Abstract
Alginate is a naturally occurring polymer derived from brown algae biomass, which has numerous applications in various fields. Chemical modification of alginate is widely used to improve alginate’s physicochemical properties and provide new potential for multiple applications. In this article, we modified alginate with L-DOPA, using periodate oxidation and reductive amination, to obtain more suitable biopolymer for biocatalyst immobilization and hydrogel formation. Obtained modified alginate was used for the immobilization of laccase on cell walls. For this purpose, laccase from Streptomyces cyaneus was expressed on the surface of Saccharomyces cerevisiae EBY100 cells. The obtained cell wall laccase was immobilized within L-DOPA-alginate beads by crosslinking the L-DOPA-alginate with calcium ions and laccase. The effect of additional crosslinking of beads by green light-induced photopolymerization with eosin Y was investigated. The immobilized laccase systems were used for dye decolorization and investigated in multiple treatment processes. Beads with L-DOPA-alginate with a higher degree of modification (5.0 mol%) showed higher enzymatic activity and better decolorization efficiency than those with a lower degree of modification (2.5 mol%). Obtained immobilized biocatalysts are suitable for decolorizing dye Evans Blue due to their high efficiency and reusability.
Keywords:
algae biomass; alginate; yeast cells; laccase; surface display; hydrogel; photopolymerization; bioremediation 1. Introduction
Alginate belongs to a group of binary heteropolysaccharides that are constituted of 1, 4-linked β-D-mannuronic (M) and α-L-guluronic acid (G) [1]. Structural characteristics of the alginate chains depend on the M/G ratio because this ratio plays an important role in the physicochemical properties of this polymer [1]. As alginate possesses numerous free hydroxyl and carboxyl groups, it is a great candidate for chemical modifications [2]. Several types of chemical modification can be performed on alginate, such as oxidation, reductive amination of oxidized alginate, sulfation, copolymerization, esterification, Ugi reaction, and amidation [2,3]. Due to favorable characteristics of alginate, like biodegradability, renewability, and biocompatibility, it is used in various industrial fields, for example, bioengineering, biotechnology, biomedicine, clinical applications, etc. [4]. It is known that enzymes are very attractive biocatalysts because of their advantageous properties (environmentally friendly, biodegradable, relatively low-priced, highly specific). Still, all these positive aspects are affected by enzyme instability in long-term storage, very narrow acceptable conditions, and difficult reusability. One way to overcome these problems is enzyme immobilization. There are two groups of immobilization methods, irreversible and reversible, and both have advantages and disadvantages. The mechanism of irreversible immobilization represents the attachment of an enzyme to a carrier so that it cannot be detached, while reversible immobilization represents the type of enzyme attachment that can be easily separated from the carrier [5]. Lately, light-induced photocrosslinking has gained popularity because of the obtained material’s rapid gelation time and tunable physical properties. Generally, ultraviolet (UV) light is used for photocrosslinking. Still, some negative aspects of UV crosslinking, like generating reactive oxygen species and DNA oxidative damage, make it undesirable, so visible light crosslinking is preferred [6]. Many photoinitiation systems exist, such as riboflavin, carboxylated camphoquinone, and eosin Y [7]. A highly reactive free radical is formed by photocleavage during the photocrosslinking reaction. The light of the photon is absorbed by the photoinitiator, where the light energy is transferred into chemical energy; after that, intra- or extra-molecular groups are covalently crosslinked by free radicals [8,9,10]. As stated before, the catalytic and biochemical properties of enzymes can be improved by immobilization, and this action can be eased if the protein is expressed on the cell surface [11,12,13]. Cell-surface display represents the type of expression in which target peptides or proteins are attached to the cell surface of bacteria, yeast, insect, or mammalian cells. The connection of targeted protein to a cell is mediated by anchor protein [13,14,15]. The research represents that yeast surface display expression has various advantages, with the main one being easy and cheap production in large quantities of immobilized laccase within cell walls without the need for purification. It includes numerous yeast strains (Saccharomyces cerevisiae, Pichia pastoris, and Yarrowia lipolytica), the possibility of post-translational modifications in yeast cells, and relatively facile genetic manipulation [13,15,16]. Laccases (p-diphenol oxidase EC 1.10.3.2) belong to a family of multicopper oxidases found in fungi, bacteria, plants, and animals, and their biological function depends on the enzyme origin [17,18]. The reactions catalyzed by laccases include oxidation of broad spectra of compounds (o- and p-diphenols, aminophenol, polyphenols, polyamines, arylamines, and several other phenolic compounds) whereby molecular oxygen is the final electron acceptor. As the only byproduct is the water molecule, laccases are considered “eco-friendly” [19]. Due to broad substrate spectra, these enzymes have distinctive applications in industry and biotechnology, such as the forest production industry, the pulp and paper industry, the food industry, the pharmaceutical and cosmetic industry, bioremediation, organic synthesis, bio-bleaching, dye decolorization, and juice and wine clarification [20]. An emerging problem in recent decades is environmental contamination by wastewater released in large quantities daily. Besides the textile industry, synthetic dyes are also used in the paper, printing, cosmetic, and pharmaceutical industries [21]. Numerous research papers are focused on the use of laccase for dye decolorization because of its high potential for that use. Degradation of colors and other resistant pollutants can be effected by different chemical and physical methods (adsorption, ion exchange, oxidation, nanofiltration, etc.), and although these advanced physicochemical methods are effective, they can be very expensive [21,22]. While biological methods that use cells could be sensitive to toxic pollutants, enzymes like laccase are more stable. They can be used for the enzymatic degradation of dyes, whereby the immobilized enzyme is a better choice due to the higher stability and possibility of multiple uses.
In this article, we have used biomass (alginate and yeast cells)-derived biocatalysts for dye decolorization. We have prepared a new modified alginate with L-DOPA using periodate oxidation and reductive amination. Modified alginate was then used to immobilize yeast cell-wall-bound laccase from Streptomyces cyaneus obtained by expression on the surface of Saccharomyces cerevisiae EBY100 cells and toluene lysis. After immobilization within L-DOPA-alginate beads, the hydrogel was, for the first time, additionally crosslinked by visible light photopolymerization that, due to new bonds, produced biocatalysts with higher thermostability and better decolorization efficiency (reusability) compared to those biocatalysts not crosslinked additionally by photopolymerization. The obtained biocatalyst was suitable for the decolorization of the dye Evans Blue.
2. Materials and Methods
2.1. Chemicals
Alginic acid sodium salt from brown algae was purchased from Sigma–Aldrich (St. Louis, MO, USA), and l-3,4-dihydroxyphenylalanine (L-DOPA) was purchased from Tokyo Chemical Industry (Tokyo, Japan). Sodium cyanoborohydride was purchased from Merck (Darmstadt, Germany) and sodium metaperiodate from VWR (Leuven, Belgium). 2,2-azino-bis-(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) was purchased from AppliChem (Darmstadt, Germany), while calcium chloride and sodium chloride were purchased from Betahem (Belgrade, Serbia). Used synthetic dyes were purchased from Roth (Karlsruhe, Germany) and Sigma (St. Louis, MO, USA). The Saccharomyces cerevisiae strain EBY100 was kindly provided by Prof. Dane Wittrup (Boston, MA, USA) and was used in the present study as the carrier host cell for laccase.
2.2. Modification of Alginate with L-DOPA
Sodium alginate was dissolved in distilled water for a final concentration of 1.12% (w/v). Sodium metaperiodate was then added to the alginate solution at final concentrations of 1.25 mM and 2.5 mM, resulting in molarity ratios of periodate to alginate glycoside units in the alginate of 2.5 mol% and 5.0 mol%, respectively. The periodate oxidation reaction was conducted in the dark for 24 h at +4 °C and was terminated by adding glycerol to a final concentration of 500 mM, followed by stirring for 30 min. The oxidized alginate was precipitated from the reaction mixture using 1% sodium chloride (w/v) and two volumes of 96% ethanol (v/v). This precipitation procedure was repeated twice using the same method. The resulting precipitate was separated, air-dried, and dissolved in 0.1 M NaAc buffer pH 4.5 to achieve a final concentration of 1% (w/v). Solid l-3,4-dihydroxyphenylalanine (L-DOPA) was added to the solution at a final concentration of 1.5% (w/v) (79 mM), and simultaneously sodium cyanoborohydride was added at a final concentration of 0.5% (w/v). The reaction mixture was then left in the dark, stirring for 24 h at room temperature. The modified alginate was precipitated by adding sodium chloride to achieve a final concentration of 1 M and two volumes of 96% ethanol (v/v). Subsequently, the precipitate was dissolved in water (1%, w/v) and subjected to precipitation twice using the same method. The modified L-DOPA-alginate was then lyophilized and stored at −20 °C.
2.3. Spectral Characterization of Modified Alginate
UV–Vis, NMR, and FTIR spectra were obtained for modified alginates. Both modified and unmodified alginates were prepared for UV–Vis spectra by dissolving them in distilled water to a final concentration of 0.1% (w/v). UV–Vis spectra were acquired using a LLG-uniSPEC2 Spectrophotometer covering a range of 200 to 330 nm. Biopolymer samples were prepared by dissolving in deuterium oxide for NMR spectra, and the NMR spectra of L-DOPA-alginate were acquired on a Bruker Avance III 500 MHz instrument. (Chemical shifts provided in parts per million (δ) referenced downfield from tetramethyl silane as the internal standard). IR spectra of alginate were recorded using 32 scans with resolution four on a Thermo Scientific Nicolet 6700 FTIR spectrometer, using the attenuated total reflectance (ATR) technique. Spectral data were gathered within the mid-IR range (1800–600 cm−1).
2.4. Expression of Laccase on Cell Surface of Saccharomyces cerevisiae
Laccase from Streptomyces cyaneus CECT 3335 was cloned into a pCTcon2 vector by a previously reported protocol [20]. Transformation of the competent Saccharomyces cerevisiae EBY100 cells with pCTcon2-lac construct and empty pCTcon2 plasmid was done according to the LiAc/PEG method [23]. Yeast cells were transformed with the empty pCTcon2 plasmid to obtain the control stain. Transformed cells were grown in YNB-CAA (yeast nitrogen base-casamino acid) glucose medium (2% (w/v) glucose) at 30 °C, 160 rpm. When the final OD600 reached 5, cells were transferred to YNB-CAA galactose medium (2% (w/v) galactose) to final OD600 0.5 to induce laccase expression. Expression was performed for 24 h at 30 °C, 160 rpm; afterward, cells were harvested by centrifugation (3000× g, 15 min, 25 °C) and washed three times with Na-acetate buffer (pH 4.5, 100 mM).
2.5. Cell Lysis and Cell Wall Laccase Preparation
Cell lysis was performed with 3% toluene in distilled water (v/v). The obtained cells were resuspended in 3% toluene and mixed at 170 rpm at room temperature for 24 h. The optimal time of cell lysis was 48 h [20]. The obtained yeast cell walls were diluted with water to eliminate toluene and separated by centrifugation at 3000 rpm for 5 min.
2.6. Determination of Enzyme Activity of Whole Cells and Cell Walls with Laccase
We determined the enzyme activity of yeast cells and yeast cell walls. The enzyme activity of whole yeast cells was determined with ABTS assay: 8 μL of the cells in NaAc buffer pH 4.5 (at a concentration of 250 mg/mL) were mixed with 200 μL of 20 mM CuCl2 solution, 1092 μL of NaAc buffer pH 4.5. 200 μL of this mixture was transferred into the well of a microtiter plate, and 50 μL of 20 mM ABTS solution was added. The change in absorbance was monitored using an Elisa reader at a wavelength of 405 nm every 5 min for 1 h.
