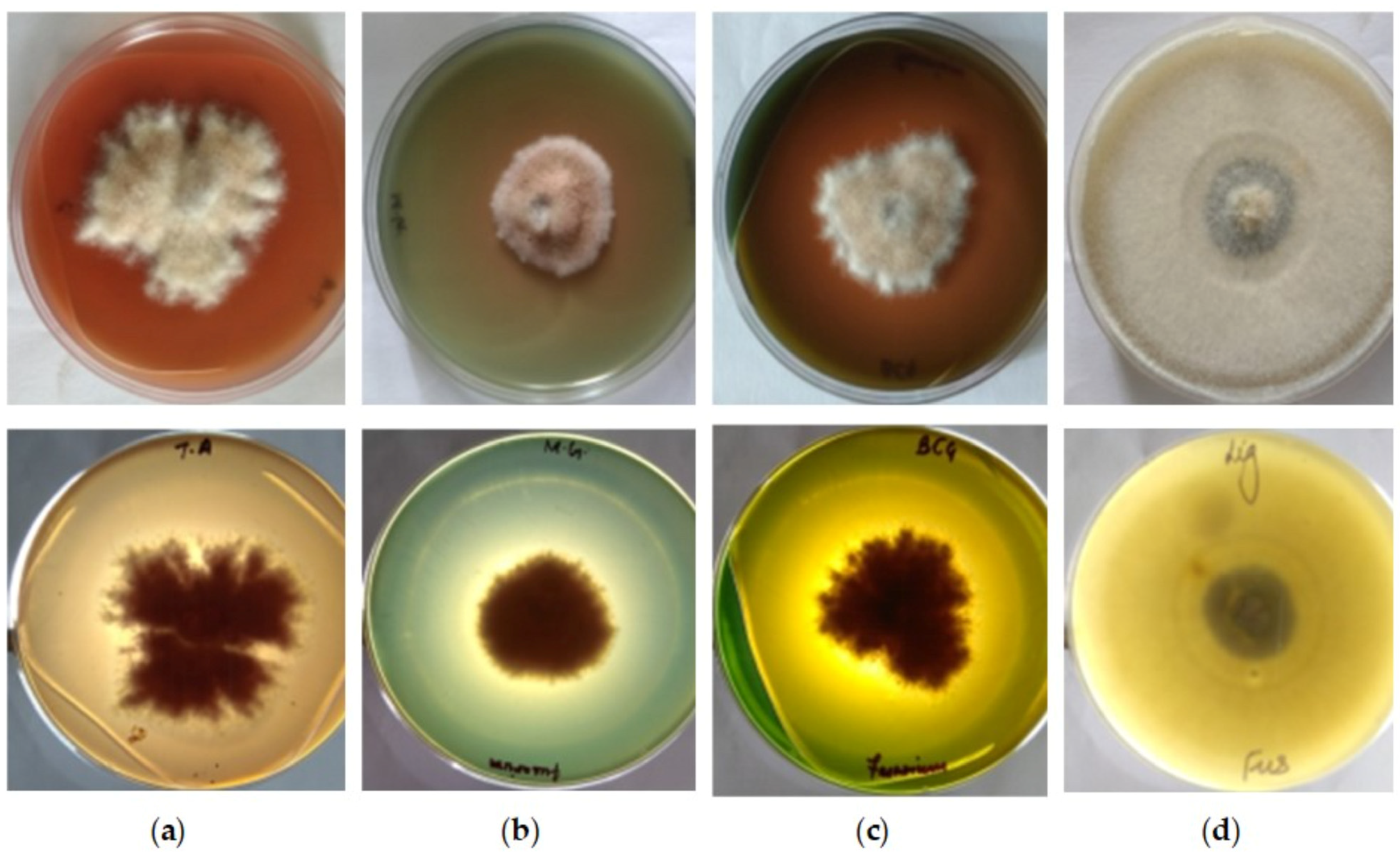
3.1. Qualitative Estimation of Enzyme Production
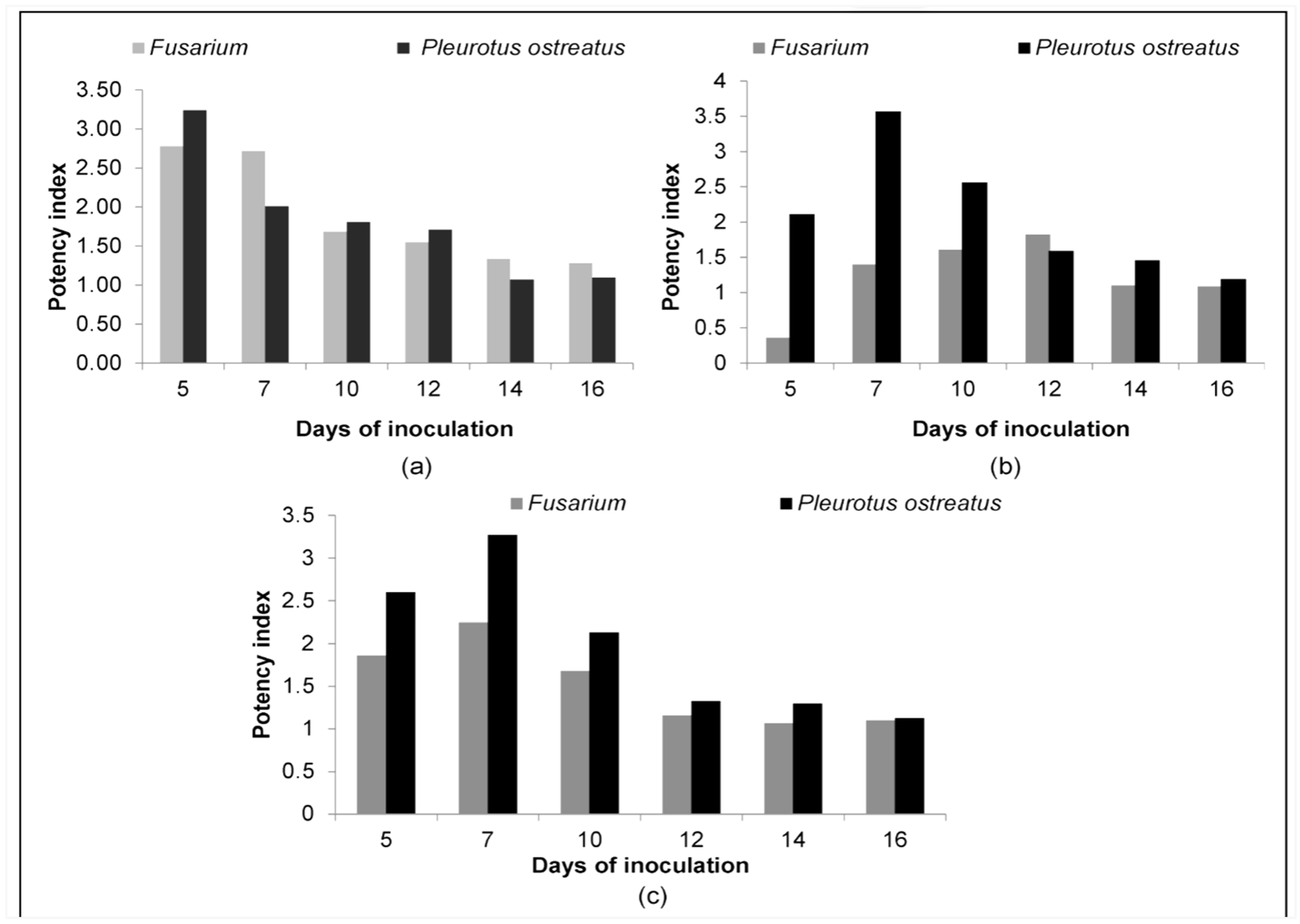
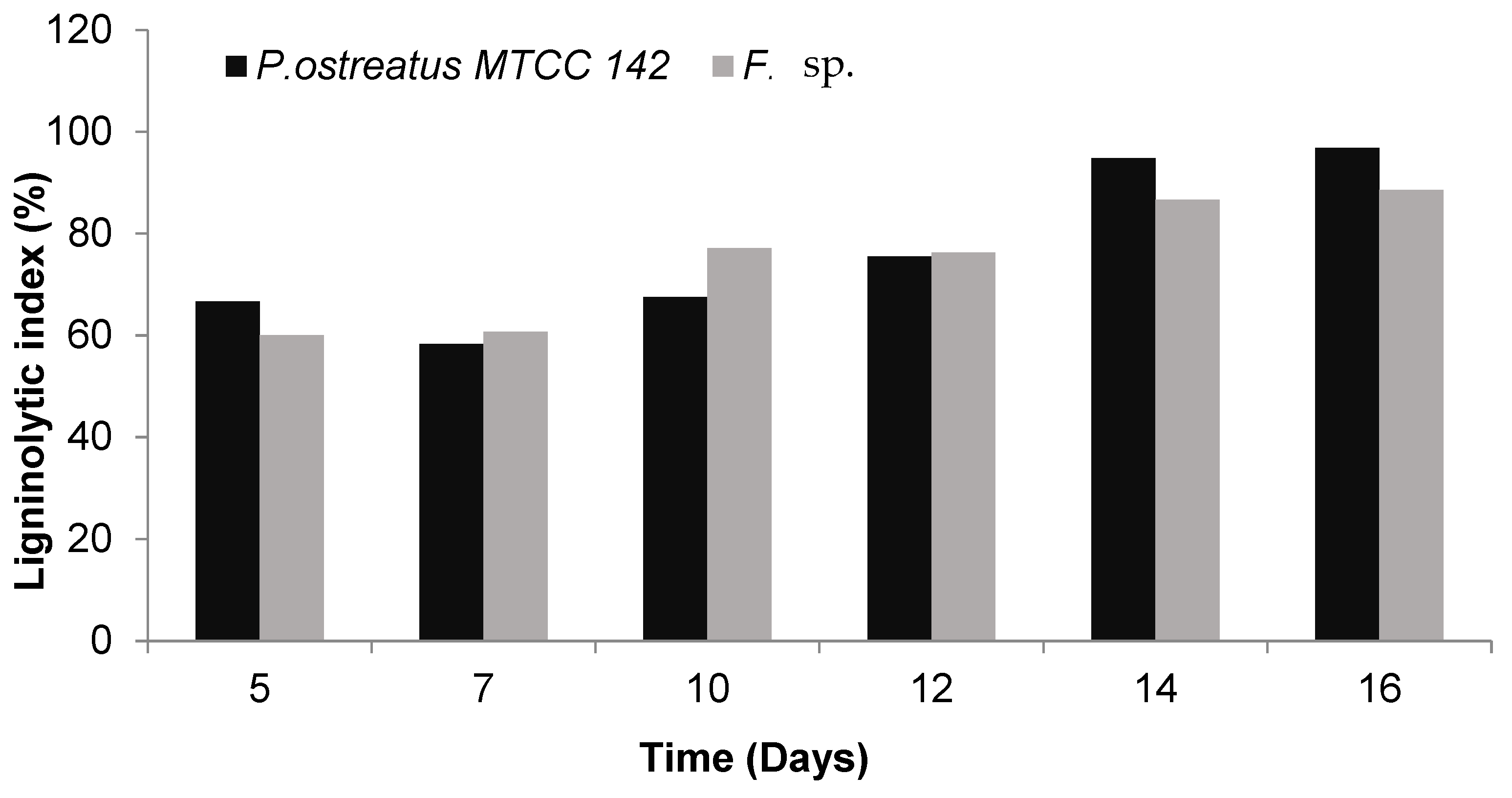
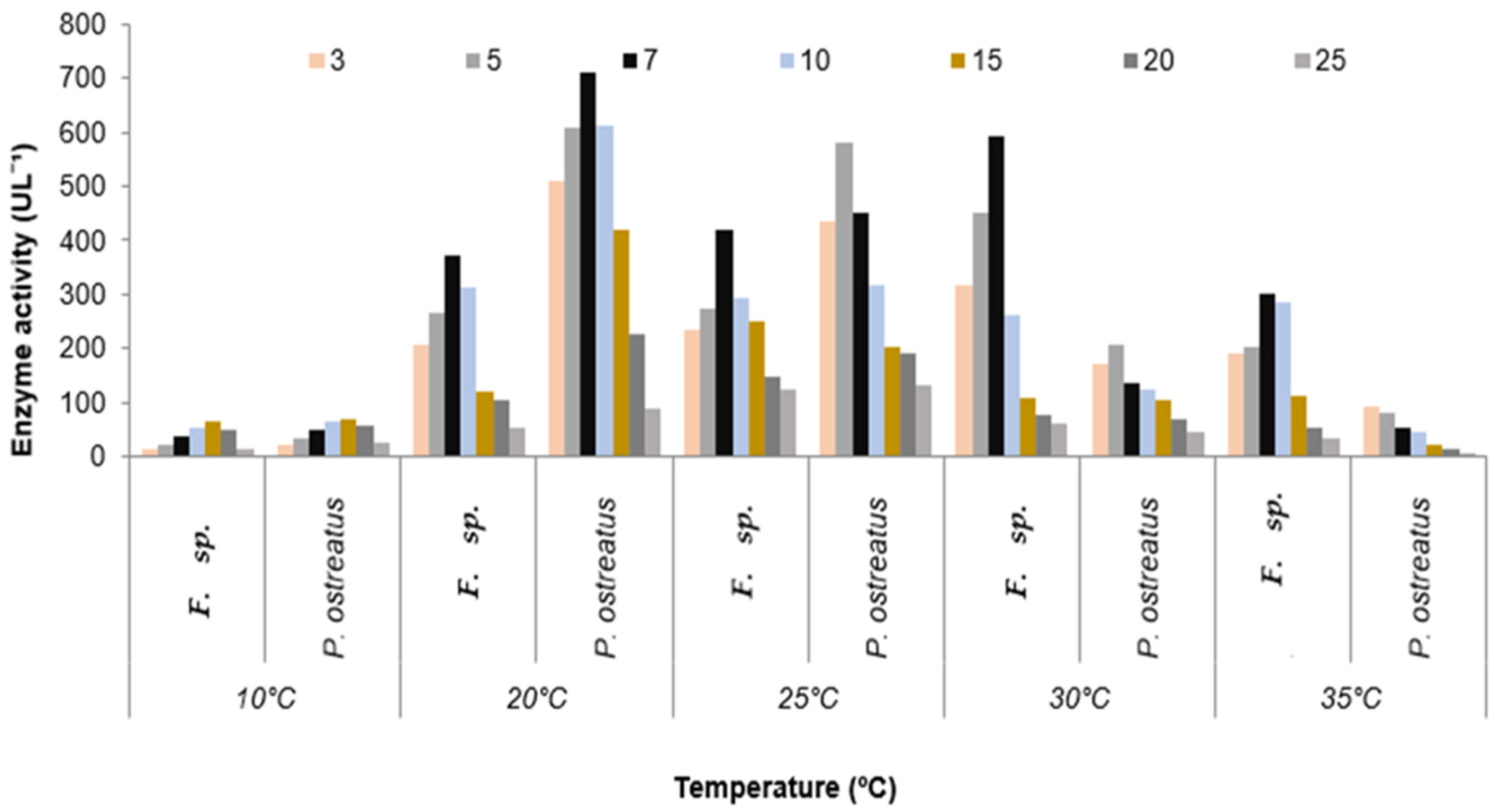
3.2. Quantitative Estimation of Enzyme Production
3.3. Confirmation of VP Gene Specific Sequences
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Marriott, P.E.; Gómez, L.D.; McQueen-Mason, S.J. Unlocking the Potential of Lignocellulosic Biomass through Plant Science. New Phytol. 2015, 209, 1366–1381. [Google Scholar] [CrossRef] [PubMed]
- Ahorsu, R.; Medina, F.; Constantí, M. Significance and Challenges of Biomass as a Suitable Feedstock for Bioenergy and Biochemical Production: A Review. Energies 2018, 11, 3366. [Google Scholar] [CrossRef]
- Satlewal, A.; Agrawal, R.; Bhagia, S.; Das, P.; Ragauskas, A.J. Rice Straw as a Feedstock for Biofuels: Availability, Recalcitrance, and Chemical Properties. Biofuels Bioprod. Biorefining 2017, 12, 83–107. [Google Scholar] [CrossRef]
- Tu, W.-C.; Hallett, J.P. Recent Advances in the Pretreatment of Lignocellulosic Biomass. Curr. Opin. Green Sustain. Chem. 2019, 20, 11–17. [Google Scholar] [CrossRef]
- Pandey, P. RRI’s Commitment to Care and Vulnerability of Agrarian Systems: The Problem of Rice Straw Burning in India. Sci. Technol. 2020, 25, 240–255. [Google Scholar] [CrossRef]
- Anwar, Z.; Gulfraz, M.; Irshad, M. Agro-Industrial Lignocellulosic Biomass a Key to Unlock the Future Bio-Energy: A Brief Review. J. Radiat. Res. Appl. Sci. 2014, 7, 163–173. [Google Scholar] [CrossRef]
- Kaur, K.; Phutela, U.G. Enhancement of Paddy Straw Digestibility and Biogas Production by Sodium Hydroxide-Microwave Pretreatment. Renew. Energy 2016, 92, 178–184. [Google Scholar] [CrossRef]
