Design of a Cyclodextrin Bioproduction Process Using Bacillus pseudofirmus and Paenibacillus macerans
Abstract
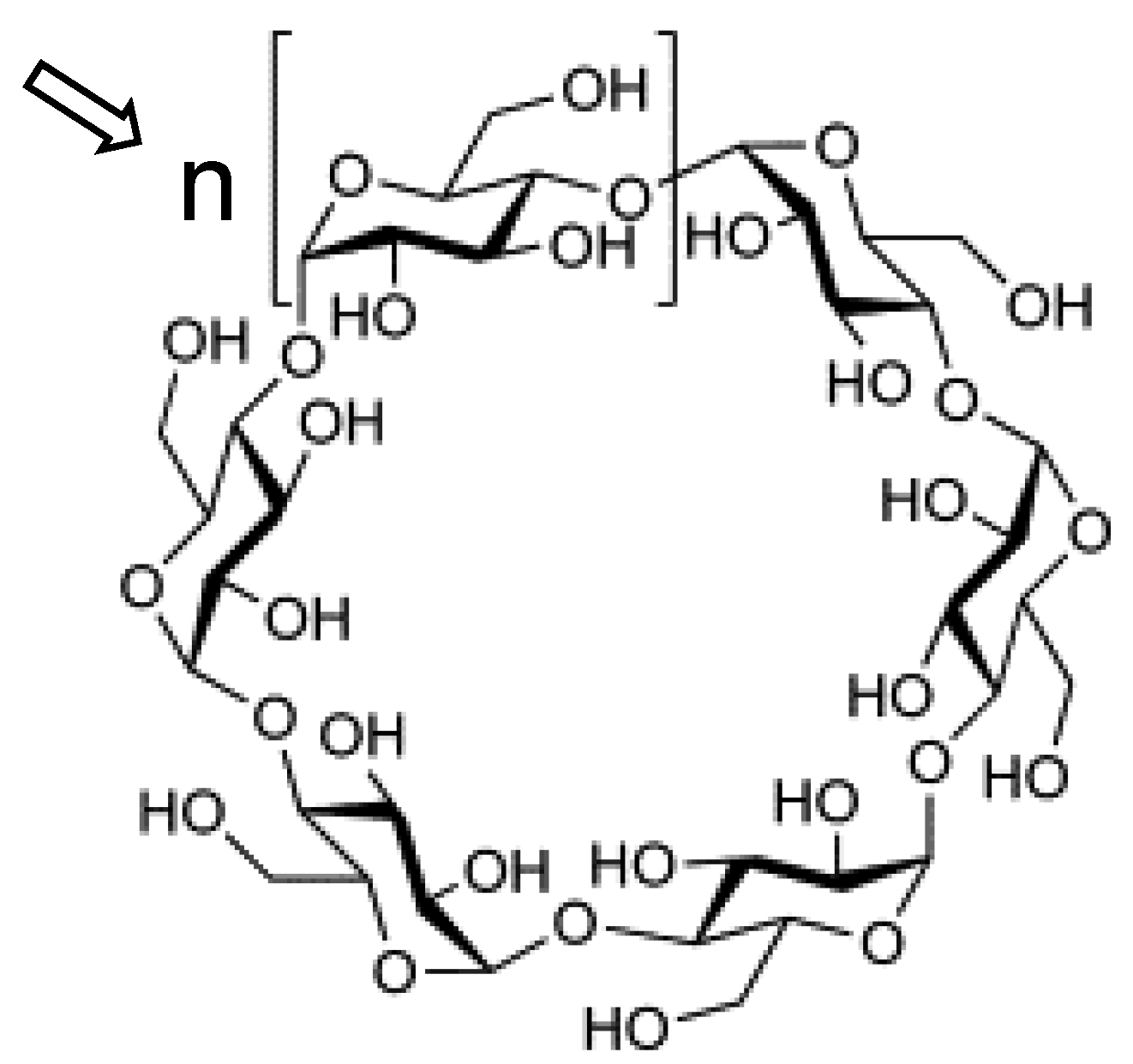
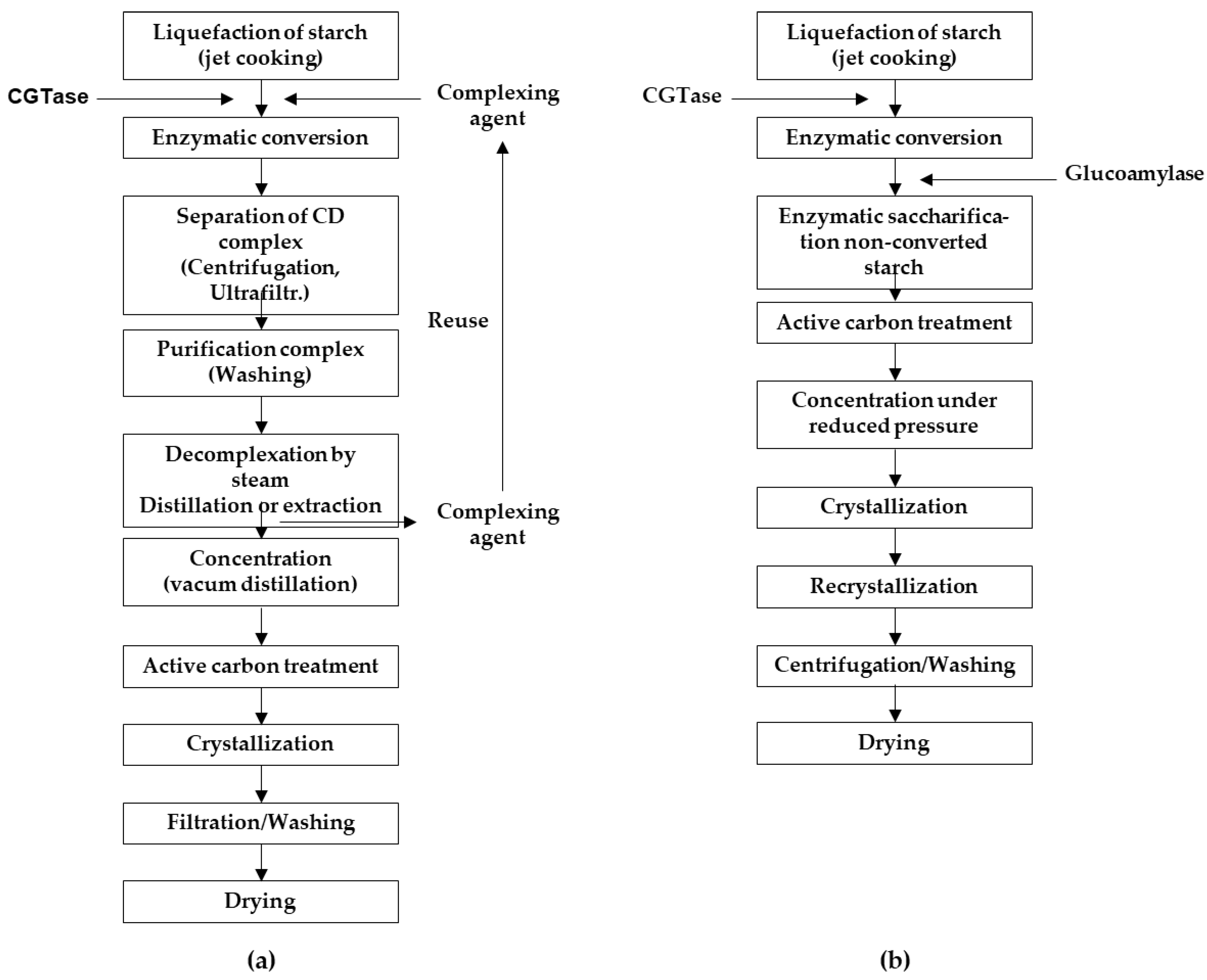
1. Introduction
2. Materials and Methods
2.1. Microrganisms
2.2. CGTase Production Media
- (i)
- Nutrient solutions
- (ii)
- Preparation of the saline and buffer solutions:
2.3. CGTase Production Assays
2.4. Analytical Methods
2.5. Analysis
3. Results and Discussion
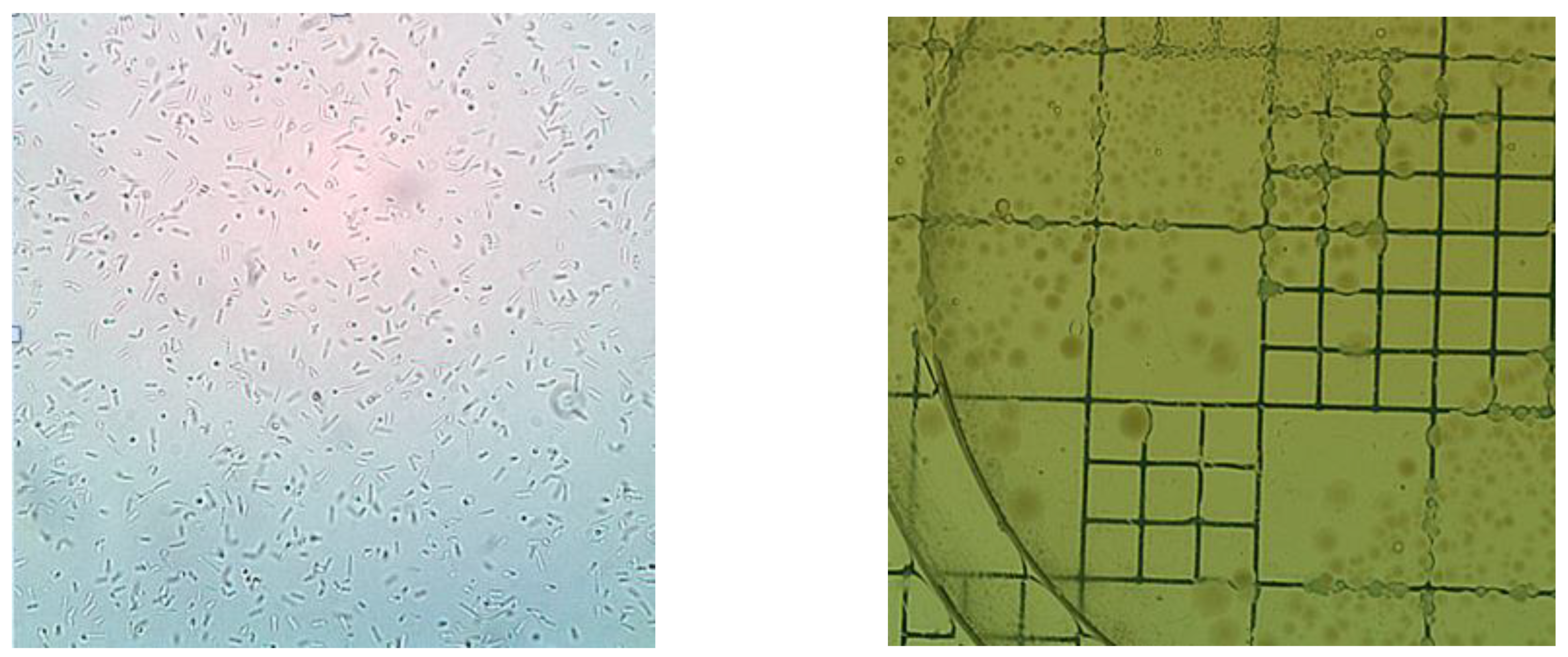
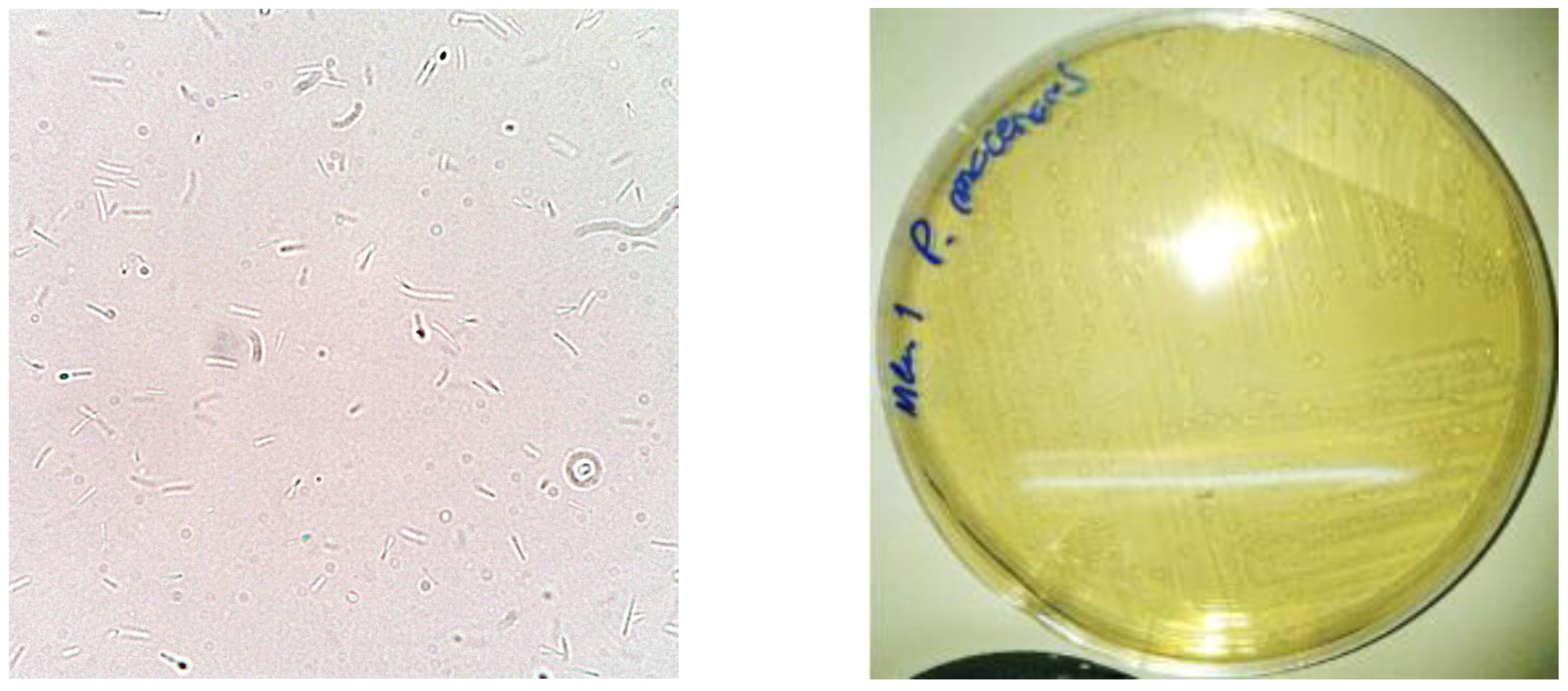
3.1. Macroscopic and Microscopic Characterization
3.2. Small Size Bioprocessing
3.3. Intermediate Assays of CGTase Production
3.4. Bioprocessing Scale up on the Bioreactor
3.5. CGTase Activity and β-CD Production
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Loftsson, T.; Brewster, M.E. Pharmaceutical Applications of Cyclodextrins: Basic Science and Product Development. J. Pharm. Pharmacol. 2010, 62, 1607–1621. [Google Scholar] [CrossRef] [PubMed]
- Mario, J. Chapter Two-Cyclodextrin-Based Drug Delivery Systems. In Nanomaterials for Clinical Applications; Elsevier: Amsterdam, The Netherlands, 2020; pp. 29–69. [Google Scholar] [CrossRef]
- Adeoye, O.; Cabral-Marques, H. Cyclodextrin Nanosystems in Oral Drug Delivery: A Mini Review. Int. J. Pharm. 2017, 531, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Szejtli, J.Z. Past, Present, and Future of Cyclodextrin Research. Pure Appl. Chem. 2004, 76, 1825–1845. [Google Scholar] [CrossRef]
