Dynamics of Water and Biofilm Bacterial Community Composition in a Mediterranean Recirculation Aquaculture System
Abstract
:1. Introduction
2. Materials and Methods
2.1. RAS Setup General Information
2.2. Water Parameter Analyses
2.3. Bacterial Abundance Assessment in Water and Biofilms
2.4. Identification of Dominant Bacterial Colonies
2.5. Monitoring of Bacterial Dynamics in Tank Water Samples and Biofilms
2.6. DNA Extraction
2.7. PCR-DGGE
2.8. Excision of Bands and Sequencing Data Analysis
2.9. Statistical Analysis
3. Results
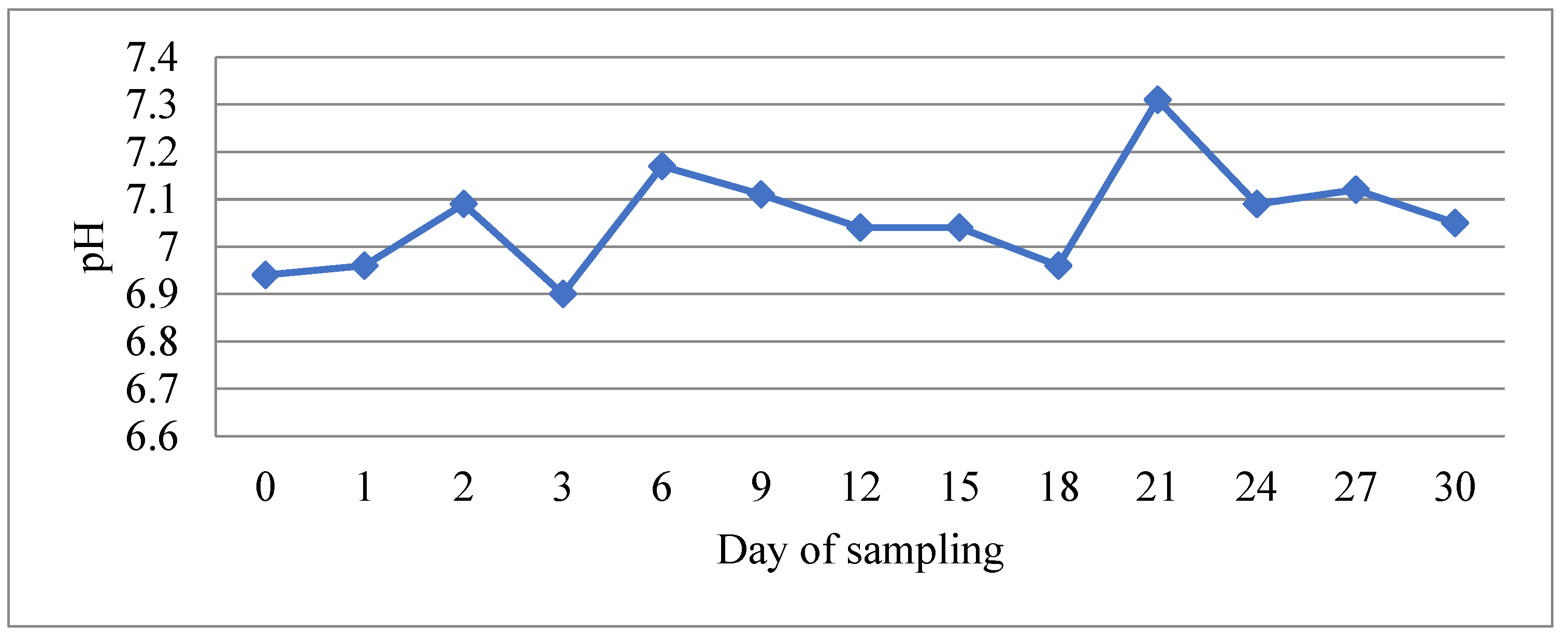
3.1. Determination of Physicochemical Parameters
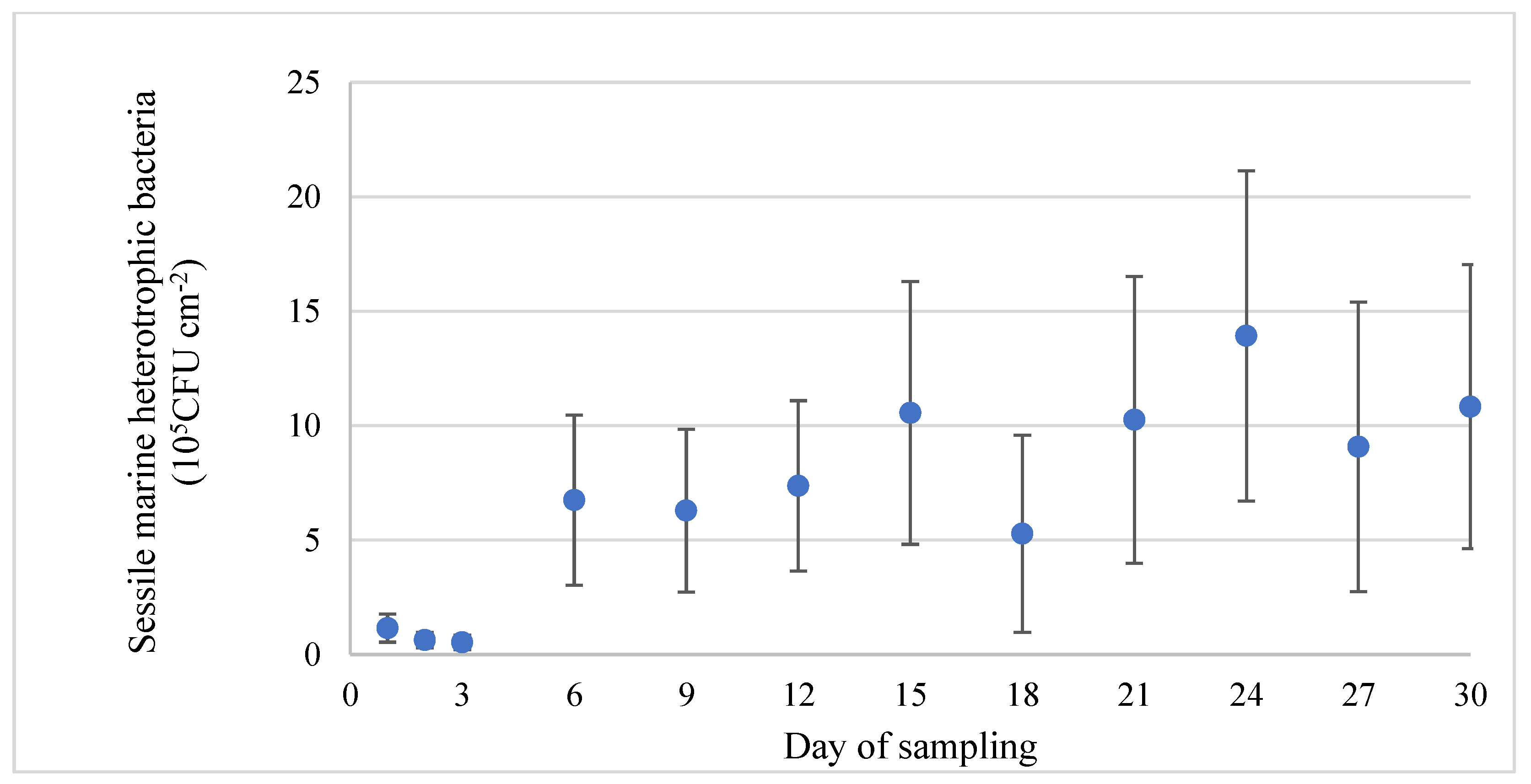
3.2. Assessment of Bacterial Population in Tank Water and on Stainless-Steel Surfaces
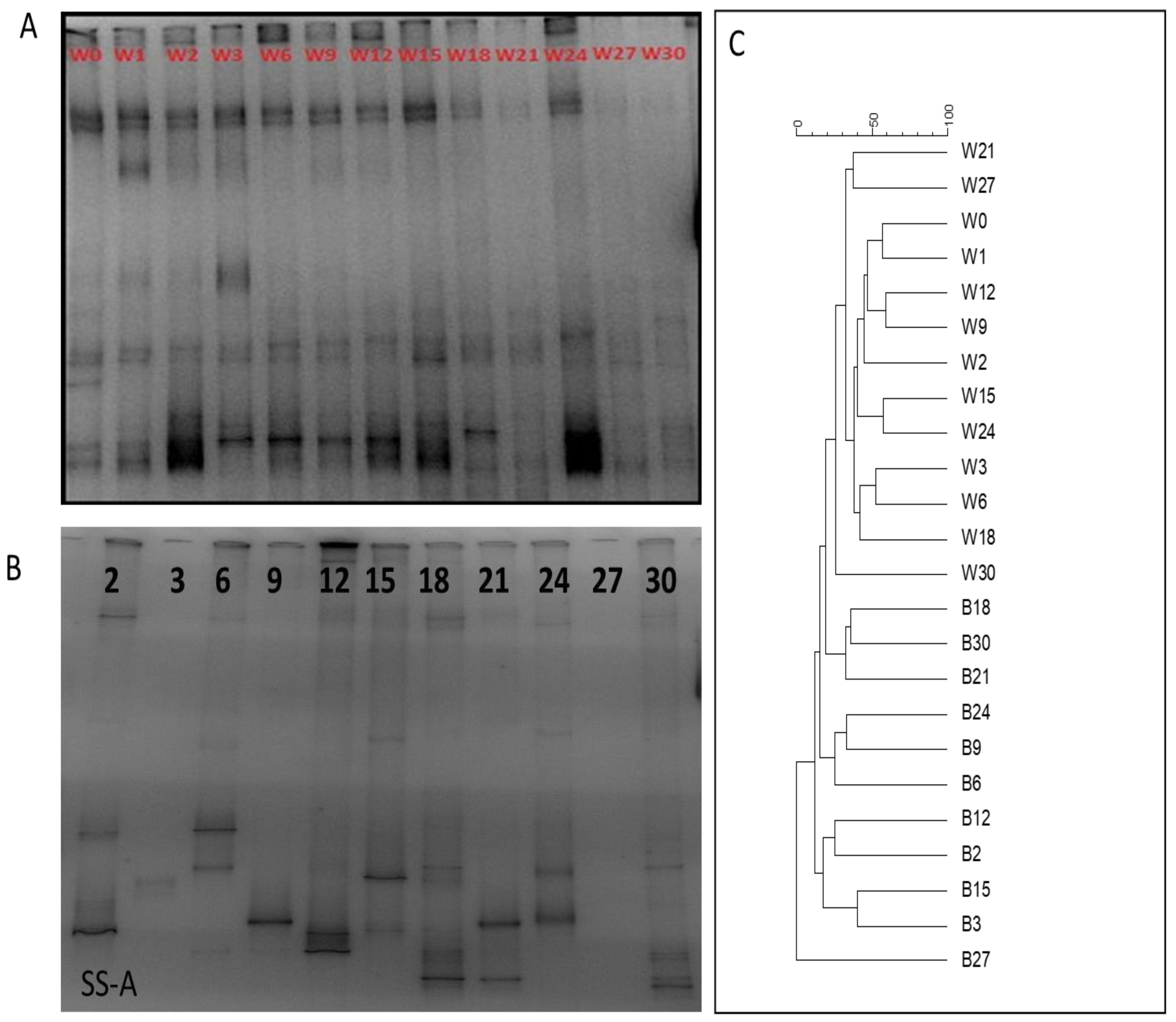
3.3. Assessment of Microbiota in Water Column and on Stainless-Steel Surfaces
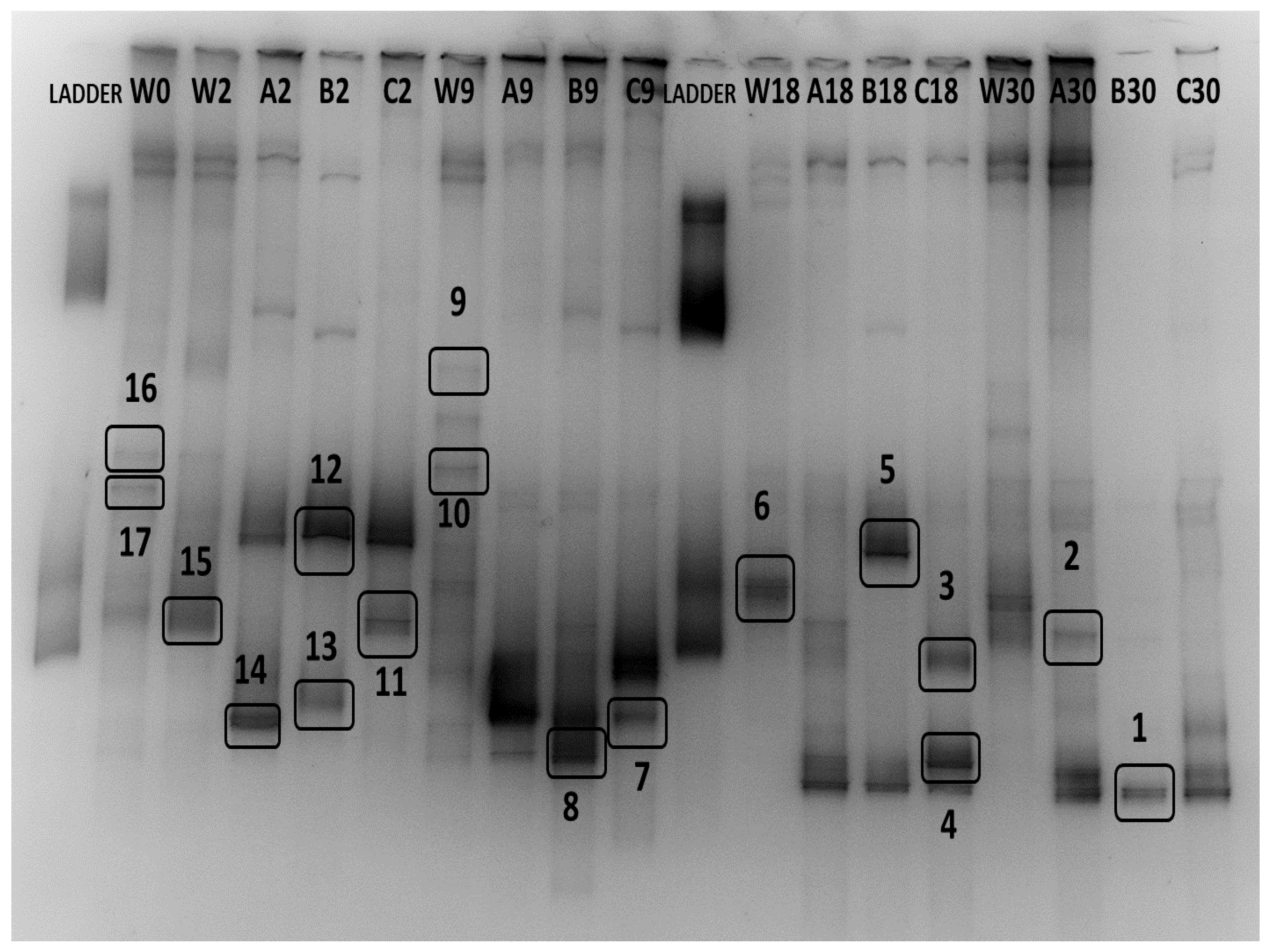
3.4. Identification of Bacterial Communities
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture 2020; FAO: Rome, Italy, 2020; Volume 32, ISBN 978-92-5-132692-3. [Google Scholar]
- Ahmed, N.; Turchini, G.M. Recirculating Aquaculture Systems (RAS): Environmental Solution and Climate Change Adaptation. J. Clean. Prod. 2021, 297, 126604. [Google Scholar] [CrossRef]
- Bregnballe, J. A Guide to Recirculation Aquaclture: An Introduction to the New Environmentlly Friendly and Highly Productive Closed Fish Farming Systems. FAO Eurofish Rep. 2015, 10–11. Available online: http://www.fao.org/3/a-i4626e.pdf (accessed on 1 March 2022).