To measure the activity of the harvested yeast cell walls, 13 μL of the cell walls in NaAc buffer pH 4.5 (at a concentration of 150 mg/mL) was mixed with 200 μL of 20 mM CuCl2 solution and 1087 μL of NaAc buffer pH 4.5, 200 μL of this mixture was dispensed into the well of a microtiter plate, and 50 μL of 20 mM ABTS solution was added. The change in absorbance was monitored using an Elisa reader at a wavelength of 405 nm every 30 s for 5 min.
2.7. Immobilization of Cell Wall Laccase in L-DOPA-Alginate
The cell walls with laccase were previously incubated in 2 mM CuCl2 solution, stirring for 1 h before immobilization. Beads were prepared with 2.5 mol% L-DOPA-alginate and 5.0 mol% L-DOPA-alginate with cell walls with laccase and cell walls with empty pCTcon2 vector. The mixture of 2.7% L-DOPA-alginate (w/v) and 150 mg/mL cell walls in 0.1 M NaAc buffer pH 4.5 were prepared and dropped in 6% CaCl2 (w/v) solution with continuous stirring. The obtained beads were washed two times with 6% CaCl2 (w/v) solution and kept in 0.1 M NaAc buffer (pH 4.5) containing 5 mM CaCl2 (w/v), at 4 °C. The beads with eosin Y were also prepared. During the preparation of the L-DOPA-alginate-cell wall laccase mixture, eosin Y was added at final concentrations of 0.001% and 0.0001%.
2.8. Incubation of Beads in Eosin Y and Photopolymerization
Immobilized cell wall laccase in 2.5 mol% and 5.0 mol% L-DOPA-alginate beads were incubated in solutions of photoinitiator eosin Y. For incubation, 0.005% and 0.01% solutions of eosin Y in PBS (1×, pH 7.4) were used, and the beads were incubated for 5 or 30 min. After incubation, the beads were removed from the solution and transferred into a customized photoreactor (a glass wrapped with green LED strips) for photopolymerization and additional hydrogel crosslinking. The beads were exposed to green light for 15 min, rinsed with NaAc buffer (0.1 M, pH 4.5), and stored in a 5 mM solution of CaCl2 in NaAc buffer (0.1 M, pH 4.5) at +4 °C.
Beads with 0.0001% and 0.001% eosin Y, where eosin Y was added during the beads’ formation, were exposed to green light for 15 min in a customized photoreactor and then stored in a 5 mM solution of CaCl2 in NaAc buffer (0.1 M, pH 4.5) at +4 °C.
2.9. Determination of Enzyme Activity of Immobilized Cell Wall Laccase
The spectrophotometric assay to measure the enzyme activity of L-DOPA-alginate beads with cell wall laccase involved utilizing beads (one bead per reaction) with 200 μL of 0.1 M NaAc buffer at pH 4.5, 50 μL of 20 mM CuCl2, and 250 μL of 20 mM ABTS (final concentration of 7 mM). ABTS concentration was increased compared to the assay with non-immobilized enzyme due to diffusion limitations [24]. After 200 μL of prepared mixture was dispensed from Eppendorf tubes with beads into the wells of a microtiter plate, the change in absorbance was monitored using an Elisa reader at a wavelength of 405 nm. Immediately after measuring the absorbance, 200 μL of the solution was transferred back into Eppendorf tubes.
2.10. Determination of Thermal Stability of Immobilized Cell Wall Laccase
The thermostability of immobilized cell wall laccase in L-DOPA-alginate beads was determined by incubating cell wall laccase beads for 1 h at 60 and 70 °C in optimal buffer. (0.1 M NaAc buffer pH 4.5) with 5 mM CaCl2. Afterward, samples were cooled on ice for 5 min, and enzyme activity was determined according to the mentioned assay. Before determining the enzyme activity, all beads were rinsed twice with NaAc buffer (0.1 M, pH 4.5) to remove excess Ca2+. The relative enzyme activity at different temperatures for each type of bead was determined by defining the highest activity as 100% and calculating the relative enzyme activity at other temperatures relative to it.
2.11. Dye Decolorization
Four different dyes were chosen to investigate the decolorization capacity of yeast cell wall laccase immobilized in L-DOPA-alginate beads. The following dyes were utilized: Evans Blue, Remazol Brilliant Blue, Amido Black 10B, and Reactive Black 5. All dyes were prepared in 0.1 M NaAc buffer at pH 5 with 2 mM Cu2+ and 5 mM Ca2+. Each dye’s concentration was prepared to achieve approximately 0.8 absorbance units at its respective maximum wavelength (all dyes have a maximum wavelength of 620 nm). The final concentrations for Amido Black 10B, Reactive Black 5, Evans Blue, and Remazol Brilliant Blue were 0.027, 0.129, 0.008, and 0.038 mM, respectively.
Decolorization was tested with various alginate beads with cell wall laccase: 2.5 mol% L-DOPA-alginate beads, 5.0 mol% L-DOPA-alginate beads, 2.5 mol% L-DOPA-alginate beads incubated in 0.01% eosin Y, 5.0 mol% L-DOPA-alginate beads incubated in 0.01% eosin Y, 2.5 mol% L-DOPA-alginate beads with 0.0001% eosin Y, and 5.0 mol% L-DOPA-alginate beads with 0.0001% eosin Y.
For decolorization, around 20 mg of alginate beads and 1 mL of each dye solution were used. Subsequently, the samples were wrapped in aluminum foil to protect them from light exposure. The reaction mixtures were incubated for 48 h at room temperature without shaking. As a control reaction of the decolorization test, 2.5 mol% L-DOPA-alginate and 5.0 mol% L-DOPA-alginate with cell walls with empty pCTcon2 were prepared under the same conditions to detect possible removal of dye due to dye adsorption onto the alginate beads and cell walls, and an additional control reaction of the decolorization test was used for each dye in the mentioned buffer, without beads. Decolorization tests were conducted in triplicates. The change in absorbance at 620 nm was monitored at 0, 4, 20, 24, and 48 h after adding the dye solution. The following equation calculated relative decolorization (%):
The relative decolorization of the selected dyes, including control beads, was calculated. The calculated value for control beads was subtracted from the decolorization value for beads with cell wall laccase.
2.12. Reusability
The reusability of the immobilized biocatalyst was tested in repeated cycles of dye decolorization. Calcium-L-DOPA-alginate beads with cell wall laccase were used in eight cycles of decolorization. Cell wall laccase immobilized in 5.0 mol% L-DOPA-alginate beads without EY, incubated in 0.01% EY and with 0.0001% EY, and cell walls with empty pCTcon2 were tested. Beads were used for eight decolorization cycles of 48 h. Reusability was investigated in 0.129 mM Evans Blue solution in 100 mM NaAc buffer pH 5. After each cycle, the beads were filtrated and washed three times with 100 mM NaAc buffer pH 5 and replaced with a fresh solution of dyes. The initial and final absorbance were measured spectrophotometrically at 620 nm for every cycle. The initial activity of freshly prepared dye during the first run was 100%. This study was conducted in triplicate, too.
3. Results and Discussion
3.1. Modification of Alginate with L-DOPA and Its Characterization
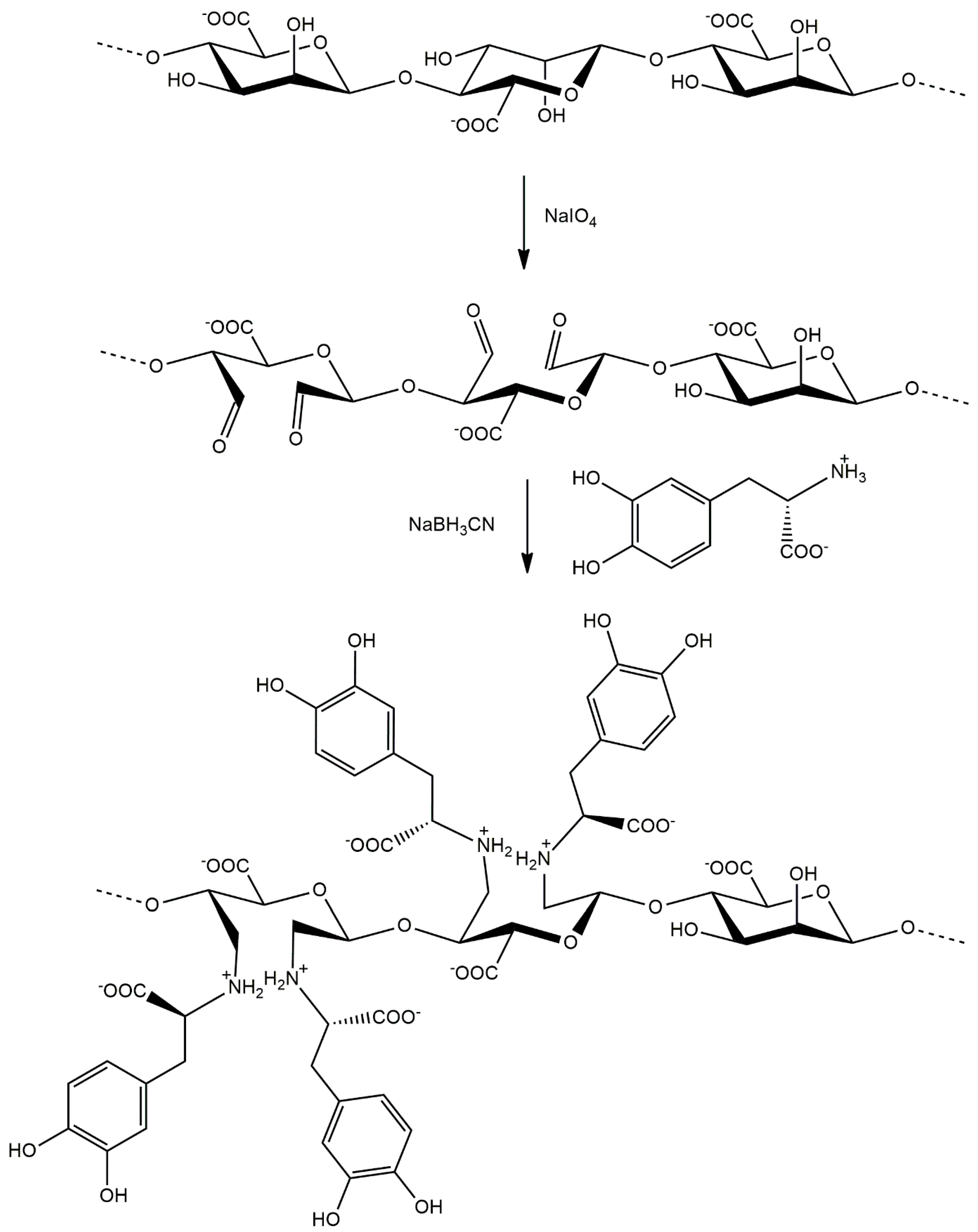
In this study, a modification of alginate is presented for the first time with L-DOPA using periodate oxidation and reductive amination. We introduced a new functional group into the alginate molecule using periodate oxidation and reductive amination in the presence of L-DOPA. This resulted in obtaining a product with stable secondary amino groups (Figure 1).