- Oulson, B.G.; Alsulami, Q.A.; Sharfalddin, A.; El Agammy, E.F.; Mouffouk, F.; Emwas, A.-H.; Jaremko, L.; Jaremko, M. Cyclodextrins: Structural, Chemical, and Physical Properties, and Applications. Polysaccharides 2022, 3, 1–31. [Google Scholar] [CrossRef]
- Duchêne, D.; Bochot, A. Thirty Years with Cyclodextrins. Int. J. Pharm. 2016, 514, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Szejtli, J. Introduction and General Overview of Cyclodextrin Chemistry. Chem. Rev. 1998, 98, 1743–1754. [Google Scholar] [CrossRef]
- Szente, L.; Szemán, J.; Sohajda, T. Analytical Characterization of Cyclodextrins: History, Official Methods and Recommended New Techniques. J. Pharm. Biomed. Anal. 2016, 130, 347–365. [Google Scholar] [CrossRef]
- Saokham, P.; Muankaew, C.; Jansook, P.; Loftsson, T. Solubility of Cyclodextrins and Drug/Cyclodextrin Complexes. Molecules 2018, 23, 1161. [Google Scholar] [CrossRef]
- Khalid, S.H.; Bashir, M.; Asghar, S.; Mallhi, T.H.; Khan, I.U. Effect of Cyclodextrin Derivatization on Solubility and Efficacy of Drugs. In Colloid Science in Pharmaceutical Nanotechnology; Intechopen: London, UK, 2019. [Google Scholar]
- Fujinami, S.; Fujisawa, M. Industrial Applications of Alkaliphiles and Their Enzymes--Past, Present and Future. Environ. Technol. 2010, 31, 845–856. [Google Scholar] [CrossRef]
- Leemhuis, H.; Kelly, R.M.; Dijkhuizen, L. Engineering of Cyclodextrin Glucanotransferases and the Impact for Biotechnological Applications. Appl. Microbiol. Biotechnol. 2010, 85, 823–835. [Google Scholar] [CrossRef]