- Giummarella, N.; Pu, Y.; Ragauskas, A.J.; Lawoko, M. A Critical Review on the Analysis of Lignin Carbohydrate Bonds. Green Chem. 2019, 21, 1573–1595. [Google Scholar] [CrossRef]
- Ayeni, A.O.; Adeeyo, O.A.; Oresegun, O.M.; Oladimeji, T.E. Compositional Analysis of Lignocellulosic Materials: Evaluation of an Economically Viable Method Suitable for Woody and Non-Woody Biomass. Am. J. Eng. Res. 2015, 4, 14–19. [Google Scholar]
- Alvira, P.; Tomás-Pejó, E.; Ballesteros, M.; Negro, M.J. Pretreatment Technologies for an Efficient Bioethanol Production Process Based on Enzymatic Hydrolysis: A Review. Bioresour. Technol. 2010, 101, 4851–4861. [Google Scholar] [CrossRef]
- Mosier, N.; Wyman, C.; Dale, B.; Elander, R.; Lee, Y.Y.; Holtzapple, M.; Ladisch, M. Features of Promising Technologies for Pretreatment of Lignocellulosic Biomass. Bioresour. Technol. 2005, 96, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Sindhu, R.; Binod, P.; Pandey, A. Biological Pretreatment of Lignocellulosic Biomass—An Overview. Bioresour. Technol. 2016, 199, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Saritha, M.; Arora, A. Lata Biological Pretreatment of Lignocellulosic Substrates for Enhanced Delignification and Enzymatic Digestibility. Indian J. Microbiol. 2011, 52, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Sahrawat, R.; Garg, A.P. Rice Straw Degradation with Mixed Cultures of Microfungi and Their Enzymes. Int. J. Environ. Clim. Chang. 2023, 13, 2097–2104. [Google Scholar] [CrossRef]
- Shi, J.; Chinn, M.; Sharmashivappa, R. Microbial Pretreatment of Cotton Stalks by Solid State Cultivation of Phanerochaete Chrysosporium. Bioresour. Technol. 2008, 99, 6556–6564. [Google Scholar] [CrossRef]
