Classification of Verticillium dahliae Vegetative Compatibility Groups (VCGs) with Machine Learning and Hyperspectral Imagery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Verticillium Dahliae Isolate Preparation and Culture
2.2. Hyperspectral Image Acquisition
2.3. Hyperspectral Image Preprocessing and Feature Extraction
2.4. Dimensionality Reduction and Feature Selection
2.5. Machine Learning Models Fitting and Prediction
3. Results
3.1. Spectral, Textural, and Morphological Features
3.2. Dimension Reduction
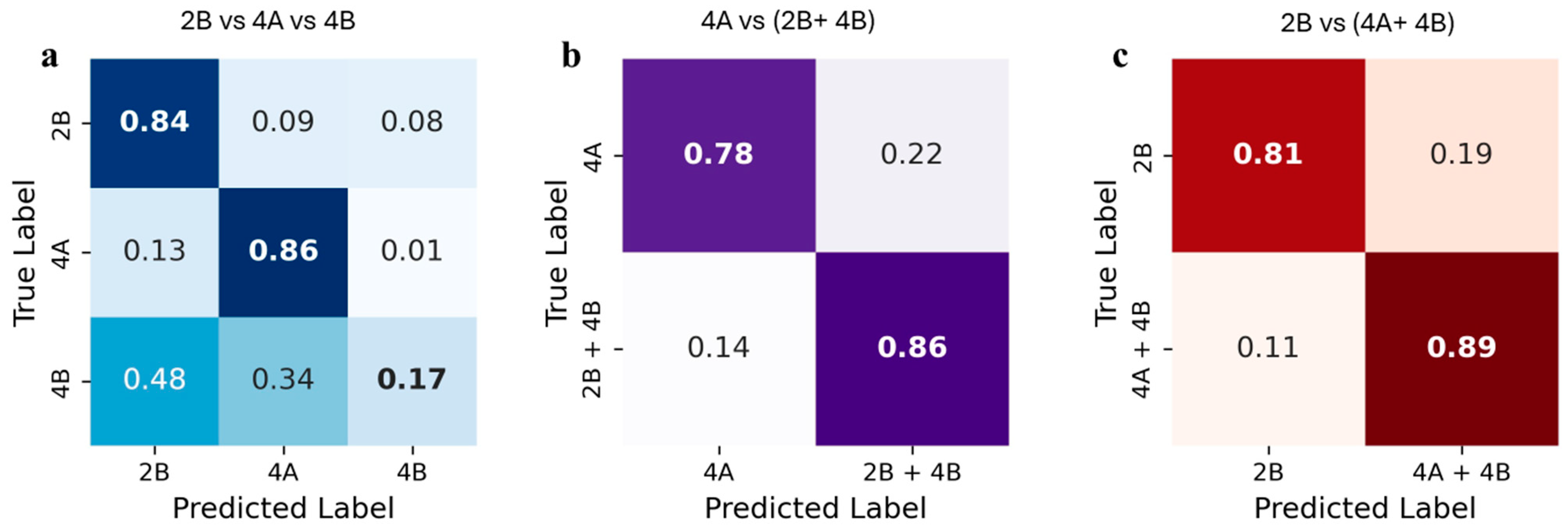
3.3. Classification
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Glass, N.L.; Kaneko, I. Fatal attraction: Nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot. Cell 2003, 2, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Saupe, S.J.; Glass, N.L. Evidence for balancing selection operating at the het-c heterokaryon incompatibility locus in a group of filamentous fungi. Proc. Natl. Acad. Sci. USA 1998, 95, 12398–12403. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Dementhon, K. Non-self recognition and programmed cell death in filamentous fungi. Curr. Opin. Microbiol. 2006, 9, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Jacobson, D.J.; Shiu, P.K. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annu. Rev. Genet. 2000, 34, 165–186. [Google Scholar] [CrossRef]
- Saupe, S.J. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 2000, 64, 489–502. [Google Scholar] [CrossRef]
- Bastiaans, E.; Debets, A.J.; Aanen, D.K. Experimental demonstration of the benefits of somatic fusion and the consequences for allorecognition. Evolution 2015, 69, 1091–1099. [Google Scholar] [CrossRef]
- Debets, F.; Yang, X.; Griffiths, A.J. Vegetative incompatibility in Neurospora: Its effect on horizontal transfer of mitochondrial plasmids and senescence in natural populations. Curr. Genet. 1994, 26, 113–119. [Google Scholar] [CrossRef]
- Cortesi, P.; McCulloch, C.E.; Song, H.; Lin, H.; Milgroom, M.G. Genetic control of horizontal virus transmission in the chestnut blight fungus, Cryphonectria parasitica. Genetics 2001, 159, 107–118. [Google Scholar] [CrossRef]
- Gonçalves, A.P.; Heller, J.; Rico-Ramírez, A.M.; Daskalov, A.; Rosenfield, G.; Glass, N.L. Conflict, competition, and cooperation regulate social interactions in filamentous fungi. Annu. Rev. Microbiol. 2020, 74, 693–712. [Google Scholar] [CrossRef]
- Moore, D.; Robson, G.D.; Trinci, A.P. 21st Century Guidebook to Fungi; Cambridge University Press: Cambridge, UK, 2020. [Google Scholar]
- Leslie, J.F. Fungal vegetative compatibility. Annu. Rev. Phytopathol. 1993, 31, 127–150. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- Punja, Z.K.; Li-Juan, S. Genetic diversity among mycelial compatibility groups of Sclerotium rolfsii (teleomorph Athelia rolfsii) and S. delphinii. Mycol. Res. 2001, 105, 537–546. [Google Scholar] [CrossRef]
- Groenewald, S.; Van Den Berg, N.; Marasas, W.F.; Viljoen, A. The application of high-throughput AFLP’s in assessing genetic diversity in Fusarium oxysporum f. sp. cubense. Mycol. Res. 2006, 110, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Milgroom, M.G.; Jimenez-Gasco, M.D.M.; Olivares-García, C.; Drott, M.T.; Jimenez-Diaz, R.M. Recombination between clonal lineages of the asexual fungus Verticillium dahliae detected by genotyping by sequencing. PLoS ONE 2014, 9, e106740. [Google Scholar] [CrossRef]