- Schöffer, J.N.; Klein, M.P.; Rodrigues, R.C.; Hertz, P.F. Continuous Production of -Cyclodextrin from Starch by Highly Stable Cyclodextrin Glycosyltransferase Immobilized on Chitosan. Carbohydr. Polym. 2013, 98, 1311–1316. [Google Scholar] [CrossRef] [PubMed]
- Biwer, A.; Antranikian, G.; Heinzle, E. Enzymatic Production of Cyclodextrins. Appl. Microbiol. Biotechnol. 2002, 59, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Rather, M.Y.; Ara, K.Z.G.; Nordberg Karlsson, E.; Adlercreutz, P. Characterization of Cyclodextrin Glycosyltransferases (Cgtases) and Their Application for Synthesis of Alkyl Glycosides with Oligomeric Head Group. Process Biochem. 2015, 50, 722–728. [Google Scholar] [CrossRef]
- Veen, B.A.V.D.; Uitdehaag, J.C.; Dijkstra, B.W.; Dijkhuizen, L. Engineering of Cyclodextrin Glycosyltransferase Reaction and Product Specificity. Biochim. Biophys. Acta 2000, 1543, 336–360. [Google Scholar] [CrossRef]
- Klein, C.; Schulz, G.E. Structure of Cyclodextrin Glycosyltransferase Refined at 2.0 Å Resolution. J. Mol. Biol. 1991, 217, 737–750. [Google Scholar] [CrossRef]
- Bender, H. Cyclodextrin Glucanotransferase from Klebsiella Pneumoniae. 2. Significance of the Enzyme for the Metabolism of Cyclodextrins by Klebsiella Pneumoniae M 5 Al (Author’s Transl). Arch. Microbiol. 1977, 113, 49–56. [Google Scholar] [CrossRef]
- Lee, J.-H.; Choi, K.; Choi, J.; Lee, I.-S.; Kwon, I.; Yu, J.-H. Enzymatic Production of A-Cuclodextrin with the Cyclomaltodextrin Gluconotransferase of Klebsiella Oxytoca 19–1. Enzym. Microb. Technol. 1992, 14, 1017–1020. [Google Scholar] [CrossRef]