- Janusz, G.; Pawlik, A.; Sulej, J.; Świderska-Burek, U.; Jarosz-Wilkołazka, A.; Paszczyński, A. Lignin Degradation: Microorganisms, Enzymes Involved, Genomes Analysis and Evolution. FEMS Microbiol. Rev. 2017, 41, 941–962. [Google Scholar] [CrossRef]
- Sani, R.K.; Krishnaraj, R.N. Extremophilic Enzymatic Processing of Lignocellulosic Feedstocks to Bioenergy; Springer International Publishing: Cham, Switzerland, 2017. [Google Scholar]
- Kumar, A.; Chandra, R. Ligninolytic Enzymes and Its Mechanisms for Degradation of Lignocellulosic Waste in Environment. Heliyon 2020, 6, e03170. [Google Scholar] [CrossRef]
- Shahid, A.; Singh, J.; Bisht, S.; Teotia, P.; Kumar, V. Biodegradation of Textile Dyes by Fungi Isolated from North Indian Field Soil. EnvironmentAsia 2013, 6, 51–57. [Google Scholar]
- Ruiz-Dueñas, F.J.; Camarero, S.; Pérez-Boada, M.; Martínez, M.J.; Martínez, A.T. A New Versatile Peroxidase from Pleurotus. Biochem. Soc. Trans. 2001, 29, 116. [Google Scholar] [CrossRef]
- Pessôa, M.G.; Paulino, B.N.; Mano, M.C.R.; Neri-Numa, I.A.; Molina, G.; Pastore, G.M. Fusarium Species—A Promising Tool Box for Industrial Biotechnology. Appl. Microbiol. Biotechnol. 2017, 101, 3493–3511. [Google Scholar] [CrossRef]
- Gordon, T.R.; Martyn, R.D. The Evolutionary Biology of Fusarium oxysporum. Annu. Rev. Phytopathol. 1997, 35, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Escobosa, A.R.C.; Figueroa, J.A.L.; Corona, J.F.G.; Wrobel, K.; Wrobel, K. Effect of Fusarium Oxysporum f. Sp. Lycopersici on the Degradation of Humic Acid Associated with Cu, Pb, and Ni: An in Vitro Study. Anal. Bioanal. Chem. 2009, 394, 2267–2276. [Google Scholar] [CrossRef] [PubMed]