- Martins, C.I.M.; Eding, E.H.; Verdegem, M.C.J.; Heinsbroek, L.T.N.; Schneider, O.; Blancheton, J.P.; d’Orbcastel, E.R.; Verreth, J.A.J. New Developments in Recirculating Aquaculture Systems in Europe: A Perspective on Environmental Sustainability. Aquac. Eng. 2010, 43, 83–93. [Google Scholar] [CrossRef] [Green Version]
- Dalsgaard, J.; Pedersen, L.-F.; Pedersen, P.B. Recirculation Technology: Science Meets Practice. Aquac. Eng. 2013, 53, 1. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Li, G.; Wu, H.B.; Liu, X.G.; Yao, Y.H.; Tao, L.; Liu, H. An Integrated Recirculating Aquaculture System (RAS) for Land-Based Fish Farming: The Effects on Water Quality and Fish Production. Aquac. Eng. 2011, 45, 93–102. [Google Scholar] [CrossRef]
- Dahle, S.W.; Attramadal, K.J.K.; Vadstein, O.; Hestdahl, H.I.; Bakke, I. Microbial Community Dynamics in a Commercial RAS for Production of Atlantic Salmon Fry (Salmo salar). Aquaculture 2022, 546, 737382. [Google Scholar] [CrossRef]
- Bugten, A.V.; Attramadal, K.J.K.; Fossmark, R.O.; Rosten, T.W.; Vadstein, O.; Bakke, I. Changes in Rearing Water Microbiomes in RAS Induced by Membrane Filtration Alters the Hindgut Microbiomes of Atlantic Salmon (Salmo salar) Parr. Aquaculture 2022, 548, 737661. [Google Scholar] [CrossRef]
- Pulkkinen, J.T.; Kiuru, T.; Aalto, S.L.; Koskela, J.; Vielma, J. Startup and Effects of Relative Water Renewal Rate on Water Quality and Growth of Rainbow Trout (Oncorhynchus mykiss) in a Unique RAS Research Platform. Aquac. Eng. 2018, 82, 38–45. [Google Scholar] [CrossRef]
- Rojas-Tirado, P.; Pedersen, P.B.; Vadstein, O.; Pedersen, L.F. Changes in Microbial Water Quality in RAS Following Altered Feed Loading. Aquac. Eng. 2018, 81, 80–88. [Google Scholar] [CrossRef]
- McIntosh, D.; Ji, B.; Forward, B.S.; Puvanendran, V.; Boyce, D.; Ritchie, R. Culture-Independent Characterization of the Bacterial Populations Associated with Cod (Gadus morhua L.) and Live Feed at an Experimental Hatchery Facility Using Denaturing Gradient Gel Electrophoresis. Aquaculture 2008, 275, 42–50. [Google Scholar] [CrossRef]
- Attramadal, K.J.K.; Truong, T.M.H.; Bakke, I.; Skjermo, J.; Olsen, Y.; Vadstein, O. RAS and Microbial Maturation as Tools for K-Selection of Microbial Communities Improve Survival in Cod Larvae. Aquaculture 2014, 432, 483–490. [Google Scholar] [CrossRef]
- Powell, A.; Chingombe, P.; Lupatsch, I.; Shields, R.J.; Lloyd, R. The Effect of Ozone on Water Quality and Survival of Turbot (Psetta maxima) Maintained in a Recirculating Aquaculture System. Aquac. Eng. 2015, 64, 20–24. [Google Scholar] [CrossRef]
- Leonard, N.; Blancheton, J.P.; Guiraud, J.P. Populations of Heterotrophic Bacteria in an Experimental Recirculating Aquaculture System. Aquac. Eng. 2000, 22, 109–120. [Google Scholar] [CrossRef]
- Almeida, D.B.; Magalhães, C.; Sousa, Z.; Borges, M.T.; Silva, E.; Blanquet, I.; Mucha, A.P. Microbial Community Dynamics in a Hatchery Recirculating Aquaculture System (RAS) of Sole (Solea senegalensis). Aquaculture 2021, 539, 736592. [Google Scholar] [CrossRef]
- Blancheton, J.P.; Attramadal, K.J.K.; Michaud, L.; d’Orbcastel, E.R.; Vadstein, O. Insight into Bacterial Population in Aquaculture Systems and Its Implication. Aquac. Eng. 2013, 53, 30–39. [Google Scholar] [CrossRef]
- Attramadal, K.J.K.; Salvesen, I.; Xue, R.; Øie, G.; Størseth, T.R.; Vadstein, O.; Olsen, Y. Recirculation as a Possible Microbial Control Strategy in the Production of Marine Larvae. Aquac. Eng. 2012, 46, 27–39. [Google Scholar] [CrossRef]