- Wind, R.D.; Liebl, W.; Buitelaar, R.M.; Penningra, D.; Spreinat, A.; Dijkhuizen, L.; Bahl, H. Cyclodextrin Formation by The Thermostable Aamylase of Thermoanaerobacterium Thermosulfurigenes Em1 and Reclassification of the Enzyme as a Cyclodextrin Glycosyltransferase. Appl. Environ. Microbiol. 1995, 61, 1257–1265. [Google Scholar] [CrossRef]
- Kelly, R.M.; Dijkhuizen, L.; Leemhuis, H. The Evolution of Cyclodextrin Glucanotransferase Product Specificity. Appl. Microbiol. Biotechnol. 2009, 84, 119–133. [Google Scholar] [CrossRef]
- Zheng, J.; Li, X.; Wu, H. High-Level Extracellular Secretion and Characterization of the Thermophilic Β-Cyclodextrin Glucanotranferase from Paenibacillus Campinasensis in Escherichia Coli. 3 Biotech. 2019, 9, 372. [Google Scholar] [CrossRef]
- Preiss, L.; Hicks, D.B.; Suzuki, S.; Meier, T.; Krulwich, T.A. Alkaliphilic Bacteria with Impact on Industrial Applications, Concepts of Early Life Forms, and Bioenergetics of Atp Synthesis. Front. Bioeng. Biotechnol. 2015, 3, 75. [Google Scholar] [CrossRef] [PubMed]
- Elbaz, A.F.; Sobhi, A.; Elmekawy, A. Purification and Characterization of Cyclodextrin Β-Glucanotransferase from Novel Alkalophilic Bacilli. Bioprocess Biosyst. Eng. 2015, 38, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhou, Z.; Li, H.; Bu, R. Optimization of the Fermentation Conditions for the Mutant Strain of Β-Cyclodextrin Glycosyltransferase H167c to Produce Cyclodextrins. 3 Biotech. 2018, 8, 165. [Google Scholar] [CrossRef] [PubMed]
- Strain Passport Bacillus Pseudofirmus Dsm2517. Available online: http://www.straininfo.net/strains/119820 (accessed on 28 December 2010).