- Sellami, K.; Couvert, A.; Nasrallah, N.; Maachi, R.; Abouseoud, M.; Amrane, A. Peroxidase Enzymes as Green Catalysts for Bioremediation and Biotechnological Applications: A Review. Sci. Total Environ. 2022, 806, 150500. [Google Scholar] [CrossRef]
- Barber-Zucker, S.; Mindel, V.; Garcia-Ruiz, E.; Weinstein, J.J.; Alcalde, M.; Fleishman, S.J. Stable and Functionally Diverse Versatile Peroxidases Designed Directly from Sequences. J. Am. Chem. Soc. 2022, 144, 3564–3571. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Chundawat, T.S. Zinc Oxide Nanoparticles Synthesized Using Fusarium Oxysporum to Enhance Bioethanol Production from Rice-Straw. Biomass Bioenergy 2020, 143, 105840. [Google Scholar] [CrossRef]
- Alsohaili, S.A.; Bani-Hasan, B.M. Morphological and Molecular Identification of Fungi Isolated from Different Environmental Sources in the Northern Eastern Desert of Jordan. Jordan J. Biol. Sci. 2018, 3, 329–337. [Google Scholar]
- Choi, Y.W.; Hyde, K.D.; Ho, W.H. Single Spore Isolation of Fungi. Fungal Divers. 1999, 3, 29–38. [Google Scholar]
- Nelson, P.E.; Toussoun, T.A.; Marasas, W.F.O. Fusarium Species. In An Illustrated Manual for Identification; Pennsylvania State University Press: University Park, PA, USA, 1983. [Google Scholar]
- Knop, D.; Levinson, D.; Makovitzki, A.; Agami, A.; Lerer, E.; Mimran, A.; Yarden, O.; Hadar, Y. Limits of Versatility of Versatile Peroxidase. Appl. Environ. Microbiol. 2016, 82, 4070–4080. [Google Scholar] [CrossRef]
- Leslie, J.F.; Summerell, B.A. Fusarium Laboratory Workshops—A Recent History. Mycotoxin Res. 2006, 22, 73–74. [Google Scholar] [CrossRef]