- Bourne, D.G.; Høj, L.; Webster, N.S.; Swan, J.; Hall, M.R. Biofilm Development within a Larval Rearing Tank of the Tropical Rock Lobster, Panulirus ornatus. Aquaculture 2006, 260, 27–38. [Google Scholar] [CrossRef]
- Mizan, M.F.R.; Jahid, I.K.; Ha, S. Do Microbial Biofilms in Seafood: A Food-Hygiene Challenge. Food Microbiol. 2015, 49, 41–55. [Google Scholar] [CrossRef]
- Fletcher, M. Bacterial biofilms and biofouling. Curr. Opin. Biotechnol. 1994, 5, 302–306. [Google Scholar] [CrossRef]
- Schäfer, H.; Muyzer, G. Denaturing Gradient Gel Electrophoresis in Marine Microbial Ecology. In Methods in Microbiology; Academic Press Inc.: Cambridge, MA, USA, 2001; Volume 30, pp. 425–468. ISBN 0-12-521530-4. [Google Scholar]
- Muyzer, G.; de Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes encoding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef] [Green Version]
- Rombaut, G.; Suantika, G.; Boon, N.; Maertens, S.; Dhert, P.; Top, E.; Sorgeloos, P.; Verstraete, W. Monitoring of the Evolving Diversity of the Microbial Community Present in Rotifer Cultures. Aquaculture 2001, 198, 237–252. [Google Scholar] [CrossRef]
- Sandaa, R.A.; Magnesen, T.; Torkildsen, L.; Bergh, Ø. Characterisation of the Bacterial Community Associated with Early Stages of Great Scallop (Pecten maximus), Using Denaturing Gradient Gel Electrophoresis (DGGE). Syst. Appl. Microbiol. 2003, 26, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Wietz, M.; Hall, M.R.; Høj, L. Effects of Seawater Ozonation on Biofilm Development in Aquaculture Tanks. Syst. Appl. Microbiol. 2009, 32, 266–277. [Google Scholar] [CrossRef] [PubMed]
- van der Meeren, T.; Brunvold, L.; Sandaa, R.A.; Bergh, Ø.; Castberg, T.; Thyrhaug, R.; Mangor-Jensen, A. Water Quality and Microbial Community Structure in Juvenile Atlantic Cod (Gadus morhua L.) Cultures. Aquaculture 2011, 316, 111–120. [Google Scholar] [CrossRef] [Green Version]
- Attramadal, K.J.K.; Minniti, G.; Øie, G.; Kjørsvik, E.; Østensen, M.A.; Bakke, I.; Vadstein, O. Microbial Maturation of Intake Water at Different Carrying Capacities Affects Microbial Control in Rearing Tanks for Marine Fish Larvae. Aquaculture 2016, 457, 68–72. [Google Scholar] [CrossRef]
- Attramadal, K.J.K.; Øien, J.V.; Kristensen, E.; Evjemo, J.O.; Kjørsvik, E.; Vadstein, O.; Bakke, I. UV Treatment in RAS Influences the Rearing Water Microbiota and Reduces the Survival of European Lobster Larvae (Homarus gammarus). Aquac. Eng. 2021, 94, 102176. [Google Scholar] [CrossRef]
- Brunvold, L.; Sandaa, R.A.; Mikkelsen, H.; Welde, E.; Bleie, H.; Bergh, Ø. Characterisation of Bacterial Communities Associated with Early Stages of Intensively Reared Cod (Gadus morhua) Using Denaturing Gradient Gel Electrophoresis (DGGE). Aquaculture 2007, 272, 319–327. [Google Scholar] [CrossRef]
- Tal, Y.; Watts, J.E.M.; Schreier, S.B.; Sowers, K.R.; Schreier, H.J. Characterization of the Microbial Community and Nitrogen Transformation Processes Associated with Moving Bed Bioreactors in a Closed Recirculated Mariculture System. Aquaculture 2003, 215, 187–202. [Google Scholar] [CrossRef]
- Standard Method 4500-NH3-H; Standard Methods for The Examination of Water and Wastewater. American Public Health Association, American Water Works Association, Water Environment Federation: Washington, DC, USA, 1992; pp. 4–84.
- Standard Method 4500-NO2-B; Standard Methods for The Examination of Water and Wastewater. American Public Health Association, American Water Works Association, Water Environment Federation: Washington, DC, USA, 1992; pp. 4–85.