- Omer, M.A.; Johnson, D.A.; Douhan, L.I.; Hamm, P.B.; Rowe, R.C. Detection, quantification, and vegetative compatibility of Verticillium dahliae in potato and mint production soils in the Columbia Basin of Oregon and Washington. Plant Dis. 2008, 92, 1127–1131. [Google Scholar] [CrossRef]
- Mehl, H.L.; Cotty, P.J. Variation in competitive ability among isolates of Aspergillus flavus from different vegetative compatibility groups during maize infection. Phytopathology 2010, 100, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Collado-Romero, M.; Mercado-Blanco, J.; Olivares-García, C.; Jiménez-Díaz, R.M. Phylogenetic analysis of Verticillium dahliae vegetative compatibility groups. Phytopathology 2008, 98, 1019–1028. [Google Scholar] [CrossRef]
- Jiménez-Díaz, R.M.; Olivares-García, C.; Landa, B.B.; del Mar Jiménez-Gasco, M.; Navas-Cortés, J.A. Region-wide analysis of genetic diversity in Verticillium dahliae populations infecting olive in southern Spain and agricultural factors influencing the distribution and prevalence of vegetative compatibility groups and pathotypes. Phytopathology 2011, 101, 304–315. [Google Scholar] [CrossRef]
- Woolliams, G.E. Host range and symptomatology of Verticillium dahliae in economic, weed, and native plants in interior British Columbia. Can. J. Plant Sci. 1966, 46, 661–669. [Google Scholar] [CrossRef]
- Malik, N.K.; Milton, J.M. Survival of Verticillium in monocotyledonous plants. Trans. Br. Mycol. Soc. 1980, 75, 496–498. [Google Scholar] [CrossRef]
- Pegg, G.F.; Brady, B.L. Verticillium Wilts; CABI Publishing: Wallingford, UK, 2002. [Google Scholar]
- Wheeler, D.L.; Dung, J.K.S.; Johnson, D.A. From pathogen to endophyte: An endophytic population of Verticillium dahliae evolved from a sympatric pathogenic population. New Phytol. 2019, 222, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Puhalla, J.E. Classification of isolates of Verticillium dahliae based on heterokaryon incompatibility. Phytopathology 1979, 69, 1186–1189. [Google Scholar] [CrossRef]
- Joaquim, T.R.; Rowe, R.C. Vegetative compatibility and virulence of strains of Verticillium dahliae from soil and potato plants. Phytopathology 1991, 81, 552–558. [Google Scholar] [CrossRef]
- Strausbaugh, C.A. Assessment of vegetative compatibility and virulence. Phytopathology 1993, 83, 1253–1258. [Google Scholar] [CrossRef]
- Bhat, R.G.; Smith, R.F.; Koike, S.T.; Wu, B.M.; Subbarao, K.V. Characterization of Verticillium dahliae isolates and wilt epidemics of pepper. Plant Dis. 2003, 87, 789–797. [Google Scholar] [CrossRef]
- Joaquim, T.R.; Rowe, R.C. Reassessment of vegetative compatibility relationships among strains of Verticillium dahliae using nitrate-nonutilizing mutants. Reactions 1990, 4, 41. [Google Scholar]
- Jiménez-Gasco, M.D.M.; Malcolm, G.M.; Berbegal, M.; Armengol, J.; Jiménez-Díaz, R.M. Complex molecular relationship between vegetative compatibility groups (VCGs) in Verticillium dahliae: VCGs do not always align with clonal lineages. Phytopathology 2014, 104, 650–659. [Google Scholar] [CrossRef]
- Collado-Romero, M.; Berbegal, M.; Jiménez-Díaz, R.M.; Armengol, J.; Mercado-Blanco, J. A PCR-based ‘molecular toolbox’ for in planta differential detection of Verticillium dahliae vegetative compatibility groups infecting artichoke. Plant Pathol. 2009, 58, 515–526. [Google Scholar] [CrossRef]