- Fernández-Fueyo, E.; Castanera, R.; Ruiz-Dueñas, F.J.; López-Lucendo, M.F.; Ramírez, L.; Pisabarro, A.G.; Martínez, A.T. Ligninolytic Peroxidase Gene Expression by Pleurotus Ostreatus: Differential Regulation in Lignocellulose Medium and Effect of Temperature and pH. Fungal Genet. Biol. 2014, 72, 150–161. [Google Scholar] [CrossRef]
- Wu, Z.H.; Wang, T.H.; Huang, W.; Qu, Y.B. A Simplified Method for Chromosome DNA Preparation from Filamentous Fungi. Mycosystema 2001, 20, 575. [Google Scholar]
- Doyle, J.; Doyle, J. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Eichlerová, I.; Baldrian, P. Ligninolytic Enzyme Production and Decolorization Capacity of Synthetic Dyes by Saprotrophic White Rot, Brown Rot, and Litter Decomposing Basidiomycetes. J. Fungi 2020, 6, 301. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Sinha, N.; Sen, M.; Ghosh, D. Isolation and Screening of Dye Degrading Lignocellulolytic Bacteria from Sundarban Mangrove Ecosystem, West Bengal, India. J. Pure Appl. Microbiol. 2023, 17, 609–626. [Google Scholar] [CrossRef]
- Kunjadia, P.D.; Sanghvi, G.V.; Kunjadia, A.P.; Mukhopadhyay, P.N.; Dave, G.S. Role of Ligninolytic Enzymes of White Rot Fungi (Pleurotus spp.) Grown with Azo Dyes. SpringerPlus 2016, 5, 1487. [Google Scholar] [CrossRef]
- Ashktorab, H.; Cohen, R.J. Facile Isolation of Genomic DNA from Filamentous Fungi. PubMed 1992, 13, 198–200. [Google Scholar]
- Huy, N.D.; Ha, D.T.T.; Khoo, K.S.; Lan, P.T.N.; Quang, H.T.; Loc, N.H.; Park, S.-M.; Veeramuthu, A.; Show, P.L. Synthetic Dyes Removal by Fusarium Oxysporum HUIB02 and Stimulation Effect on Laccase Accumulation. Environ. Technol. Innov. 2020, 19, 101027. [Google Scholar] [CrossRef]