- Bower, C.E.; Bidwell, J.P. Ionization of ammonia in seawater—Effects of temperature, pH and salinity. J. Fish. Res. Board Can. 1978, 35, 1012–1016. [Google Scholar] [CrossRef]
- Kostaki, M.; Chorianopoulos, N.; Braxou, E.; Nychas, G.J.; Giaouris, E. Differential Biofilm Formation and Chemical Disinfection Resistance of Sessile Cells of Listeria monocytogenes Strains under Monospecies and Dual-Species (with Salmonella enterica) Conditions. Appl. Environ. Microbiol. 2012, 78, 2586–2595. [Google Scholar] [CrossRef] [Green Version]
- Lindsay, D.; von Holy, A. Evaluation of Dislodging Methods for Laboratory-Grown Bacterial Biofilms. Food Microbiol. 1997, 14, 383–390. [Google Scholar] [CrossRef]
- Lytou, A.E.; Schoina, E.; Liu, Y.; Michalek, K.; Stanley, M.S.; Panagou, E.Z.; Nychas, G.E. Quality and Safety Assessment of Edible Seaweeds Alaria esculenta and Saccharina latissima Cultivated in Scotland. Foods 2021, 10, 2210. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; Smalla, K. Application of Denaturing Gradient Electrophoresis (DGGE) and Temperature Gradient Electrophoresis (TGGE) in Microbial Ecology. Antonie Van Leeuwenhoek 1998, 73, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Doulgeraki, A.I.; Paramithiotis, S.; Nychas, G.J.E. Characterization of the Enterobacteriaceae Community That Developed during Storage of Minced Beef under Aerobic or Modified Atmosphere Packaging Conditions. Int. J. Food Microbiol. 2011, 145, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams, 2007–2015. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 1 March 2022).
- Fossmark, R.O.; Vadstein, O.; Rosten, T.W.; Bakke, I.; Košeto, D.; Bugten, A.V.; Helberg, G.A.; Nesje, J.; Jørgensen, N.O.G.; Raspati, G.; et al. Effects of Reduced Organic Matter Loading through Membrane Filtration on the Microbial Community Dynamics in Recirculating Aquaculture Systems (RAS) with Atlantic Salmon Parr (Salmo Salar). Aquaculture 2020, 524, 735268. [Google Scholar] [CrossRef]
- Holan, A.B.; Wold, P.A.; Leiknes, T.O. Intensive Rearing of Cod Larvae (Gadus morhua) in Recirculating Aquaculture Systems (RAS) Implementing a Membrane Bioreactor (MBR) for Enhanced Colloidal Particle and Fine Suspended Solids Removal. Aquac. Eng. 2014, 58, 52–58. [Google Scholar] [CrossRef]
- Olsen, L.M.; Hernández, K.L.; Van Ardelan, M.; Iriarte, J.L.; Bizsel, K.C.; Olsen, Y. Responses in Bacterial Community Structure to Waste Nutrients from Aquaculture: An in situ Microcosm Experiment in a Chilean Fjord. Aquac. Environ. Interact. 2017, 9, 21–32. [Google Scholar] [CrossRef] [Green Version]
- Dahle, S.W.; Bakke, I.; Birkeland, M.; Nordøy, K.; Dalum, A.S.; Attramadal, K.J.K. Production of Lumpfish (Cyclopterus lumpus L.) in RAS with Distinct Water Treatments: Effects on Fish Survival, Growth, Gill Health and Microbial Communities in Rearing Water and Biofilm. Aquaculture 2020, 522, 735097. [Google Scholar] [CrossRef]
- Gullian, M.; Espinosa-Faller, F.J.; Núñez, A.; López-Barahona, N. Effect of Turbidity on the Ultraviolet Disinfection Performance in Recirculating Aquaculture Systems with Low Water Exchange. Aquac. Res. 2012, 43, 595–606. [Google Scholar] [CrossRef]
- Song, T.; Zhang, X.; Li, J.; Wu, X.; Feng, H.; Dong, W. A Review of Research Progress of Heterotrophic Nitrification and Aerobic Denitrification Microorganisms (HNADMs). Sci. Total Environ. 2021, 801, 149319. [Google Scholar] [CrossRef]
- Luo, G.; Xu, J.; Meng, H. Nitrate Accumulation in Biofloc Aquaculture Systems. Aquaculture 2020, 520, 734675. [Google Scholar] [CrossRef]
- Rejish Kumar, V.J.; Sukumaran, V.; Achuthan, C.; Joseph, V.; Philip, R.; Bright Singh, I.S. Molecular Characterization of the Nitrifying Bacterial Consortia Employed for the Activation of Bioreactors Used in Brackish and Marine Aquaculture Systems. Int. Biodeterior. Biodegrad. 2013, 78, 74–81. [Google Scholar] [CrossRef]
- Liu, Y.; Ai, G.M.; Miao, L.L.; Liu, Z.P. Marinobacter Strain NNA5, a Newly Isolated and Highly Efficient Aerobic Denitrifier with Zero N2O Emission. Bioresour. Technol. 2016, 206, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Anh, H.T.H.; Shahsavari, E.; Bott, N.J.; Ball, A.S. The Application of Marinobacter hydrocarbonoclasticus as a Bioaugmentation Agent for the Enhanced Treatment of Non-Sterile Fish Wastewater. J. Environ. Manag. 2021, 291, 112658. [Google Scholar] [CrossRef] [PubMed]