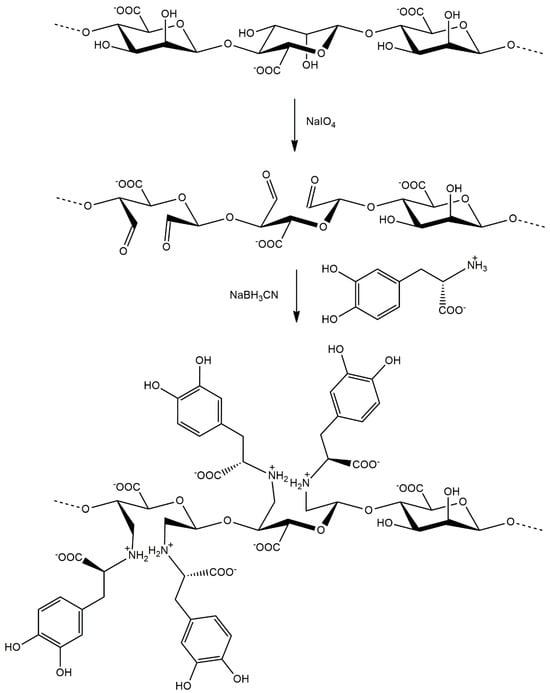
Figure 1.
Synthesis of L-DOPA-alginate in the reaction of periodate oxidation and reductive amination.
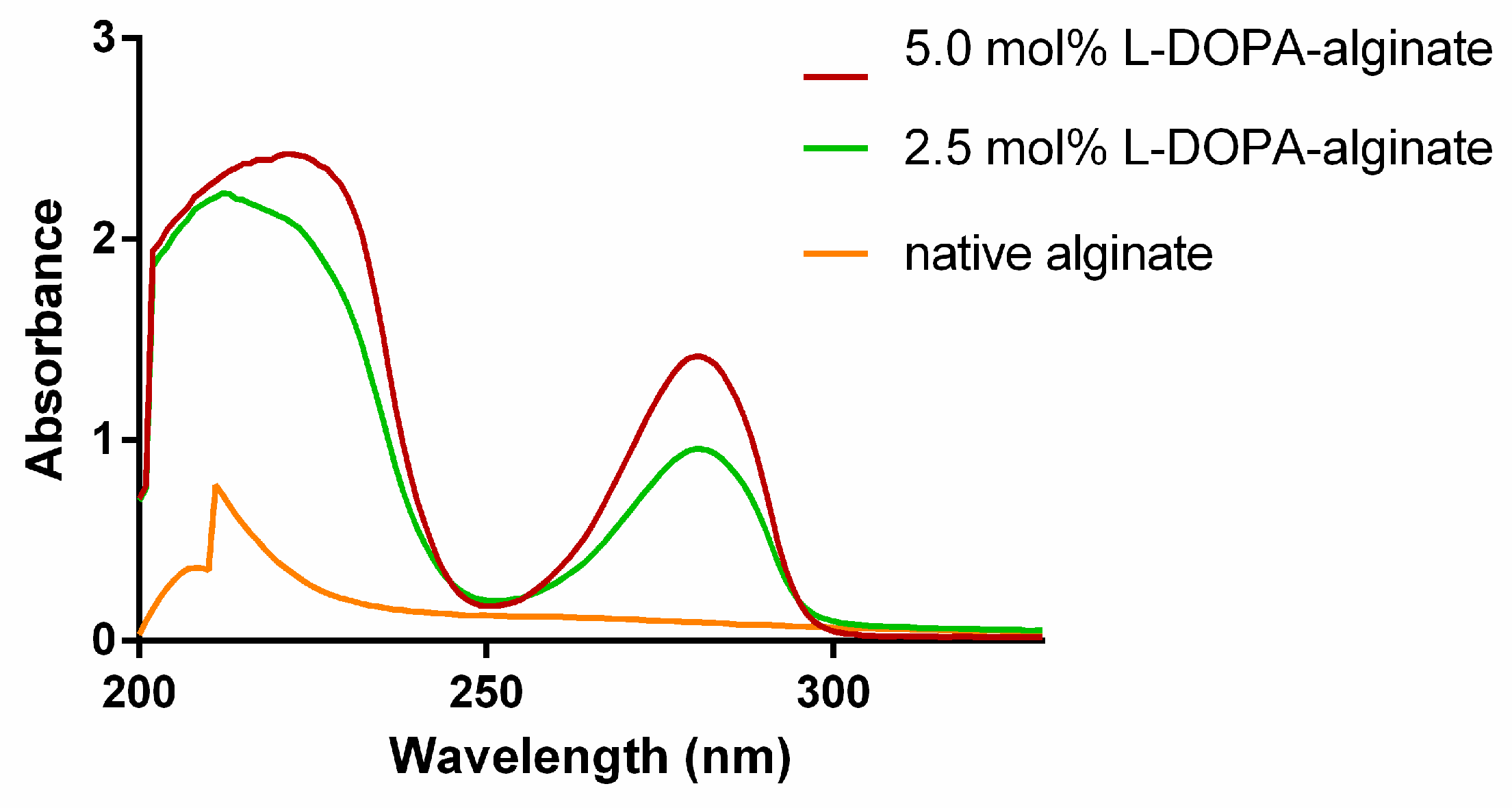
The prepared alginate derivates were characterized spectroscopically using UV–Vis, 1H NMR, and FTIR spectroscopy. UV–Vis spectra of modified alginate show a characteristic absorption peak at 280 nm due to the presence of L−DOPA phenolic groups. In contrast, this peak is absent in the spectra of native alginate (Figure 2). Additionally, it can be observed that the absorption intensity at 280 nm is higher in the case of modified alginate with a higher degree of oxidation, confirming the introduction of more L-DOPA units into the alginate structure in this manner.
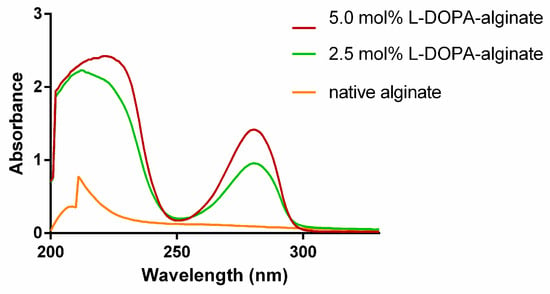
Figure 2.
UV–Vis spectra of native and modified L-DOPA-alginate.
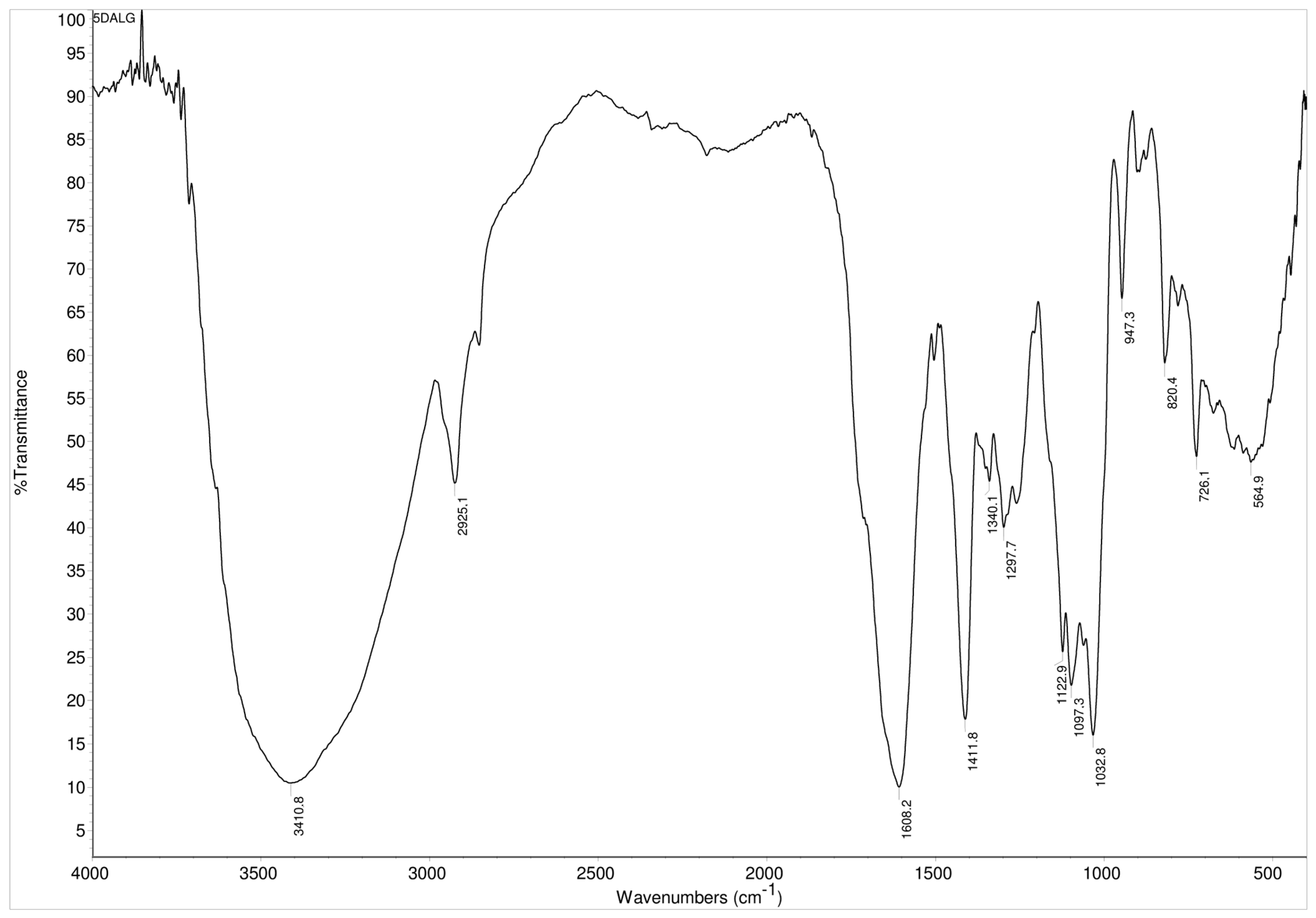
The modified 5.0 mol% L-DOPA-alginate was characterized also by FTIR spectroscopy. FTIR spectra of L-DOPA-alginate showed L-DOPA aromatic structure within modified alginate by the appearance of a peak around 1500 cm−1 originating from C–C in-ring aromatic stretching vibrations (Figure 3).
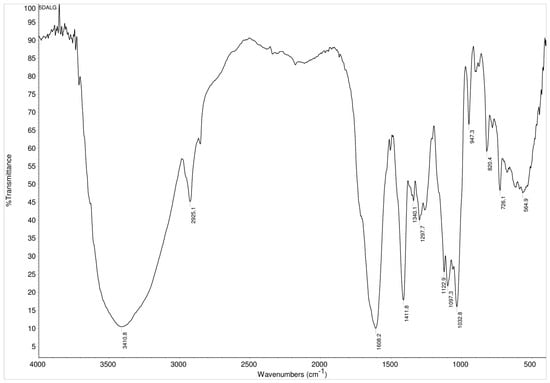
Figure 3.
FTIR spectra of 5.0 mol% L-DOPA-alginate.