- El-Bebany, A.F.; Rampitsch, C.; Daayf, F. Proteomic analysis of Verticillium dahliae: Deacetylation of specific fungal protein during interaction with resistant and susceptible potato varieties. J. Plant Pathol. 2013, 95, 239–248. [Google Scholar]
- Sankaran, S.; Mishra, A.; Ehsani, R.; Davis, C. A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 2010, 72, 1–13. [Google Scholar] [CrossRef]
- Feng, Y.Z.; Sun, D.W. Application of hyperspectral imaging in food safety inspection and control: A review. Crit. Rev. Food Sci. Nutr. 2012, 52, 1039–1058. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Asaari, M.S.M.; Herrero-Langreo, A.; Lohumi, S.; Diezma, B.; Scheunders, P. Close range hyperspectral imaging of plants: A review. Biosyst. Eng. 2017, 164, 49–67. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, W.; Sankaran, S. High-throughput field phenotyping of Ascochyta blight disease severity in chickpea. Crop Prot. 2019, 125, 104885. [Google Scholar] [CrossRef]
- Bonah, E.; Huang, X.; Aheto, J.H.; Osae, R. Application of hyperspectral imaging as a nondestructive technique for foodborne pathogen detection and characterization. Foodborne Pathog. Dis. 2019, 16, 712–722. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, W.; Huang, M.; Ni, X.; Chu, X.; Li, C. Evaluation and classification of five cereal fungi on culture medium using Visible/Near-Infrared (Vis/NIR) hyperspectral imaging. Infrared Phys. Technol. 2020, 105, 103206. [Google Scholar] [CrossRef]
- Williams, P.J.; Geladi, P.; Britz, T.J.; Manley, M. Near-infrared (NIR) hyperspectral imaging and multivariate image analysis to study growth characteristics and differences between species and strains of members of the genus Fusarium. Anal. Bioanal. Chem. 2012, 404, 1759–1769. [Google Scholar] [CrossRef] [PubMed]
- Salman, A.; Shufan, E.; Lapidot, I.; Tsror, L.; Moreh, R.; Mordechai, S.; Huleihel, M. Assignment of Colletotrichum coccodes isolates into vegetative compatibility groups using infrared spectroscopy: A step towards practical application. Analyst 2015, 140, 3098–3106. [Google Scholar] [CrossRef]
- Dung, J.K.; Peever, T.L.; Johnson, D.A. Verticillium dahliae populations from mint and potato are genetically divergent with predominant haplotypes. Phytopathology 2013, 103, 445–459. [Google Scholar] [CrossRef]
- Boggs, T. Spectral Python. Software. 2016. Available online: https://github.com/spectralpython/spectral (accessed on 22 January 2024).
- Van der Walt, S.; Schönberger, J.L.; Nunez-Iglesias, J.; Boulogne, F.; Warner, J.D.; Yager, N.; Gouillart, E.; Yu, T. scikit-image: Image processing in Python. PeerJ 2014, 2, e453. [Google Scholar] [CrossRef]
- Marzougui, A.; Ma, Y.; Zhang, C.; McGee, R.J.; Coyne, C.J.; Main, D.; Sankaran, S. Advanced imaging for quantitative evaluation of Aphanomyces root rot resistance in lentil. Front. Plant Sci. 2019, 10, 383. [Google Scholar] [CrossRef]
- Harris, C.R.; Millman, K.J.; Van Der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- McKinney, W. pandas: A foundational Python library for data analysis and statistics. Python for High Performance and Scientific Computing 2011, 14, 1–9. [Google Scholar]
- Mordvintsev, A.; Abid, K. OpenCV-Python Tutorials Documentation. 2014. Available online: https://media.readthedocs.org/pdf/opencv-python-tutroals/latest/opencv-python-tutroals.pdf (accessed on 22 January 2024).