- Gordon, R.E.; Hyde, J.L. The Bacillusfirmus-Bacillus Lentus Complex and Ph 7.0 Variants Of Some Alkalophilic Strains. Microbiology 1982, 128, 1109–1116. [Google Scholar]
- Fritze, D.; Flossdorf, J.; Claus, D. Taxonomy of Alkaliphilic Bacillus Strains. Int. J. Syst. Bacteriol. 1990, 40, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Nieisen, P.; Fritze, D.; Priest, F.G. Phenetic Diversity of Alkaliphilic Bacillus Strains: Proposal for Nine New Species. Microbiology 1995, 141, 1745–1761. [Google Scholar] [CrossRef]
- Moriwaki, C.; Mazzer, C.; Pazzetto, R.; Matioli, G. Produção, Purificação e Aumento da Performance de Ciclodextrina Glicosiltransferase Para Produção de Ciclodextrinas. Quim. Nova 2009, 32, 2360–2366. [Google Scholar] [CrossRef]
- Rendleman, J.A. The Production of Cyclodextrins Using Cgtase from Bacillus Macerans. In Carbohydrate Biotechnology Protocols; Methods in Biotechnology; Springer: Berlin/Heidelberg, Germany, 2008; pp. 89–101. [Google Scholar]
- Yang, Y.-N.; Shan, W.-X.; Wang, P.-W. Upscale Production of a Recombinant Cyclodextrin Glycosyltransferase from Paenibacillus Macerans in Escherichia Coli. 3 Biotech. 2017, 7, 207. [Google Scholar] [CrossRef]
- Schardinger, F. Bacillus Macerans Ein Aceton Bildender Rottebacillus. Zbl Bakteriol Parasitenkd Infektionskr Hyg Abt Ii 1905, 14, 772–781. [Google Scholar]
- Ash, C.; Priest, F.G.; Collins, M.D. Molecular Identification Of Rrna Group 3 Bacilli (Ash, Farrow, Wallbanks And Collins) Using A Pcr Probe Test. Antonie Van Leeuwenhoek 1993, 64, 253–260. [Google Scholar] [CrossRef]
- Fujiwara, S.; Kakihara, H.; Woo, K.B.; Lejeune, A.; Kanemoto, M.; Sakaguchi, K.; Imanaka, T. Cyclization Characteristics of Cyclodextrin Glucanotransferase Are Conferred by the Nh2-Terminal Region of the Enzyme. Appl. Environ. Microbiol. 1992, 58, 4016–4025. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhou, J.; Wu, R.; Xin, F.; Zhang, W.; Fang, Y.; Ma, J.; Dong, W.; Jiang, M. Heterologous Expression of Cyclodextrin Glycosyltransferase from Paenibacillus Macerans in Escherichia Coli and Its Application in 2-O-A-D-Glucopyranosyl-Lascorbic Acid Production. Bmc Biotechnol. 2018, 18, 53. [Google Scholar] [CrossRef] [PubMed]
- Qi, Q.; Zimmermann, W. Cyclodextrin Glucanotransferase: From Gene to Applications. Appl. Microbiol. Biotechnol. 2005, 66, 475–485. [Google Scholar] [PubMed]
- Dsmz Catalogue Paenibacillus Macerans (Schardinger 1905), Ash Et Al. 1994. Available online: http://www.dsmz.de/catalogues/details/culture/DSM-1574.html (accessed on 28 December 2010).
- Nakamura, N.; Horikoshi, K. Characterization and Some Cultural Conditions of a Cyclodextrin Glycosyltransferase-Producing Alkalophilic Bacillus Sp. Agr. Bioi. Chem. 1976, 40, 753–775. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Nunes, M.A.P.; Vila-Real, H.; Fernandes, P.C.B.; Ribeiro, M.H.L. Immobilization of Naringinase in Pva–Alginate Matrix Using an Innovative Technique. Appl. Biochem. Biotechnol. 2010, 160, 2129–2147. [Google Scholar] [CrossRef]
- Kuwabara, T.; Takamura, M.; Matsushita, A.; Ikeda, H.; Nakamura, A.; Ueno, A.; Toda, F. Phenolphthalein-Modifíed Β-Cyclodextrin as A Molecule-Responsive Colorless-to-Color Change Indicator. J. Org. Chem. 1998, 63, 8729–8735. [Google Scholar] [CrossRef]
- Nakatani, H.; Hiromi, K. Kinetic Study of Beta-Cyclodextrin-Dye System by High-Pressure Temperature-Jump Method. J. Biochem. 1984, 96, 69–72. [Google Scholar] [CrossRef]
- Higuti, I.H.; Anunciação Da Silva, P.; Papp, J.; Okiyama, V.M.E.; Andrade, E.A.; Aluizio De Abreu Marcondes, A.A.; Nascimento, A.J. Colorimetric Determination of α and β-Cyclodextrins and Studies on Optimization OF Cgtase Production from B. Firmus Using Factorial Designs. Braz. Arch. Biol. Technol. 2004, 47, 837–841. [Google Scholar] [CrossRef]
- Dsmz Catalogue Bacillus Pseudofirmus Nielsen Et Al. 1995. Available online: http://www.dsmz.de/catalogues/details/culture/DSM-2517.html (accessed on 28 December 2010).