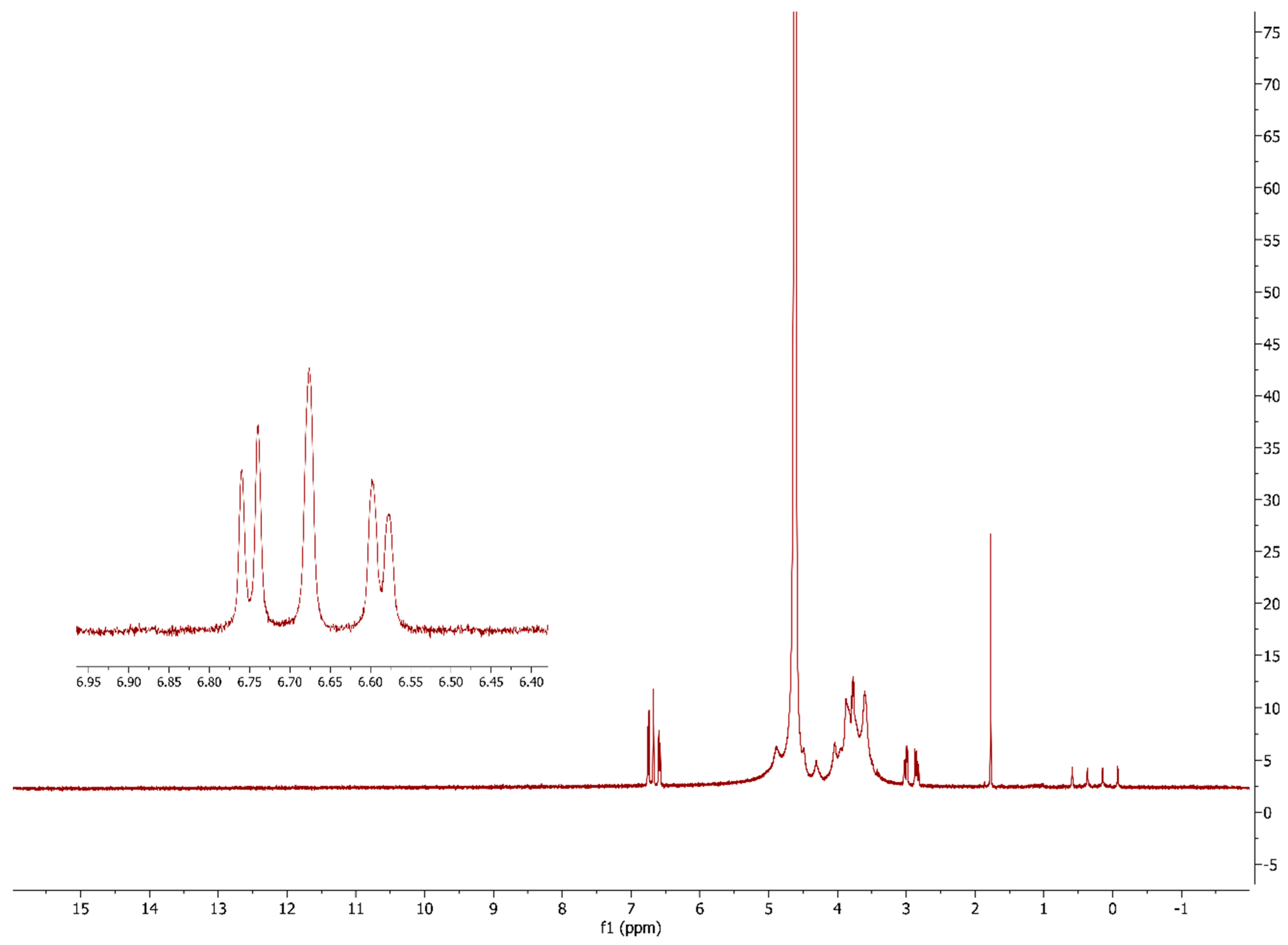
The success of the alginate modification was further confirmed by recording the 1H NMR spectra of the 5.0 mol% L-DOPA-alginate (Figure 4). Peaks at chemical shifts of 6.59, 6.68, and 6.75 ppm originate from three aromatic H atoms of the L-DOPA that were not present in the FTIR spectra of native alginate from our previous work [20]. This confirms the successful incorporation of L-DOPA into the alginate structure. The absence of peaks between 9 and 10 ppm confirms that all previously formed aldehyde groups by periodate oxidation have reacted in the reductive amination reaction.
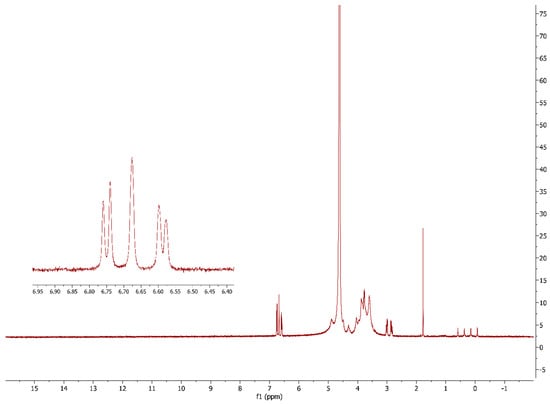
Figure 4.
1H NMR spectra of 5.0 mol% L-DOPA-alginate.
3.2. Yeast Surface Display of Laccase
Cell-surface display enables the expression of target peptides or proteins on the cell surface of various cells, including bacteria, yeast, insects, or mammalian cells, achieved by linking a protein of interest with an anchor protein. This technique combines precise gene expression and protein immobilization, with the advantages of a purification process, reusability, and recovery of biocatalyst.
In this research, we successfully achieved the functional expression and surface display of laccase from Streptomyces cyaneus CECT 3335 in a strain of S. cerevisiae EBY 100. The activity of recombinant laccase was detected using ABTS as a substrate in the activity assay with the addition of copper. The addition of copper was necessary for the activation of the enzyme since copper ions are present in the active center of laccase. In the absence of copper in the reaction mixture, laccase activity was not detectable, consistent with findings from a previous report on the expression of the same enzyme in E. coli [25]. The surface displayed laccase showed enzyme activity of 0.062 ± 0.013 IU/g wet cell biomass. Control cells, with empty pCTcon2 vector, did not show enzyme activity. Since the recombinant laccase was obtained by yeast surface display technology, the laccase was anchored on the cell surface, which can be classified as a method of laccase immobilization. This technique combines precise gene expression and protein immobilization, with the advantages of the purification process, reusability, and recovery of biocatalyst from biomass. Due to these advantages, this technology has been used to produce various enzymes, including laccase. In previous studies, laccase from different sources has been expressed on the surface of yeast cells [20,26,27,28,29,30], and it demonstrates improvements in characteristics such as reusability towards phenolic substrates and excellent ethanol tolerance [26], as well as reusability in the process of removing emerging contaminant acetaminophen (APAP) [30], etc. Summary results show that the advantages of recombinant laccase on the surface of yeast cells include more straightforward and more efficient purification, enhanced enzyme stability, longevity, reusability, and ease of enzyme manipulation and engineering.
3.3. Preparation of Cell Wall Laccase
The obtained cells were lysed with an organic solvent, 3% toluene (v/v), in order to remove ballast proteins by disrupting yeast cells and releasing their cellular content, but not to remove laccase from the cell wall. In previous research, the optimal time of lysis was determined as 24 h [20]. The specific activity of cell wall laccase was 2.062 ± 0.445 IU/g, which is a significant improvement in comparison to the activity of whole cells, 0.062 ± 0.013 IU/g. This could result from a better accessibility of enzyme to substrates, compared to intact cells, or from a decreased mass of cell walls compared to yeast cells after releasing of non-secreted enzyme from the cell wall’s internal cell compartments.
3.4. Immobilization of Cell Wall Laccase in L-DOPA-Alginate
Despite the fact that laccase attached to the cell wall exhibited characteristics of immobilized enzyme, such as increased enzyme stability and reusability, additional immobilization was performed. Immobilization was done by using the entrapment method with calcium ions and alginate. Calcium was used as crosslinking agent and spherical regular shaped beads were obtained after adding the immobilization mixture dropwise to a CaCl2 solution. The conditions of immobilization were previously determined in our study, and beads were prepared using a mixture of 2.7% L-DOPA-alginate (w/v) and 150 mg/mL cell walls with laccase and dropped in 2% CaCl2 (w/v) solution [20]. In this study, the same concentration of cell walls and L-DOPA-alginate was found to be optimal, with an increase in the calcium chloride concentration to 6%, to achieve enhanced mechanical stability of the beads. We used L-DOPA-alginate with a lower (2.5 mol%) and higher (5.0 mol%) degree of modification.
3.5. Photopolymerization of L-DOPA-Alginate Beads with Immobilized Cell Wall Laccase
In order to further crosslink the obtained beads and improve enzyme stability, photo-crosslinking of L-DOPA groups on the alginate chain was investigated in the presence of a photoinitiator. There has been an increasing interest in a visible light-induced photo-crosslinking, in comparison to conventional UV crosslinking. Previous research has demonstrated adverse effects of UV crosslinking, and in order to avoid the damaging effects, including DNA damage, cancer effects, protein damage resulting in reduced protein activity, etc., visible light photo-crosslinking has been investigated [31].
Eosin Y is known for its high reactivity in initiating photopolymerization reactions, leading to efficient crosslinking or polymerization processes. In a previous study, modified hyaluronan with tyramine was photo-crosslinked in the presence of eosin Y as a photoinitiator in a non-toxic, rapid photosensitized process under visible light illumination [32]. It was also described as a crosslinking of modified polyethylene glycol (PEG) with eosin Y as a photoinitiator [33].
In a subset of the beads, the photoinitiator was added during the formation of beads, in a final concentration of 0.0001% and 0.001% eosin Y, while another subset of beads was subsequently incubated in eosin Y solutions (0.005% and 0.01%) for 5 or 30 min. The color of the obtained beads varied depending on the preparation method. Beads prepared with eosin Y were light pink, with the shade depending on the concentration. Beads without eosin Y were white, and beads incubated in eosin Y were of intense pink color whose shade depended on the concentration of eosin Y solution and the incubation time (Figure 5).