- Goud, B.S.; Cha, H.L.; Koyyada, G.; Kim, J.H. Augmented Biodegradation of Textile Azo Dye Effluents by Plant Endophytes: A Sustainable, Eco-Friendly Alternative. Curr. Microbiol. 2020, 77, 3240–3255. [Google Scholar] [CrossRef]
- Eichlerová, I.; Homolka, L.; Nerud, F. Ability of Industrial Dyes Decolorization and Ligninolytic Enzymes Production by Different Pleurotus Species with Special Attention on Pleurotus Calyptratus, Strain CCBAS 461. Process Biochem. 2006, 41, 941–946. [Google Scholar] [CrossRef]
- Neelamegam, R.; Baskaran, V.; Dhanasekar, R.; Viruthagiri, T. Decolourization of Synthetic Dyes Using Rice Straw Attached Pleurotus Ostreatus. Indian J. Chem. Technol. 2004, 11, 622–625. [Google Scholar]
- Lim, S.-H.; Lee, Y.-H.; Kang, H.-W. Efficient Recovery of Lignocellulolytic Enzymes of Spent Mushroom Compost from Oyster Mushrooms, Pleurotus spp., and Potential Use in Dye Decolorization. Mycobiology 2013, 41, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Singh, M. Bioremediation of Synthetic Dyes Pollution by White Rot Fungi (Pleurotus spp.); Reference No. (F. No. 41-1126/2012 (SR)); UGC: New Delhi, India, 2012. [Google Scholar]
- Singh, L. Biodegradation of Synthetic Dyes: A Mycoremediation Approach for Degradation/Decolourization of Textile Dyes and Effluents. J. Appl. Biotechnol. Bioeng. 2017, 3, 430–435. [Google Scholar] [CrossRef]
- Matjuškova, N.; Okmane, L.; Zaļā, D.; Rozenfelde, L.; Puķe, M.; Krūma, I.; Vederņikovs, N.; Rapoport, A. Effect of Lignin-Containing Media on Growth of Medicinal Mushroom Lentinula Edodes. Proc. Latvian Acad. Sci. Sect. B 2017, 71, 38–42. [Google Scholar] [CrossRef]
- Wu, F.; Wang, H.; Chen, Q.; Pang, X.; Jing, H.; Yin, L.; Zhang, X. Lignin Promotes Mycelial Growth and Accumulation of Polyphenols and Ergosterol in Lentinula Edodes. J. Fungi 2023, 9, 237. [Google Scholar] [CrossRef] [PubMed]