- Cytryn, E.; Minz, D.; Gelfand, I.; Neori, A.; Gieseke, A.; De Beer, D.; Van Rijn, J. Sulfide-Oxidizing Activity and Bacterial Community Structure in a Fluidized Bed Reactor from a Zero-Discharge Mariculture System. Environ. Sci. Technol. 2005, 39, 1802–1810. [Google Scholar] [CrossRef] [PubMed]
- Cytryn, E.; Van Rijn, J.; Schramm, A.; Gieseke, A.; De Beer, D.; Minz, D. Identification of Bacteria Potentially Responsible for Oxic and Anoxic Sulfide Oxidation in Biofilters of a Recirculating Mariculture System. Appl. Environ. Microbiol. 2005, 71, 6134–6141. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.H.; Park, S.; Jung, Y.T.; Lee, J.S.; Lee, K.C. Paraperlucidibaca wandonensis Sp. Nov., Isolated from Seawater, and Emended Description of the Genus Paraperlucidibaca Oh et al. 2011. Int. J. Syst. Evol. Microbiol. 2013, 63, 4113–4117. [Google Scholar] [CrossRef]
- Soininen, L.; Grönroos, M.; Roslund, M.I.; Sinkkonen, A. Long-Term Storage Affects Resource Availability and Occurrence of Bacterial Taxa Linked to Pollutant Degradation and Human Health in Landscaping Materials. Urban For. Urban Green. 2021, 60, 127065. [Google Scholar] [CrossRef]
- Lofthus, S.; Bakke, I.; Tremblay, J.; Greer, C.W.; Brakstad, O.G. Biodegradation of Weathered Crude Oil in Seawater with Frazil Ice. Mar. Pollut. Bull. 2020, 154, 111090. [Google Scholar] [CrossRef]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival Mechanisms of Clinically Revelant Microorganisms. Clin. Microbiol. Revies 2002, 15, 167–193. [Google Scholar] [CrossRef] [Green Version]
- Finore, I.; Di Donato, P.; Mastascusa, V.; Nicolaus, B.; Poli, A. Fermentation Technologies for the Optimization of Marine Microbial Exopolysaccharide Production. Mar. Drugs 2014, 12, 3005–3024. [Google Scholar] [CrossRef] [Green Version]
- Panigrahi, A.; Saranya, C.; Sundaram, M.; Vinoth Kannan, S.R.; Das, R.R.; Satish Kumar, R.; Rajesh, P.; Otta, S.K. Carbon: Nitrogen (C:N) Ratio Level Variation Influences Microbial Community of the System and Growth as Well as Immunity of Shrimp (Litopenaeus Vannamei) in Biofloc Based Culture System. Fish Shellfish Immunol. 2018, 81, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, K.; Lin, X.; He, P.; Li, G. Production and Characterization of an Extracellular Polysaccharide of Antarctic Marine Bacteria Pseudoalteromonas Sp. S-15-13. Acta Oceanol. Sin. 2006, 25, 106–115. [Google Scholar]
- Roca, C.; Lehmann, M.; Torres, C.A.V.; Baptista, S.; Gaudêncio, S.P.; Freitas, F.; Reis, M.A.M. Exopolysaccharide Production by a Marine Pseudoalteromonas Sp. Strain Isolated from Madeira Archipelago Ocean Sediments. N. Biotechnol. 2016, 33, 460–466. [Google Scholar] [CrossRef]
- Holmström, C.; Kjelleberg, S. Marine Pseudoalteromonas Species Are Associated with Higher Organisms and Produce Biologically Active Extracellular Agents. FEMS Microbiol. Ecol. 1999, 30, 285–293. [Google Scholar] [CrossRef]
- Offret, C.; Desriac, F.; Le Chevalier, P.; Mounier, J.; Jégou, C.; Fleury, Y. Spotlight on Antimicrobial Metabolites from the Marine Bacteria Pseudoalteromonas: Chemodiversity and Ecological Significance. Mar. Drugs 2016, 14, 129. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Álvarez, C.; Santos, Y. Identification and Typing of Fish Pathogenic Species of the Genus Tenacibaculum. Appl. Microbiol. Biotechnol. 2018, 102, 9973–9989. [Google Scholar] [CrossRef]
- Bourne, D.G.; Young, N.; Webster, N.; Payne, M.; Salmon, M.; Demel, S.; Hall, M. Microbial Community Dynamics in a Larval Aquaculture System of the Tropical Rock Lobster, Panulirus ornatus. Aquaculture 2004, 242, 31–51. [Google Scholar] [CrossRef]
- Giaouris, E.D.; Nychas, G.-J.E. The adherence of Salmonella Enteritidis PT4 to stainless steel: The importance of the air-liquid interface and nutrient availability. Food Microbiol. 2006, 23, 747–752. [Google Scholar] [CrossRef]



| Isolate Code | Source of Isolation | Closest Relative | Accession Number | Similarity (%) | Taxon (Class) |
|---|---|---|---|---|---|
| MB1 | Biofilm | Tenacibaculum discolor | NR_042576.1 | 100.00 | Flavobacteriia |