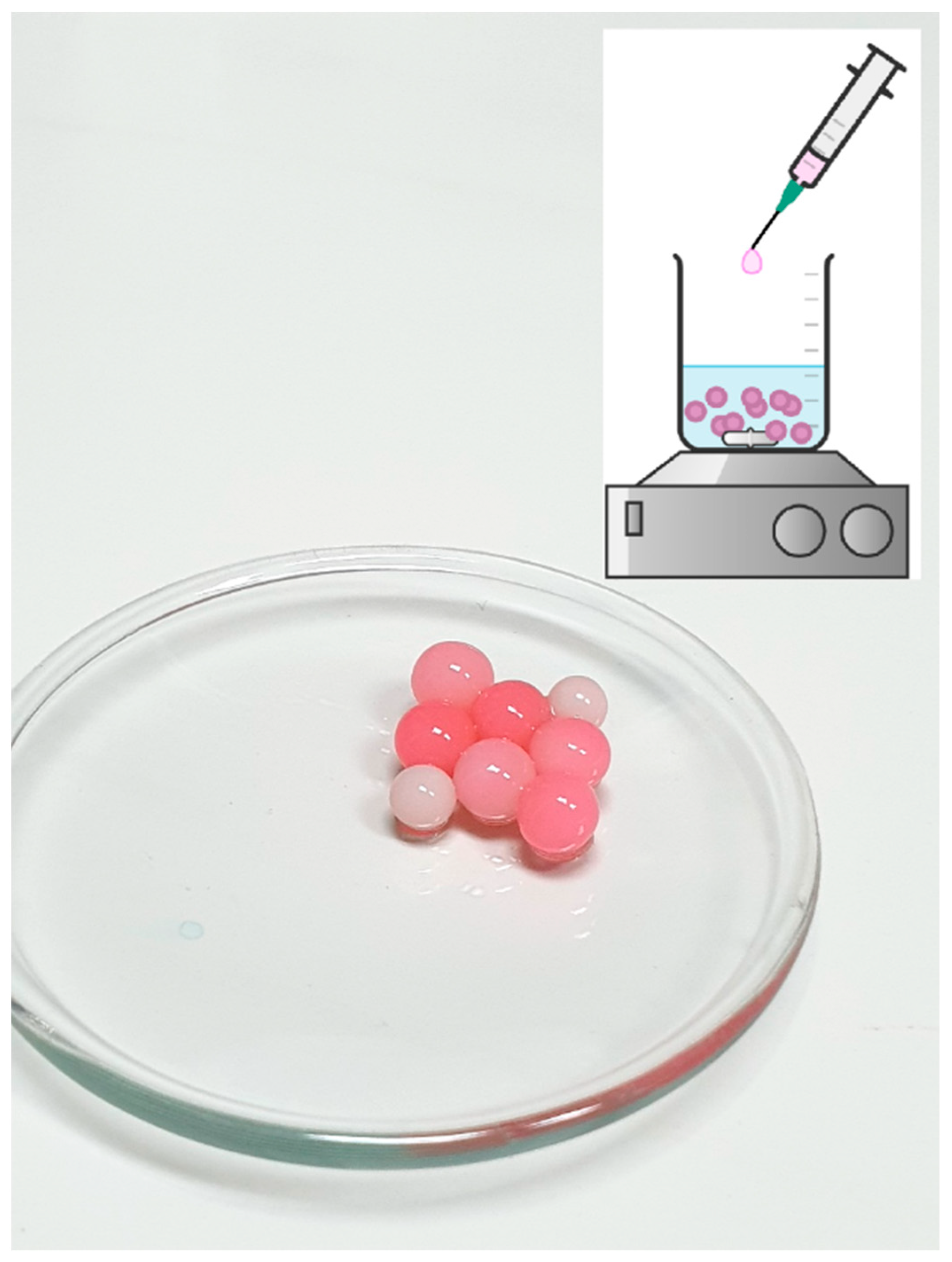
Figure 5.
Schematic representation of immobilization process and picture of obtained beads—Ca-L-DOPA-alginate beads with cell wall laccase, non-incubated with eosin Y (white beads) and incubated with eosin Y (pink beads). Bead size was 3 mm ± 0.5, while sphericity was around 0.95.
3.6. Determination of Enzyme Activity of Immobilized Cell Wall Laccase in Different Types of Beads
After measuring the enzymatic activity, the results indicate that photopolymerized beads, with 0.001% and 0.0001% eosin Y and beads incubated in 0.01% and 0.005% EY for 5 min, show higher activities than non-photopolymerized beads (Table 1). Lower enzymatic activity compared to non-photopolymerized beads was observed only at beads incubated in both concentrations of EY but over a more extended period (30 min). If we compare the activity of beads incubated in different concentrations of EY for 5 min, we can see that higher activity is obtained with beads incubated in 0.01% EY for both polymers (2.5 mol% and 5.0 mol% L-DOPA-alginate). Here, it can also be observed that beads with 5 mol% L-DOPA modification demonstrate higher activity for the same parameters than beads with 2.5 mol% L-DOPA-alginate.

Table 1.
Enzyme activity (IU/g) of cell wall laccase immobilized in 2.5 mol% and 5.0 mol% L-DOPA-alginate beads, without photopolymerization and addition of EY/photopolymerized with 0.001% or 0.0001% EY/incubated in 0.01% and 0.005% EY for 5 and 30 min. The specific activity of the cell-wall-bound laccase was 2.062 ± 0.445 IU/g.
The enzymatic activity of beads with immobilized cell walls and the empty pCTcon2 vector was also examined at room temperature. Even after several hours, the absorbance of the assay mixture did not change, confirming that these beads do not exhibit enzymatic activity.
Based on this, it was decided to conduct decolorization experiments with beads where the concentration of EY during immobilization was 0.0001% and 0.001% EY and with beads incubated in 0.01% EY for 5 min.
3.7. Determination of Temperature Stability
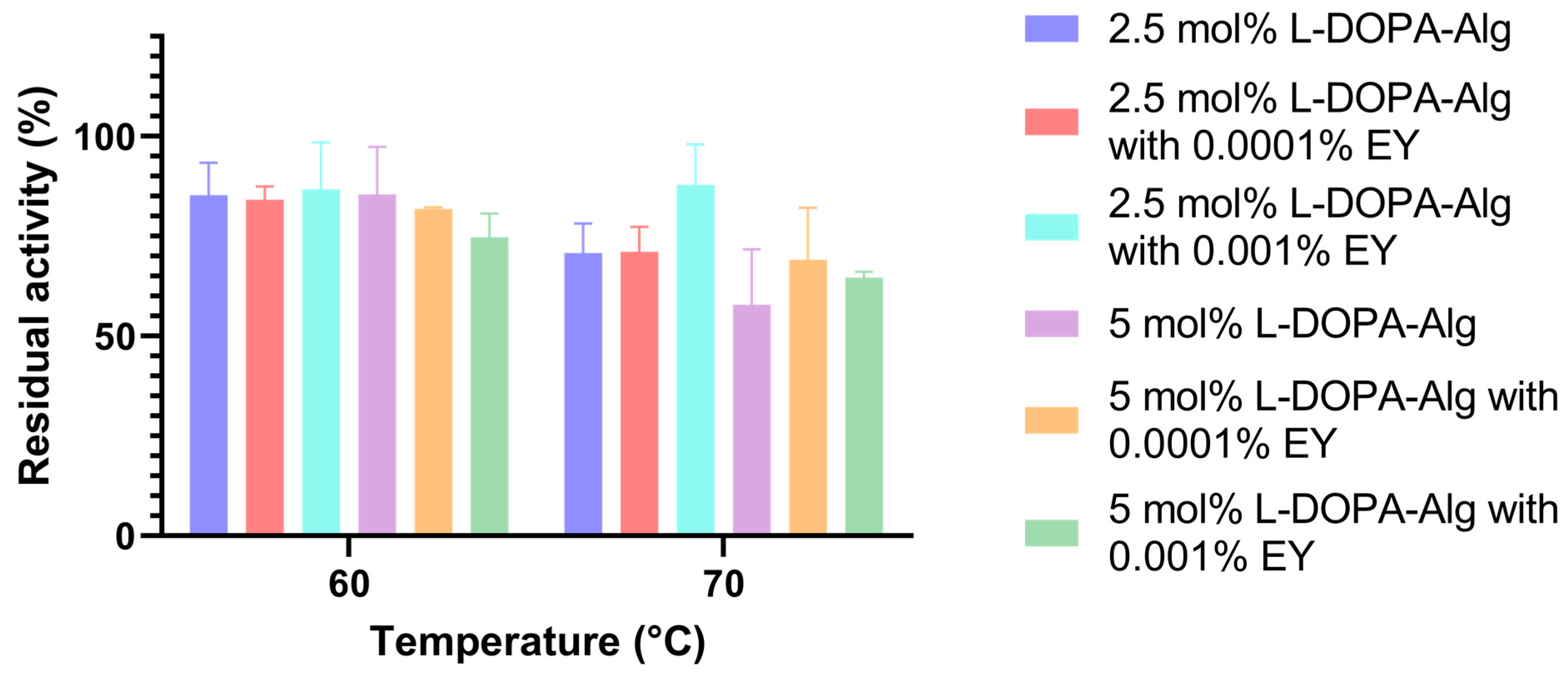
The thermostability of immobilized cell wall laccase in different L-DOPA-alginate beads was determined: 2.5 and 5.0 mol% L-DOPA-alginate beads without eosin Y, 2.5 and 5.0 mol% L-DOPA-alginate beads with 0.001% eosin Y, and 2.5 and 5.0 mol% L-DOPA-alginate beads with 0.0001% eosin Y. All beads were incubated for 1 h at 60 and 70 °C at pH 4.5. Residual activity was measured using ABTS as described above.
In a previous report, free cell wall laccase at 60 °C retained 69% enzyme activity [20], while all immobilized counterparts retained higher enzyme activity. In the present study, photopolymerized 2.5 mol% L-DOPA-alginate with 0.001% EY retained the highest enzyme activity at that temperature, 93.4%, while the same beads without EY and with 0.0001% EY retained 85.2% and 84.1% activity, respectively.
At 70 °C, photopolymerized 2.5 mol% L-DOPA-alginate beads with 0.001% EY also retain the highest activity, 93.6%, while the same beads without adding a photoinitiator retain only 66.6% activity. The 2.5 mol% L-DOPA-alginate beads with a lower concentration of EY (0.0001%) retain 74.6% activity (Figure 6).
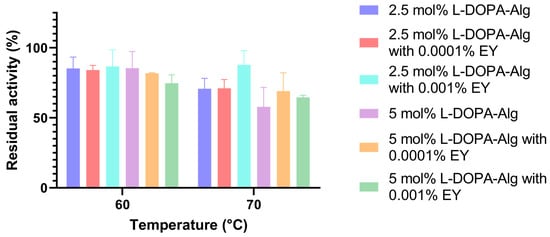
Figure 6.
The temperature stability of cell wall laccase immobilized in 2.5 and 5.0 mol% L-DOPA-alginate, without the addition of eosin Y (non-photopolymerized) and photopolymerized with 0.001% and 0.0001% of eosin Y.
The beads with 5 mol% L-DOPA-alginate did not exhibit such a significant difference in activity after 1 h of incubation at 60 °C. At 70 °C, the photopolymerized immobilizate with 0.0001% EY retained 76.3% activity, the photopolymerized immobilizate with 0.001% EY retained 64.7% activity, and the immobilizate without additional photo-crosslinking retained only 50.01% of its initial activity.
Despite the observed increase in temperature stability of enzymes obtained through yeast surface display technology compared to the free enzymes [30], it has been demonstrated that additional immobilization within cell walls further improves stability at elevated temperatures [20]. The enhanced temperature stability of the immobilized cell wall laccase was attributed to the binding of the enzyme onto the support, reducing the conformational changes induced by higher temperature.
From the presented results, we can determine that the photopolymerized beads containing 2.5 mol% L-DOPA-alginate with 0.001% eosin Y exhibited the highest temperature stability, at 60 °C and 70 °C, in comparison to other beads with cell wall laccase and non-immobilized cell wall laccase. This was especially seen at 70 °C, where after 1 h of incubation, around 90% of the original activity was preserved. It can be deduced that the highest degree of crosslinking in the resulting beads occurs at the specified concentration of introduced L-DOPA groups into the alginate molecule and at the concentration of the photoinitiator. In this case, the immobilized enzyme was most protected from external influences, resulting in the preservation of the enzyme’s native structure to the greatest extent despite the increase in temperature.
3.8. Decolorization of Dyes
Various dyes were decolorized using the obtained biocatalysts to evaluate the potential of immobilized cell wall laccase for dye decolorization. The selected dyes included the azo dye Evans Blue and the anthraquinone dye Remazol Brilliant Blue. In our study, decolorization of the dyes was investigated using various modifications of alginate, namely 2.5 mol% and 5 mol% L-DOPA-alginate, both with and without photopolymerization in the presence of eosin Y. Additionally, the concentration and method of EY addition were varied, employing beads to which 0.0001% EY was added during immobilization, as well as beads subsequently incubated in 0.01% EY. All photopolymerized beads were irradiated for the same duration of 15 min.