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R.; Taylor, J. An Introduction to Statistical Learning: With applications in Python; Springer: New York, NY, USA, 2023. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Waskom, M.L. seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Eldin, A.B. Near Infra Red Spectroscopy; INTECH Open Access Publisher: London, UK, 2011; pp. 237–248. [Google Scholar]
- Wilson, R.H.; Nadeau, K.P.; Jaworski, F.B.; Tromberg, B.J.; Durkin, A.J. Review of short-wave infrared spectroscopy and imaging methods for biological tissue characterization. J. Biomed. Opt. 2015, 20, 030901. [Google Scholar] [CrossRef]
- Manan, S.; Ullah, M.W.; Ul-Islam, M.; Atta, O.M.; Yang, G. Synthesis and applications of fungal mycelium-based advanced functional materials. J. Bioresour. Bioprod. 2021, 6, 1–10. [Google Scholar] [CrossRef]
- Xing, F.; Yao, H.; Liu, Y.; Dai, X.; Brown, R.L.; Bhatnagar, D. Recent developments and applications of hyperspectral imaging for rapid detection of mycotoxins and mycotoxigenic fungi in food products. Crit. Rev. Food Sci. Nutr. 2019, 59, 173–180. [Google Scholar] [CrossRef]
- Delwiche, S.R.; Pearson, T.C.; Brabec, D.L. High-speed optical sorting of soft wheat for reduction of deoxynivalenol. Plant Dis. 2005, 89, 1214–1219. [Google Scholar] [CrossRef]
- Zeise, K.; Von Tiedemann, A. Host specialization among vegetative compatibility groups of Verticillium dahliae in relation to Verticillium longisporum. J. Phytopathol. 2002, 150, 112–119. [Google Scholar] [CrossRef]


| Model | All Spectral Features (n = 134) | Selected Spectral Features (n = 15) | ||||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1 Score | Accuracy | Precision | Recall | F1 Score | |
| LDA | 0.693 | 0.603 | 0.606 | 0.597 | 0.739 | 0.644 | 0.597 | 0.581 |
| RF | 0.761 | 0.656 | 0.624 | 0.621 | 0.761 | 0.694 | 0.635 | 0.636 |
| SVM | 0.706 | 0.545 | 0.548 | 0.539 | 0.679 | 0.622 | 0.629 | 0.613 |
| k-NN | 0.729 | 0.817 | 0.571 | 0.539 | 0.734 | 0.711 | 0.583 | 0.561 |
| ANN | 0.711 | 0.560 | 0.570 | 0.548 | 0.734 | 0.613 | 0.593 | 0.577 |
| Dataset | Classifier | Vegetative Compatibility Groups (VCGs) | ||
|---|---|---|---|---|
| 2B | 4A | 4B | ||
| Textural (n = 70) | RF | 0.89 | 0.87 | 0.03 |
| ANN | 0.78 | 0.84 | 0.28 | |
| Morphological (n = 7) | RF | 0.71 | 0.79 | 0.00 |
| ANN | 0.82 | 0.75 | 0.00 | |
| Combined (Spectral + Textural + Morphological) (n = 211) | RF | 0.91 | 0.88 | 0.17 |
| ANN | 0.87 | 0.88 | 0.28 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Upadhaya, S.G.; Zhang, C.; Sankaran, S.; Paulitz, T.; Wheeler, D. Classification of Verticillium dahliae Vegetative Compatibility Groups (VCGs) with Machine Learning and Hyperspectral Imagery. Appl. Microbiol. 2025, 5, 41. https://doi.org/10.3390/applmicrobiol5020041
Upadhaya SG, Zhang C, Sankaran S, Paulitz T, Wheeler D. Classification of Verticillium dahliae Vegetative Compatibility Groups (VCGs) with Machine Learning and Hyperspectral Imagery. Applied Microbiology. 2025; 5(2):41. https://doi.org/10.3390/applmicrobiol5020041
Chicago/Turabian StyleUpadhaya, Sudha GC, Chongyuan Zhang, Sindhuja Sankaran, Timothy Paulitz, and David Wheeler. 2025. "Classification of Verticillium dahliae Vegetative Compatibility Groups (VCGs) with Machine Learning and Hyperspectral Imagery" Applied Microbiology 5, no. 2: 41. https://doi.org/10.3390/applmicrobiol5020041
APA StyleUpadhaya, S. G., Zhang, C., Sankaran, S., Paulitz, T., & Wheeler, D. (2025). Classification of Verticillium dahliae Vegetative Compatibility Groups (VCGs) with Machine Learning and Hyperspectral Imagery. Applied Microbiology, 5(2), 41. https://doi.org/10.3390/applmicrobiol5020041