- Atcc Catalogue, B. Pseudofirmus Atcc21593. Available online: http://www.atcc.org/ATCCAdvancedCatalogSearch/ProductDetails/tabid/452/Default.aspx?ATCCNum=21593&Template=bacteria (accessed on 4 February 2010).
- Decker, S.; Maier, S. Fine Structure of Mesosomal Involvement during Bacillus Macerans Sporulation. J. Bacteriol. 1975, 121, 363–372. [Google Scholar] [CrossRef]
- Mccormick, N.G. Kinetics of Spore Germination. J. Bacteriol. 1965, 89, 1180–1185. [Google Scholar] [CrossRef] [PubMed]
- Prescott, L.M.; Harley, J.P.; Klein, D.A. Microbiology, 6th ed.; Mcgraw-Hill Higher Education: Dubuque, IA, USA, 2005. [Google Scholar]
- Rosso, A.M.; Rosso, A.M.; Ferrarotti, S.A.; Krymkiewicz, N.; Nudel, B. Optimisation of Batch Culture Conditions for Cyclodextrin Glucanotransferase Production from Bacillus Circulans Df 9r. Microbial. Cell. Factories 2002, 1, 3. [Google Scholar] [CrossRef] [PubMed]
- Chung, B.H.; Cannon, R.Y.; Smith, R.C. Influence of Growth Temperature on Glucose Metabolism of a Psychotrophic Strain of Bacillus Cereus. Appl. Environ. Microbiol. 1976, 31, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Gawande, B.N.; Goel, A.; Patkar, A.Y.; Nene, S.N. Purification and Properties of a Novel Raw Starch Degradingcyclomaltodextrin Glucanotransferase from Bacillus Firmus. Appl. Microbiol. Biotechnol. 1999, 51, 504–509. [Google Scholar] [CrossRef]