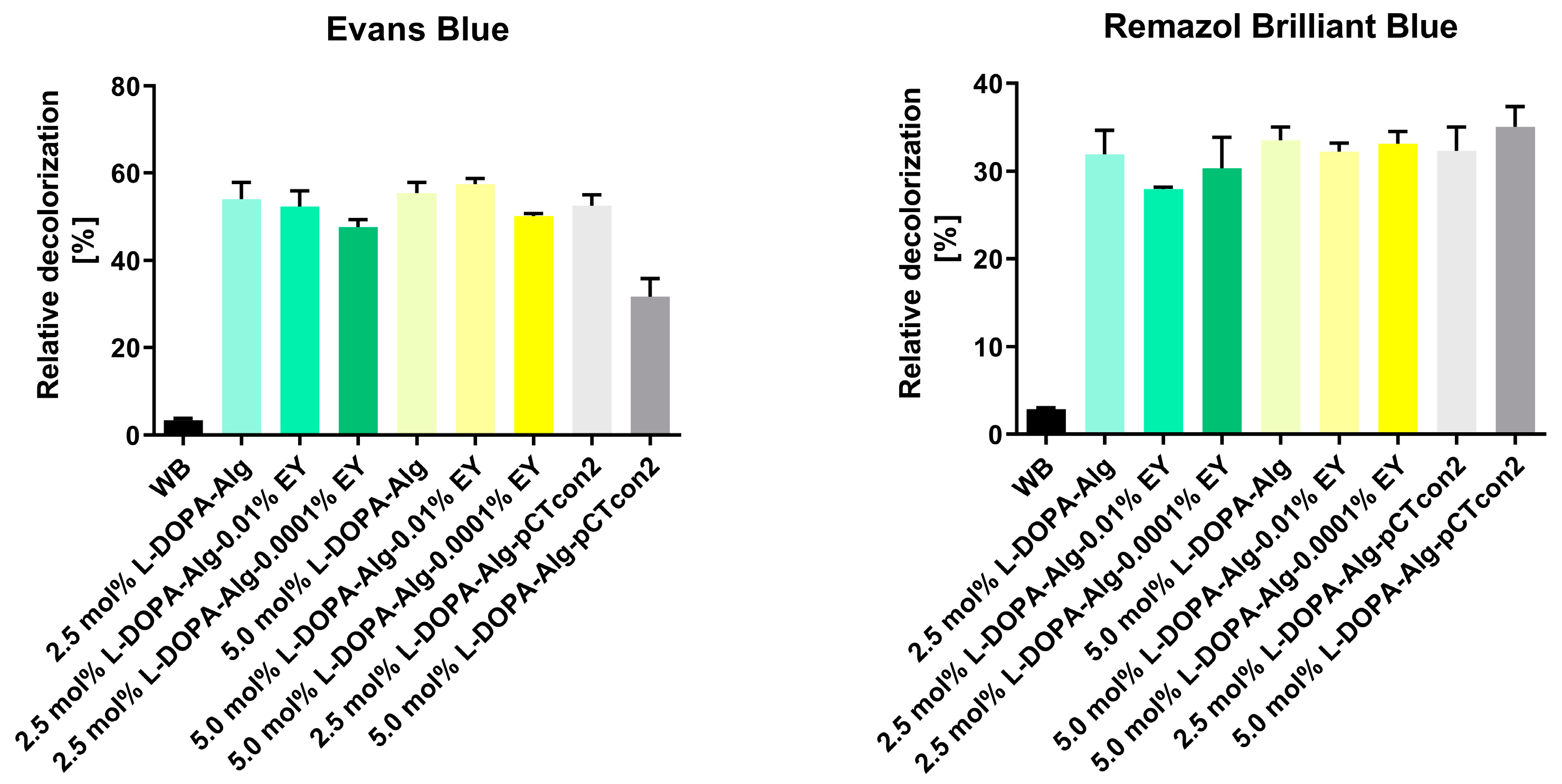
The results of Evans Blue decolorization showed that non-photopolymerized beads with both modified alginates (2.5 and 5.0 mol%) exhibited a similar percentage of relative decolorization after 48 h, 54% and 55%, respectively (Figure 7).
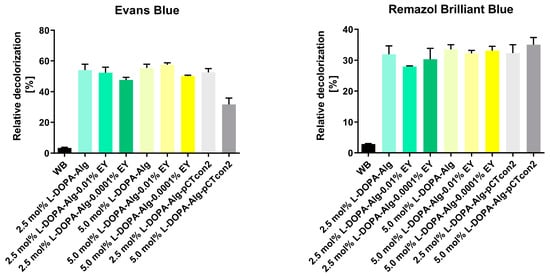
Figure 7.
Relative decolorization of dyes Evans Blue and Remazol Brilliant Blue after 48 h of dye decolorization with immobilized cell wall laccase in 2.5 and 5.0 mol% L-DOPA-alginate beads with (L-DOPA-Alg-EY) or without EY (L-DOPA-Alg) or immobilized cell walls with empty pCTcon2 (L-DOPA-Alg-pCTcon2) in 2.5 and 5.0 mol% L-DOPA-alginate beads and 0.001 and 0.0001 EY. A control reaction was performed without beads (WB).
As the blank we have used empty enzyme-free beads: samples L-DOPa-Alg-pCTcon2. When comparing the decolorization of photopolymerized beads incubated in 0.01% EY, we observe a similar degree of dye decolorization for both types of polymers in comparison to non-photopolymerized beads, with percentages of 52% and 57% for 2.5 mol% and 5.0 mol% L-DOPA-alginate, respectively. The decolorization of photopolymerized beads with the addition of a photoinitiator at a concentration of 0.0001% during immobilization was slightly less successful with both polymers, resulting in percentages of 47.6% and 50.0% for 2.5 mol% and 5.0 mol% L-DOPA-alginate beads, respectively. We can assume that in this case, there has been a higher degree of crosslinking of the beads, which could hinder the diffusion of substrate and products of enzymatic reaction. This especially applies to this reaction, with a very bulky substrate. A previous study with α-amylase showed that the enzymatic activity of immobilized enzymes is influenced by the degree of crosslinking. The enzyme’s affinity for the substrate decreases with a higher crosslinking degree owing to the slow diffusion of the substrate to the enzyme molecules inside the particles [34].
Previous studies have reported the ability of laccase to decolorize various dyes [20,21,35,36,37,38,39,40,41,42,43,44]. In certain studies, for example, laccase was successfully used to decolorize dye Evans Blue [45,46,47]. The mechanism of laccase-catalyzed degradation of Evans Blue was reported by Xia et al. [48]. The theorized mechanism for decolorization and detoxification, as deduced from LCeMS analysis, suggests that the azo bond (-N=N-) undergoes transformation into N2, with water being the only byproduct of degradation. This process ultimately leads to the decomposition of Evans Blue into several non-toxic intermediate products, resulting in a reduction in toxicity [48]. From this, we can conclude that there is a significant advantage in using laccase as a biocatalyst for removing this dye, with non-toxic intermediate products, mild conditions, etc., especially since this dye is highly prevalent in textile wastewater and is classified as a group-3 carcinogen by the International Agency for Research on Cancer (IARC) due to the potential metabolization and reduction of the azo group to cancerogenic aromatic amines [48].
Remazol Brilliant Blue decolorization results show slightly lower degrees of decolorization for all biocatalysts than the decolorization efficiency observed for Evans Blue dye. The degree of relative decolorization for all types of enzyme-incorporated beads was approximately 30% (Figure 7).
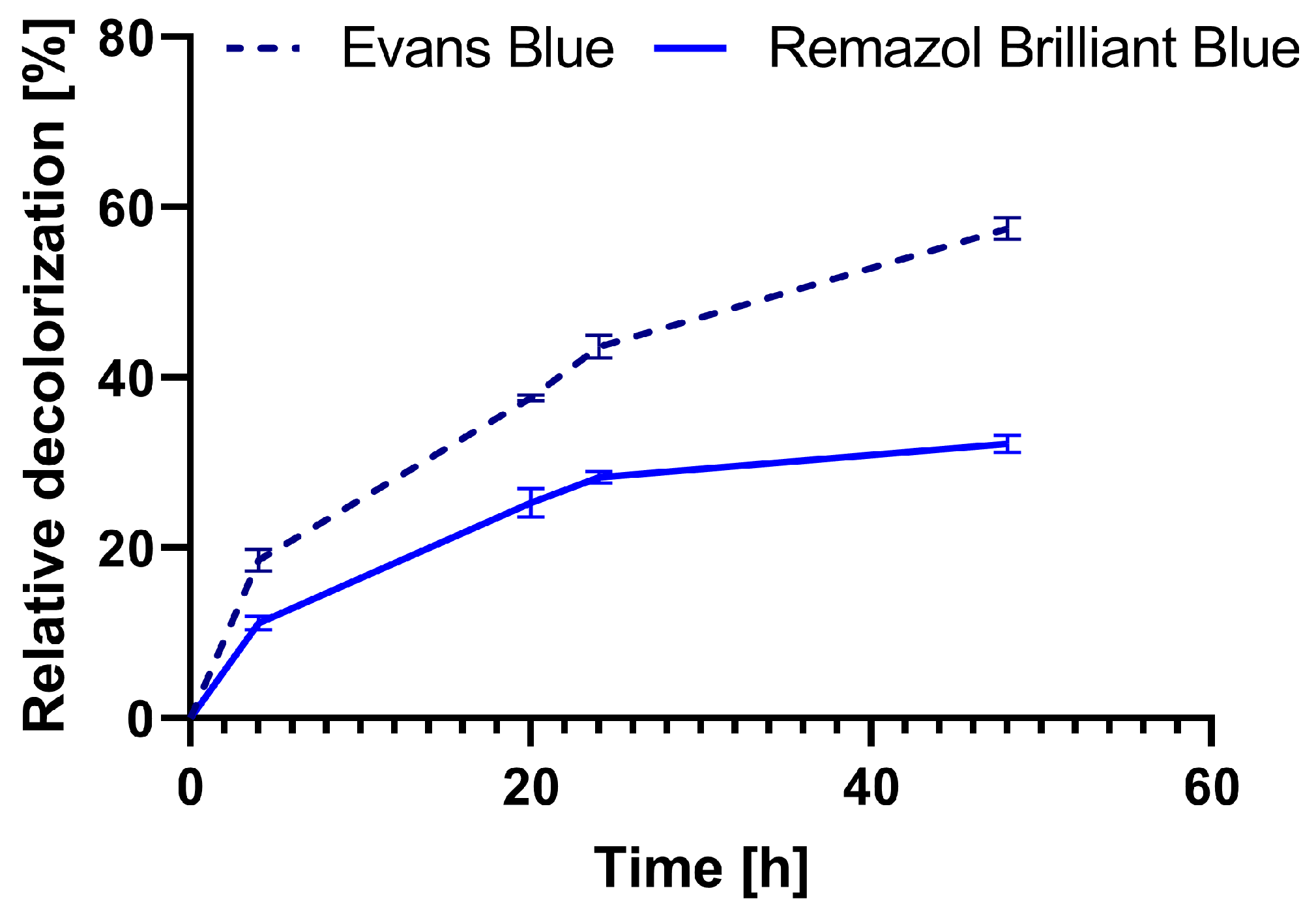
Following decolorization over time showed that after 24 h, there was no significant change in decolorization rate, so using reaction times longer than 48 h was not necessary (Figure 8). Therefore, we have used 48 h as a standard time period for one cycle of decolorization.