- Eibes, G.M.; Lú-Chau, T.A.; Ruiz-Dueñas, F.J.; Feijoo, G.; Martínez, M.J.; Martínez, A.T.; Lema, J.M. Effect of Culture Temperature on the Heterologous Expression of Pleurotus Eryngii Versatile Peroxidase in Aspergillus Hosts. Bioprocess Biosyst. Eng. 2008, 32, 129–134. [Google Scholar] [CrossRef] [PubMed]
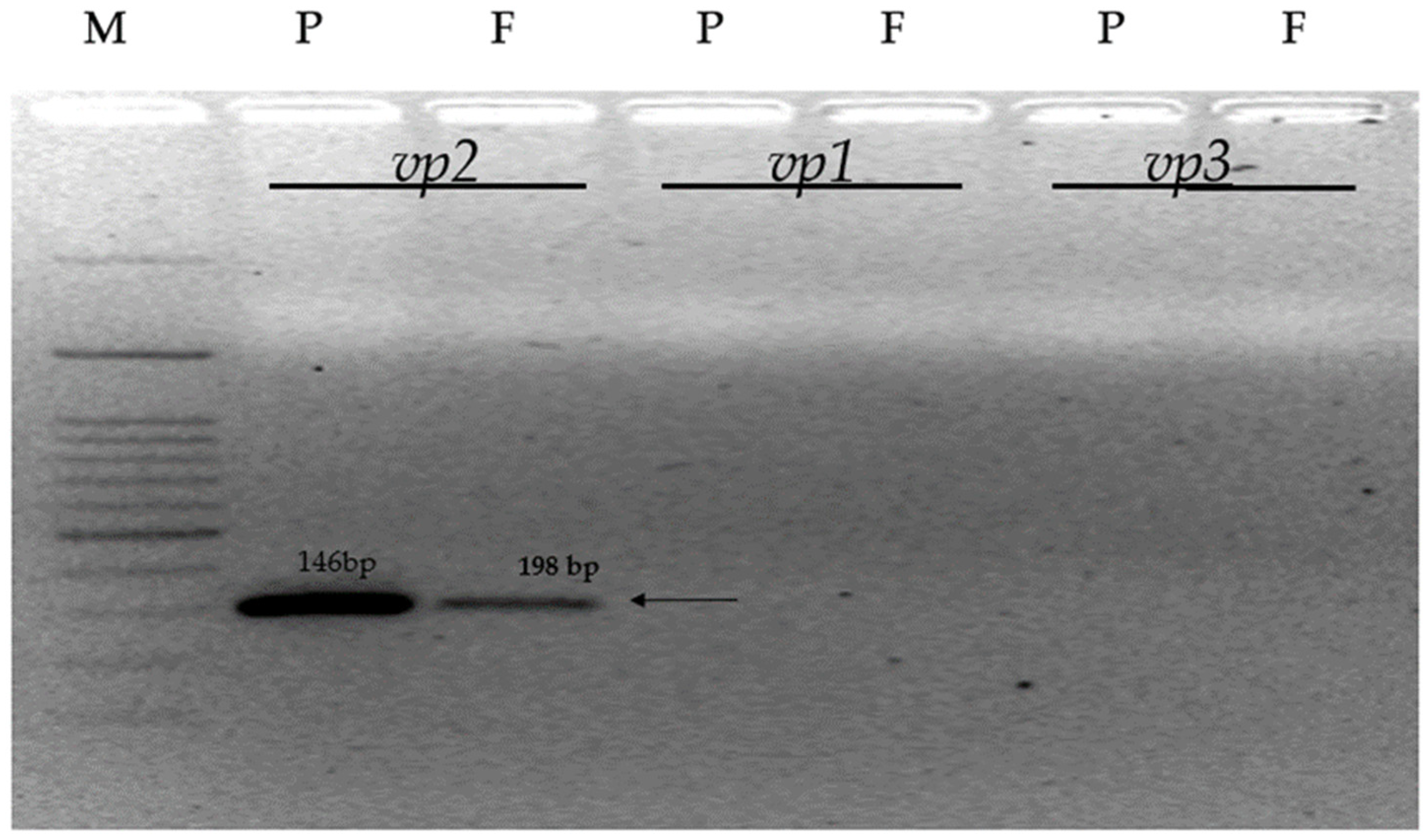
| Gene Code | Forward (5′-3′) | Reverse (5′-3′) | Amplicon Size (bp) |
|---|---|---|---|
| vp1 | CTCCTGACAACAAGGGAGAAGTCC | CAATCAGTTTGCTCTTGTCCTGG | 198 |
| vp2 | TATCGCTCGTCACAACATCAGT | CTGGGACAAGACCATCAGGTGGA | 146 |
| vp3 | TGGATTCTCTCCCACCAAAG | GGGCAGTTGGAAACACCTAA | 195 |
| Days | Potency Index | |||||
|---|---|---|---|---|---|---|
| Pleurotus ostreatus MTCC 142 | Fusarium sp. | |||||
| A1 | A2 | Index | A1 | A2 | Index | |
| 5 | 0.64 | 0.20 | 3.20 | 0.20 | 0.07 | 2.78 |
| 7 | 2.27 | 1.13 | 2.01 | 6.16 | 2.27 | 2.71 |
| 10 | 14.52 | 8.04 | 1.81 | 18.10 | 10.75 | 1.68 |
| 12 | 20.43 | 11.95 | 1.71 | 24.63 | 15.90 | 1.55 |
| 14 | 31.17 (62%) * | 29.22 (58%) | 1.07 | 28.27 (56%) | 21.24 (42%) | 1.33 |
| 16 | 36.32 (72%) | 33.18 (66%) | 1.09 | 29.22 (58%) | 22.90 (46%) | 1.28 |
| Days | Potency Index | |||||
|---|---|---|---|---|---|---|
| Pleurotus ostreatus MTCC 142 | Fusarium sp. | |||||
| A1 | A2 | Index | A1 | A2 | Index | |
| 5 | 0.68 | 0.32 | 2.11 | 0.07 | 0.20 | 0.36 |
| 7 | 6.65 | 1.86 | 3.57 | 5.31 | 3.80 | 1.40 |
| 10 | 16.19 | 6.33 | 2.56 | 17.35 | 10.75 | 1.61 |
| 12 | 34.32 | 21.57 | 1.59 | 26.42 | 14.52 | 1.82 |
| 14 | 37.39 (74%) | 25.61 (51) | 1.46 | 34.21 (68%) | 31.17 (62%) | 1.10 |
| 16 | 38.48 (77%) | 32.47 (65) | 1.19 | 39.59 (79%) | 36.32 (72%) | 1.09 |