- Ibrahim, H.M.; Yusoff, W.M.W.; Hamid, A.A.; Illias, R.M.; Hassan, O.; Omar, O. Optimization of Medium for The Production Of Β-Cyclodextrin Glucanotransferase Using Central Composite Design (Ccd). Process Biochem. 2005, 40, 753–758. [Google Scholar] [CrossRef]
- Chen, S.; Li, Z.; Gu, Z.; Ban, X.; Hong, Y.; Cheng, L.; Li, C. Immobilization of Β-Cyclodextrin Glycosyltransferase on Gelatin Enhances β-Cyclodextrin Production. Process Biochem. 2022, 113, 216–223. [Google Scholar] [CrossRef]
- Solanki, P.; Awadhiya, P.; Banerjee, T. Conventional Production Optimization of Cyclodextrin Glucosyl Transferase by a Novel Isolate of Bacillus Sp. Pbs1 from Potato Rhizosphere. J. Microbiol. Biotech. Food Sci. 2022, 12, E5130. [Google Scholar] [CrossRef]
- Salústio, P.J.; Feio, G.; Figueirinhas, J.L.; Pinto, J.F.; Cabral-Marques, H.M. The Influence of The Preparation Methods on the Inclusion of Model Drugs in a β-Cyclodextrin Cavity. Eur. J. Pharm. Biopharm. 2009, 71, 377–386. [Google Scholar] [CrossRef]
- Salústio, P.J.; Cabral-Marques, H.M.; Costa, P.C.; Pinto, J.F. Release Profiles of Indometacin IN ß-Cyclodextrin Complexes from Hpmc Capsules. J. Incl. Phenom. Macrocycl. Chem. 2013, 75, 101–109. [Google Scholar] [CrossRef]
- Vieira Da Silva, S.A.; Clemente, A.; Rocha, J.; Direito, R.; Cabral-Marques, H.M.; Sepodes, B.; Figueira, M.-E.; Ribeiro, M.H. Anti-Inflammatory Effect of Limonin from Cyclodextrin (Un)Processed Orange Juices in In Vivo Acute Inflammation and Chronic Rheumatoid Arthritis Models. J. Funct. Foods 2018, 49, 146–153. [Google Scholar] [CrossRef]