Figure 8.
Relative decolorization of dyes Evans Blue and Remazol Brilliant Blue during 48 h of dye decolorization with immobilized cell wall laccase in 5.0 mol% L-DOPA-alginate beads with 0.001% EY (L-DOPA-Alg-EY).
3.9. Reusability
The beads were utilized in decolorization cycles to examine the stability and potential for multiple uses of immobilized cell walls with laccase. Each decolorization cycle lasted 48 h, after which the beads were rinsed and mixed with fresh dye solution. Based on the results after the first 48 h, it was decided to continue the decolorization examination using beads with 5 mol% L-DOPA-alginates for Evans Blue and Remazol Brilliant Blue dyes. In the second cycle, for Remazol Brilliant Blue dye, the decolorization reached a maximum of 5% of the dye solution. Since there was no significant decolorization after reusing the beads for Remazol Brilliant Blue dye, further investigations into the potential for multiple uses of immobilized cell walls were conducted only with Evans Blue dye.
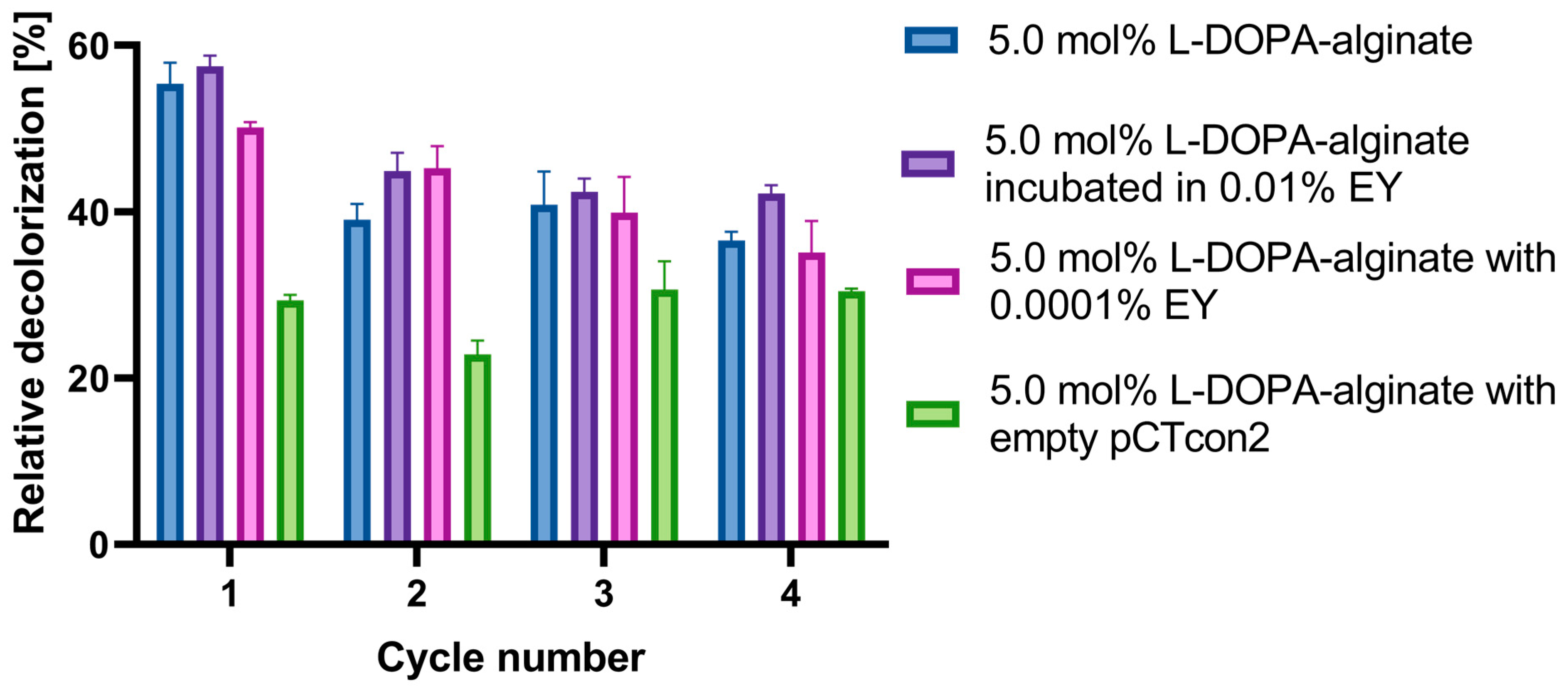
Figure 9 illustrates the results of multiple decolorization cycles of Evans Blue dye over four cycles of 48 h each. In this experiment, the following types of beads were utilized: 5 mol% L-DOPA-alginate beads with cell wall laccase without EY, beads incubated in 0.01% EY for 5 min, and beads with EY added directly during immobilization at a concentration of 0.0001%. Immobilized cell walls with the pCTcon2 vector were also employed.
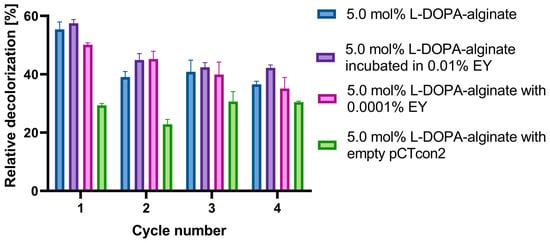
Figure 9.
Reusability of cell wall laccase immobilized in 5.0 mol% L-DOPA-alginate beads without EY/incubated in 0.01% EY/with 0.0001% EY, and cell wall with empty pCTcon2 immobilized in 5.0 mol% L-DOPA-alginate beads. Beads were used for four decolorization cycles of 48 h. After each cycle, the beads were filtrated and washed three times with 100 mM NaAc buffer pH 4.5 and replaced with a fresh solution of dyes.
The results demonstrate that the highest decolorization of Evans Blue dye in all cycles is achieved using 5 mol% L-DOPA-alginate beads incubated in 0.01% EY for 5 min and photopolymerized (Figure 9). Nearly 60% of the dye solution is decolorized in the first cycle, and 42% after the fourth cycle, making these biocatalysts a promising candidate for Evans Blue dye decolorization. The non-photopolymerized 5 mol% L-DOPA-alginate beads with cell wall laccase exhibited a slightly lower degree of decolorization, with approximately 36% of the dye removed after the fourth cycle.
4. Conclusions
From this, we can conclude that photopolymerization of the alginate polymer material containing yeast cell-wall-bound laccase led to greater enzyme stabilization, resulting in higher decolorization capability of Evans Blue dye and improved efficiency upon reuse in multiple decolorization cycles.
Reusing immobilized biocatalysts is crucial for their industrial application, significantly reducing the process cost. The laccase enzyme’s efficiency for removing and degrading dyes in multiple decolorization cycles has been previously reported and discussed [20,49,50,51,52]. Immobilized laccases present a promising, environmentally friendly, and commercially viable alternative to physical, chemical, and oxidative methods for dye decolorization.
Author Contributions
Conceptualization R.P.; methodology, A.N. and N.P.K.; validation, M.C.P. and M.S.; investigation, A.N.; writing—original draft preparation, N.P.K., M.C.P. and M.S.; writing—review and editing, R.P.; visualization, N.P.K.; supervision, N.P.K.; funding acquisition, R.P. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Ministry of Science, Technological Development, and Innovation of the Republic of Serbia, Grant No. 451-03-47/2024-01/200168 (University of Belgrade Faculty of Chemistry).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data supporting reported results can be obtained from corresponding author on request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Akshaya, S.; Nathanael, A.J. A Review on Hydrophobically Associated Alginates: Approaches and Applications. ACS Omega 2024, 9, 4246–4262. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-S.; Xie, Y.-J.; He, W. Research progress on chemical modification of alginate: A review. Carbohydr. Polym. 2011, 84, 33–39. [Google Scholar] [CrossRef]
- Tao, L.; Shi, C.; Zi, Y.; Zhang, H.; Wang, X.; Zhong, J. A review on the chemical modification of alginates for food research: Chemical nature, modification methods, product types, and application. Food Hydrocoll. 2024, 147, 109338. [Google Scholar] [CrossRef]
- Hurtado, A.; Aljabali, A.A.A.; Mishra, V.; Tambuwala, M.M.; Serrano-Aroca, A. Alginate: Enhancement Strategies for Advanced Applications. Int. J. Mol. Sci. 2022, 23, 4486. [Google Scholar] [CrossRef] [PubMed]
- Taheri-Kafrani, A.; Kharazmi, S.; Nasrollahzadeh, M.; Soozanipour, A.; Ejeian, F.; Etedali, P.; Mansouri-Tehrani, H.-A.; Razmjou, A.; Yek, S.M.-G.; Varma, R.S. Recent developments in enzyme immobilization technology for high-throughput processing in food industries. Crit. Rev. Food Sci. Nutr. 2021, 61, 3160–3196. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Rizzo, R.; Surman, F.; Zenobi-Wong, M. Guiding Lights: Tissue Bioprinting Using Photoactivated Materials. Chem. Rev. 2020, 120, 10950–11027. [Google Scholar] [CrossRef] [PubMed]
- Sharifi, S.; Sharifi, H.; Akbari, A.; Chodosh, J. Systematic optimization of visible light-induced crosslinking conditions of gelatin methacryloyl (GelMA). Sci. Rep. 2021, 11, 23276. [Google Scholar] [CrossRef] [PubMed]
- Shopperly, L.K.; Spinnen, J.; Krüger, J.; Endres, M.; Sittinger, M.; Lam, T.; Kloke, L.; Dehne, T. Blends of gelatin and hyaluronic acid stratified by stereolithographic bioprinting approximate cartilaginous matrix gradients. J. Biomed. Mater. Res. Part B Appl. Biomater. 2022, 110, 2310–2322. [Google Scholar] [CrossRef] [PubMed]
- Maiz-Fernández, S.; Pérez-Álvarez, L.; Silván, U.; Vilas-Vilela, J.L.; Lanceros-Mendez, S. Photocrosslinkable and self-healable hydrogels of chitosan and hyaluronic acid. Int. J. Biol. Macromol. 2022, 216, 291–302. [Google Scholar] [CrossRef]
- Moon, S.H.; Hwang, H.J.; Jeon, H.R.; Park, S.J.; Bae, I.S.; Yang, Y.J. Photocrosslinkable natural polymers in tissue engineering. Front. Bioeng. Biotechnol. 2023, 11, 1127757. [Google Scholar] [CrossRef]
- Lu, S.-Y.; Liu, S.; Patel, M.H.H.; Glenzinski, K.M.M.; Skory, C.D.D. Saccharomyces cerevisiae surface display of endolysin LysKB317 for control of bacterial contamination in corn ethanol fermentations. Front. Bioeng. Biotechnol. 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Moura, M.V.H.; Da Silva, G.P.; Machado, A.C.D.O.; Torres, F.A.G.; Freire, D.M.G.; Almeida, R.V. Displaying Lipase B from Candida antarctica in Pichia pastoris Using the Yeast Surface Display Approach: Prospection of a New Anchor and Characterization of the Whole Cell Biocatalyst. PLoS ONE 2015, 10, e0141454. [Google Scholar] [CrossRef] [PubMed]
- Teymennet-Ramírez, K.V.; Martínez-Morales, F.; Trejo-Hernández, M.R. Yeast Surface Display System: Strategies for Improvement and Biotechnological Applications. Front. Bioeng. Biotechnol. 2022, 9, 794742. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kondo, A. Cell-surface display of enzymes by the yeast Saccharomyces cerevisiae for synthetic biology. FEMS Yeast Res. 2015, 15, 1–9. [Google Scholar] [PubMed]
- Lim, S.; Glasgow, J.E.; Interrante, M.F.; Storm, E.M.; Cochran, J.R. Dual display of proteins on the yeast cell surface simplifies quantification of binding interactions and enzymatic bioconjugation reactions. Biotechnol. J. 2017, 12, 1600696. [Google Scholar] [CrossRef] [PubMed]
- Raeeszadeh-Sarmazdeh, M.; Greene, K.A.; Sankaran, B.; Downey, G.P.; Radisky, D.C.; Radisky, E.S. Directed evolution of the metalloproteinase inhibitor TIMP-1 reveals that its N- and C-terminal domains cooperate in matrix metalloproteinase recognition. J. Biol. Chem. 2019, 294, 9476–9488. [Google Scholar] [CrossRef]
- Piscitelli, A.; Pezzella, C.; Giardina, P.; Faraco, V.; Sannia, G. Heterologous laccase production and its role in industrial applications. Bioeng. Bugs 2010, 1, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Necochea, R.; Valderrama, B.; Dãaz-Sandoval, S.; Folch-Mallol, J.L.; Vã¡Zquez-Duhalt, R.; Iturriaga, G. Phylogenetic and biochemical characterisation of a recombinant laccase from Trametes versicolor. FEMS Microbiol. Lett. 2005, 244, 235–241. [Google Scholar] [CrossRef]