| Days | Potency Index | |||||
|---|---|---|---|---|---|---|
| Pleurotus ostreatus MTCC 142 | Fusarium sp. | |||||
| A1 | A2 | Index | A1 | A2 | Index | |
| 5 | 6.61 | 2.54 | 2.60 | 1.77 | 0.95 | 1.86 |
| 7 | 11.34 | 3.46 | 3.27 | 7.07 | 3.14 | 2.25 |
| 10 | 18.86 | 8.87 | 2.13 | 9.62 | 5.73 | 1.68 |
| 12 | 20.51 | 15.41 | 1.33 | 25.52 | 22.06 | 1.16 |
| 14 | 29.22 (58%) | 22.40 (45) | 1.30 | 29.22 (58%) | 27.34 (54%) | 1.07 |
| 16 | 37.39 (74%) | 33.18 (66) | 1.13 | 30.19 (60%) | 27.34 (54%) | 1.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parmar, M.; Patel, S.A.H.; Phutela, U.G.; Dhawan, M. Comparative Analysis of Ligninolytic Potential among Pleurotus ostreatus and Fusarium sp. with a Special Focus on Versatile Peroxidase. Appl. Microbiol. 2024, 4, 1348-1361. https://doi.org/10.3390/applmicrobiol4030093
Parmar M, Patel SAH, Phutela UG, Dhawan M. Comparative Analysis of Ligninolytic Potential among Pleurotus ostreatus and Fusarium sp. with a Special Focus on Versatile Peroxidase. Applied Microbiology. 2024; 4(3):1348-1361. https://doi.org/10.3390/applmicrobiol4030093
Chicago/Turabian StyleParmar, Manisha, Sayeed A. H. Patel, Urmila Gupta Phutela, and Manish Dhawan. 2024. "Comparative Analysis of Ligninolytic Potential among Pleurotus ostreatus and Fusarium sp. with a Special Focus on Versatile Peroxidase" Applied Microbiology 4, no. 3: 1348-1361. https://doi.org/10.3390/applmicrobiol4030093
APA StyleParmar, M., Patel, S. A. H., Phutela, U. G., & Dhawan, M. (2024). Comparative Analysis of Ligninolytic Potential among Pleurotus ostreatus and Fusarium sp. with a Special Focus on Versatile Peroxidase. Applied Microbiology, 4(3), 1348-1361. https://doi.org/10.3390/applmicrobiol4030093