| MB2 | Biofilter | Pseudoalteromonas spongiae | NR_043172.1 | 100.00 | Gamma-proteobacteria |
| MB3 | Biofilter | Tamlana crocina | NR_115857.1 | 99.62 | Flavobacteriia |
| MB4 | Biofilter | Pseudoalteromonas gelatinilytica | NR_152003.1 | 99.65 | Gamma-proteobacteria |
| MB5 | Biofilter | Mesonia algae | NR_025263.1 | 100.00 | Flavobacteriia |
| MB6 | Water column | Pseudoalteromonas shioyasakiensis | NR_125458.1 | 99.87 | Gamma-proteobacteria |
| MB7 | Water column | Algibacter lectus | NR_132290.1 | 96.71 | Flavobacteriia |
| MB8 | Water column | Vibrio atypicus | NR_116535.1 | 99.80 | Gamma-proteobacteria |
| MB9 | Water column | Roseovarius halotolerans | NR_116320.1 | 99.08 | Alpha-proteobacteria |
| MB10 | Water column | Winogradskyella sediminis | NR_151891.1 | 99.82 | Flavobacteriia |
| Band Number | Source | Closest Relative | Accession Number | Similarity (%) | Taxon (class) | Found in |
|---|---|---|---|---|---|---|
| 1 | Biofilm day 30 | Rhodococcus sp. Strain 639 | KY974223 | 100 (501/501) | Actinobacteria | Biofilm day 18, Biofilm day 30 |
| 2 | Biofilm day 30 | No significant similarity | - | - | - | Biofilm day 18, Biofilm day 30 |
| 3 | Biofilm day 18 | Marinobacter sp. | AB026946.1 | 99 (511/518) | Gamma-proteobacteria | Biofilm day 9, Biofilm day 18 |
| 4 | Biofilm day 18 | Microbacterium oxydans strain CJ-G-PYD5 | HM584226.1 | 100 (494/494) | Actinobacteria | Biofilm day 18, Biofilm day 30 |
| 5 | Biofilm day 18 | Psychrobacter sp. strain TaseBurcu001 | MN923049.1 | 100 (506/506) | Gamma-proteobacteria | Biofilm day 2, Biofilm day 18 |
| 6 | Water column day 18 | Uncultured Roseobacter sp. clone C139300178 | JX528567.1 | 98 (450/457) | Alpha-proteobacteria | Water column day 0, 9, 18, 30 |
| 7 | Biofilm day 9 | Hydrogenophaga sp. 7A-385 | KF441648.1 | 95 (430/454) | Beta-proteobacteria | Biofilm day 9, Biofilm day 30 |
| 8 | Biofilm day 9 | Halomonas neptunia strain SCA-83 | MT114601.1 | 100 (509/509) | Gamma-proteobacteria | Biofilm day 9, |
| 9 | Water column day 9 | Marine bacterium | AB377218.1 | 99 (484/487) | Flavobacteriia | Water column day 9, 30 |
| 10 | Water column day 9 | N/A | - | - | Water column day 9 | |
| 11 | Biofillm day 2 | Paraperlucidibaca wandonensis strain WT-RY4 | NR_109730.1 | 99 (515/516) | Gamma-proteobacteria | Water column day 0, 9, 30 |
| 12 | Biofilm day 2 | Psychrobacter sp. strain TaseBurcu001 | MN923049.1 | 100 (507/507) | Gamma-proteobacteria | Biofilm day 2 |
| 13 | Biofilm day 2 | Planococcus rifietoensis strain NF29 | MT269280.1 | 99 (506/507) | Bacilli | Biofilm day 2 |
| 14 | Biofilm day 2 | Hydrogenophaga laconesensis strain 0-12 | MN061010.1 | 99 (504/506) | Beta-proteobacteria | Biofilm day 2 |
| 15 | Water day 2 | Colwellia sp. R2A112631 | LR722804.1 | 100 (515/515) | Gamma-proteobacteria | Water column day 2, 9, 18 |
| 16 | Water day 0 | Uncultured bacterium isolate DGGE gel band B11 | KP966419.1 | 99 (504/509) | N/A | Water column day 0, 2, 30 Biofilm day 30 |
| 17 | Water day 0 | N/A | - | - | - | Water column day 0, 30 Biofilm day 30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schoina, E.; Doulgeraki, A.I.; Miliou, H.; Nychas, G.-J.E. Dynamics of Water and Biofilm Bacterial Community Composition in a Mediterranean Recirculation Aquaculture System. Aquac. J. 2022, 2, 164-179. https://doi.org/10.3390/aquacj2020008
Schoina E, Doulgeraki AI, Miliou H, Nychas G-JE. Dynamics of Water and Biofilm Bacterial Community Composition in a Mediterranean Recirculation Aquaculture System. Aquaculture Journal. 2022; 2(2):164-179. https://doi.org/10.3390/aquacj2020008
Chicago/Turabian StyleSchoina, Eirini, Agapi I. Doulgeraki, Helen Miliou, and George-John E. Nychas. 2022. "Dynamics of Water and Biofilm Bacterial Community Composition in a Mediterranean Recirculation Aquaculture System" Aquaculture Journal 2, no. 2: 164-179. https://doi.org/10.3390/aquacj2020008
APA StyleSchoina, E., Doulgeraki, A. I., Miliou, H., & Nychas, G.-J. E. (2022). Dynamics of Water and Biofilm Bacterial Community Composition in a Mediterranean Recirculation Aquaculture System. Aquaculture Journal, 2(2), 164-179. https://doi.org/10.3390/aquacj2020008