- Crnoglavac Popović, M.; Stanišić, M.; Prodanović, R. State of the Art Technologies for High Yield Heterologous Expression and Production of Oxidoreductase Enzymes: Glucose Oxidase, Cellobiose Dehydrogenase, Horseradish Peroxidase, and Laccases in Yeasts P. pastoris and S. cerevisiae. Fermentation 2024, 10, 93. [Google Scholar] [CrossRef]
- Popović, N.; Pržulj, D.; Mladenović, M.; Prodanović, O.; Ece, S.; Đurđić Ilić, K.; Ostafe, R.; Fischer, R.; Prodanović, R. Immobilization of yeast cell walls with surface displayed laccase from Streptomyces cyaneus within dopamine-alginate beads for dye decolorization. Int. J. Biol. Macromol. 2021, 181, 1072–1080. [Google Scholar] [CrossRef]
- Kunamneni, A.; Ghazi, I.; Camarero, S.; Ballesteros, A.; Plou, F.J.; Alcalde, M. Decolorization of synthetic dyes by laccase immobilized on epoxy-activated carriers. Process. Biochem. 2008, 43, 169–178. [Google Scholar] [CrossRef]
- Lin, S.H.; Peng, C.F. Treatment of textile wastewater by electrochemical method. Water Res. 1994, 28, 277–282. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Eldin, M.M.; Mita, D. Immobilized Enzymes: Strategies for Overcoming the Substrate Diffusion- Limitation Problem. Curr. Biotechnol. 2014, 3, 207–217. [Google Scholar] [CrossRef]
- Ece, S.; Lambertz, C.; Fischer, R.; Commandeur, U. Heterologous expression of a Streptomyces cyaneus laccase for biomass modification applications. AMB Express 2017, 7, 86. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, B.; Trejo-Hernández, M.R.; Morales-Guzmán, D.; Caspeta, L.; Rodríguez, R.S.; Martínez-Morales, F. Functional expression, production, and biochemical characterization of a laccase using yeast surface display technology. Fungal Biol. 2016, 120, 1609–1622. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Stemple, B.; Kumar, M.; Wei, N. Cell Surface Display Fungal Laccase as a Renewable Biocatalyst for Degradation of Persistent Micropollutants Bisphenol A and Sulfamethoxazole. Environ. Sci. Technol. 2016, 50, 8799–8808. [Google Scholar] [CrossRef] [PubMed]
- Bleve, G.; Lezzi, C.; Spagnolo, S.; Rampino, P.; Perrotta, C.; Mita, G.; Grieco, F. Construction of a Laccase Chimerical Gene: Recombinant Protein Characterization and Gene Expression via Yeast Surface Display. Appl. Biochem. Biotechnol. 2014, 172, 2916–2931. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.Z.; Guo, Q.; Cui, M.L.; Yang, L.; Du, S.S.; Ruan, H.; He, G.Q. Construction of a Yeast Cell-Surface Display System and Expression of Trametes sp. laccase. Adv. Mater. Res. 2012, 347–353, 3635–3640. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, Y.; Wei, N. Biocatalytic properties of cell surface display laccase for degradation of emerging contaminant acetaminophen in water reclamation. Biotechnol. Bioeng. 2019, 117, 342–353. [Google Scholar] [CrossRef]
- Nieto, D.; Corrales, J.A.M.; de Mora, A.J.; Moroni, L. Fundamentals of light-cell-polymer interactions in photo-cross-linking based bioprinting. APL Bioeng. 2020, 4, 041502. [Google Scholar] [CrossRef]
- Loebel, C.; Broguiere, N.; Alini, M.; Zenobi-Wong, M.; Eglin, D. Microfabrication of Photo-Cross-Linked Hyaluronan Hydrogels by Single- and Two-Photon Tyramine Oxidation. Biomacromolecules 2015, 16, 2624–2630. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Shih, H.; Muňoz, Z.; Kemp, A.; Lin, C.-C. Visible light cured thiol-vinyl hydrogels with tunable degradation for 3D cell culture. Acta Biomater. 2014, 10, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Iurciuc, C.E.T.; Bouhadiba, B.; Atanase, L.I.; Stan, C.S.; Popa, M.; Ochiuz, L. An Accessible Method to Improve the Stability and Reusability of Porcine Pancreatic α-Amylase via Immobilization in Gellan-Based Hydrogel Particles Obtained by Ionic Cross-Linking with Mg2+ Ions. Molecules 2023, 28, 4695. [Google Scholar] [CrossRef] [PubMed]
- Teerapatsakul, C.; Parra, R.; Keshavarz, T.; Chitradon, L. Repeated batch for dye degradation in an airlift bioreactor by laccase entrapped in copper alginate. Int. Biodeterior. Biodegrad. 2017, 120, 52–57. [Google Scholar] [CrossRef]
- Bagewadi, Z.K.; Mulla, S.I.; Ninnekar, H.Z. Purification and immobilization of laccase from Trichoderma harzianum strain HZN10 and its application in dye decolorization. J. Genet. Eng. Biotechnol. 2017, 15, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Daâssi Dalel, R.-C.S.; Nasri, M.M.T. Biodegradation of textile dyes by immobilized laccase from Coriolopsis gallica into Ca-alginate beads. Int. Biodeterior. Biodegrad. 2014, 90, 71–78. [Google Scholar] [CrossRef]
- Jaiswal, N.; Pandey, V.P.; Dwivedi, U.N. Immobilization of papaya laccase in chitosan led to improved multipronged stability and dye discoloration. Int. J. Biol. Macromol. 2016, 86, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Zhao, M.; Wang, Y. Immobilization of Laccase by Alginate–Chitosan Microcapsules and its Use in Dye Decolorization. World J. Microbiol. Biotechnol. 2007, 23, 159–166. [Google Scholar] [CrossRef]
- Sharma, D.; Goel, G.; Sud, A.; Chauhan, R.S. A novel laccase from newly isolated Cotylidia pannosa and its application in decolorization of synthetic dyes. Biocatal. Agric. Biotechnol. 2015, 4, 661–666. [Google Scholar] [CrossRef]
- Khaled, J.M.; Alyahya, S.A.; Govindan, R.; Chelliah, C.K.; Maruthupandy, M.; Alharbi, N.S.; Kadaikunnan, S.; Issac, R.; Murugan, S.; Li, W.-J. Laccase producing bacteria influenced the high decolorization of textile azo dyes with advanced study. Environ. Res. 2022, 207, 112211. [Google Scholar] [CrossRef]
- Jeon, S.-J.; Park, J.-H. Refolding, characterization, and dye decolorization ability of a highly thermostable laccase from Geobacillus sp. JS12. Protein Expr. Purif. 2020, 173, 105646. [Google Scholar] [CrossRef] [PubMed]
- Amari, A.; Alzahrani, F.M.; Alsaiari, N.S.; Katubi, K.M.; Ben Rebah, F.; Tahoon, M.A. Magnetic Metal Organic Framework Immobilized Laccase for Wastewater Decolorization. Processes 2021, 9, 774. [Google Scholar] [CrossRef]
- Chopra, N.K.; Sondhi, S. Cloning, expression and characterization of laccase from Bacillus licheniformis NS2324 in E. coli application in dye decolorization. Int. J. Biol. Macromol. 2022, 206, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Zabłocka-Godlewska, E.; Przystaś, W.; Grabińska-Sota, E. Decolourization of Diazo Evans Blue by Two Strains of Pseudomonas fluorescens Isolated from Different Wastewater Treatment Plants. Water, Air, Soil Pollut. 2012, 223, 5259–5266. [Google Scholar] [CrossRef] [PubMed]
- Pardo, I.; Chanagá, X.; Vicente, A.I.; Alcalde, M.; Camarero, S. New colorimetric screening assays for the directed evolution of fungal laccases to improve the conversion of plant biomass. BMC Biotechnol. 2013, 13, 90. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lin, Q.; Ng, T.B.; Ye, X.; Lin, J. Purification and characterization of a novel laccase from Cerrena sp. HYB07 with Dye Decolorizing Ability. PLoS ONE 2014, 9, e110834. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wang, Q.; Luo, Q.; Chen, Y.; Liao, X.; Guan, Z. Secretory expression and optimization of Bacillus pumilus CotA-laccase mutant GWLF in Pichia pastoris and its mechanism on Evans blue degradation. Process Biochem. 2019, 78, 33–41. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, J.; Cavaco-Paulo, A.; Su, J.; Wang, H. Encapsulated laccase in bimetallic Cu/Zn ZIFs as stable and reusable biocatalyst for decolorization of dye wastewater. Int. J. Biol. Macromol. 2023, 233, 123410. [Google Scholar] [CrossRef] [PubMed]
- Morsy, S.A.G.Z.; Tajudin, A.A.; Ali, M.S.M.; Shariff, F.M. Current Development in Decolorization of Synthetic Dyes by Immobilized Laccases. Front. Microbiol. 2020, 11, 572309. [Google Scholar] [CrossRef]
- Lopez-Barbosa, N.; Florez, S.L.; Cruz, J.C.; Ornelas-Soto, N.; Osma, J.F. Congo Red Decolorization Using Textile Filters and Laccase-Based Nanocomposites in Continuous Flow Bioreactors. Nanomaterials 2020, 10, 1227. [Google Scholar] [CrossRef]
- Silveira, T.R.; Ebling, C.D.; Magro, L.D.; Rodrigues, R.C.; Schneider, W.D.H.; Camassola, M.; de Menezes, E.W.; Meneguzzi, A.; Klein, M.P. An efficient decolorization of methyl orange dye by laccase from Marasmiellus palmivorus immobilized on chitosan-coated magnetic particles. Biocatal. Agric. Biotechnol. 2020, 30, 101859. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).