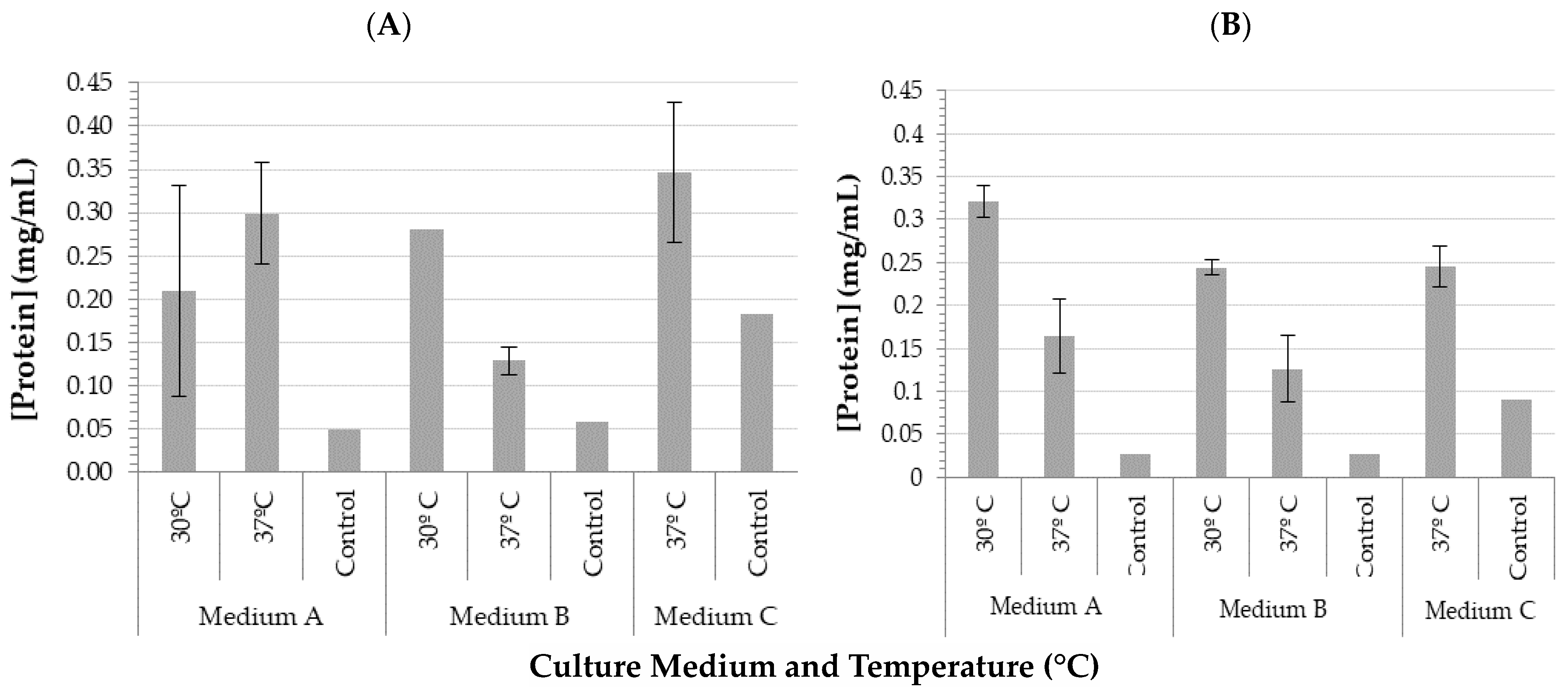
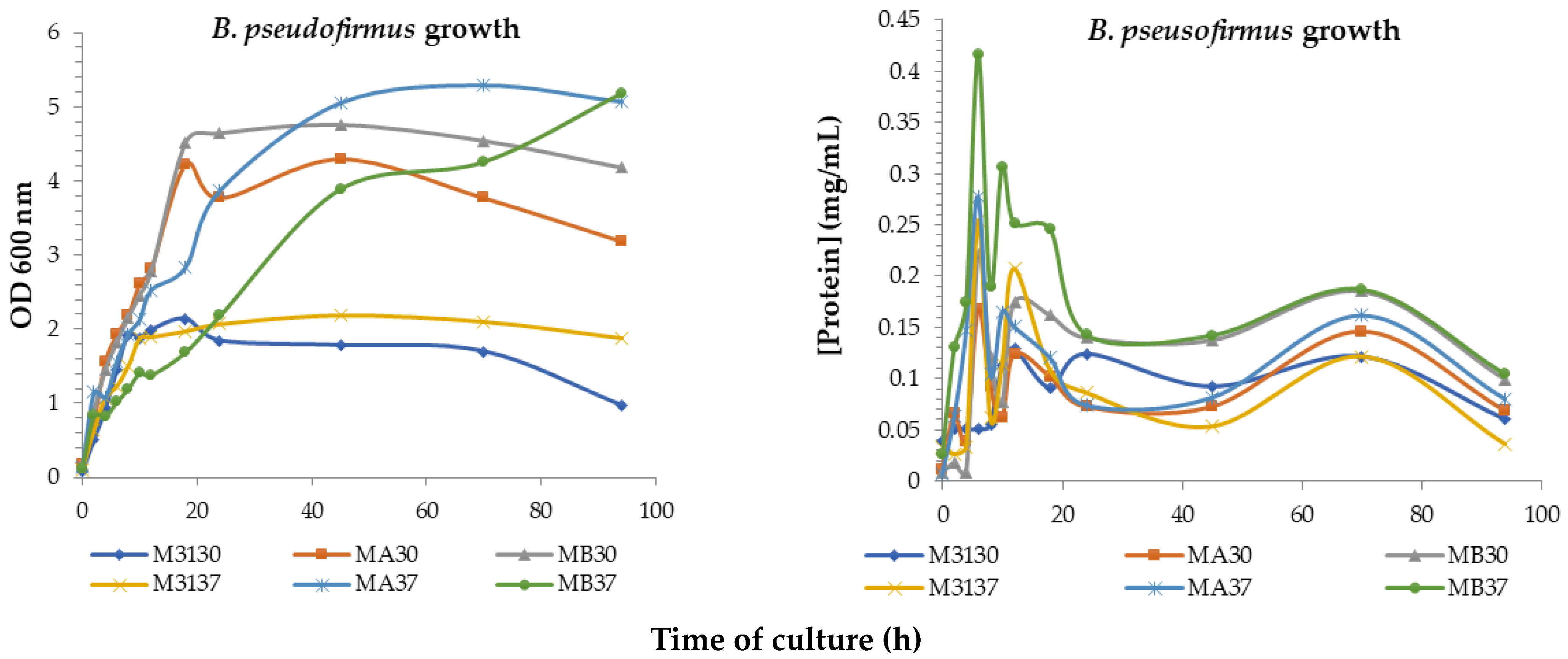


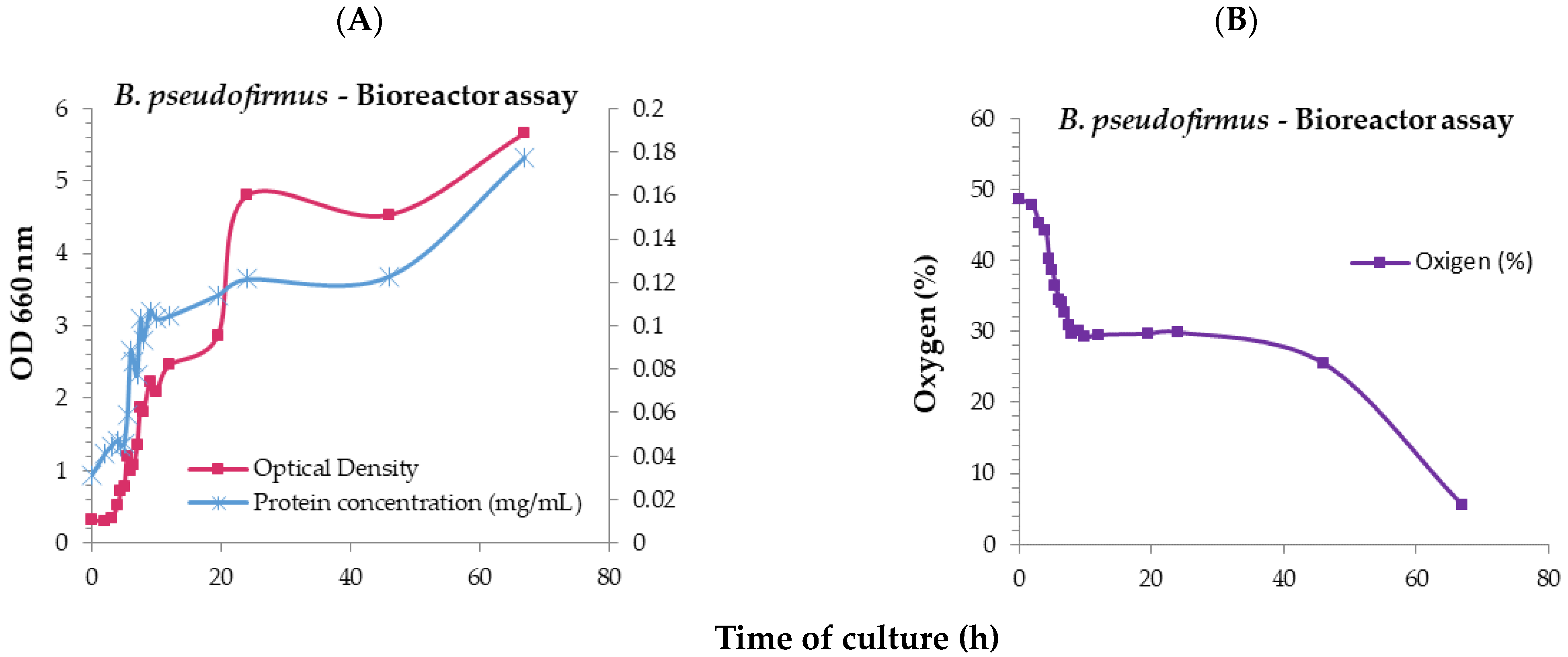

| Culture Media | μ (h−1) | Td (h) | rP (mg/mL h) | |||
|---|---|---|---|---|---|---|
| 30 °C | 37 °C | 30 °C | 37 °C | 30 °C | 37 °C | |
| Medium 31 | 0.224 | 0.126 | 3.09 | 5.50 | 0.011 | 0.017 |
| Medium A | 0.169 | 0.091 | 4.10 | 7.61 | 0.010 | 0.013 |
| Medium B | 0.145 | 0.071 | 4.78 | 9.76 | 0.014 | 0.021 |
| Culture Media | μ (h−1) | Td (h) | rP (mg/mL h) | |||
|---|---|---|---|---|---|---|
| 30 °C | 37 °C | 30 °C | 37 °C | 30 °C | 37 °C | |
| Medium 1 | 0.138 | 0.735 | 5.02 | 4.00 | 0.005 | 0.007 |
| Medium A | 0.171 | 0.217 | 4.05 | 7.61 | 0.006 | 0.013 |
| Medium B | 0.203 | 0.269 | 3.41 | 9.76 | 0.010 | 0.021 |
| Medium B | μ (h−1) | Td (h) | rP (mg/mL h) |
|---|---|---|---|
| B. pseudofirmus | 0.305 | 2.27 | 0.014 |
| P. macerans | 0.251 | 2.76 | 0.014 |
| Samples | Absorbance | [β-CD] (mmol/mL) | ||
|---|---|---|---|---|
| Blank (PHE + H2O) | 1.564 | 0 | ||
| Microrganisms | Filtrate | Concentrate | Filtrate | Concentrate |
| B. pseudofirmus T24h | 1.517 | 1.448 | 0.014 | 0.035 |
| B. pseudofirmus T48h | 1.488 | 1.464 | 0.023 | 0.030 |
| P. macerans T9h | 1.511 | 0.878 | 0.016 | 0.211 |
| P. macerans T15h | 1.473 | 0.962 | 0.028 | 0.186 |
| P. macerans T33h | - | 1.252 | - | 0.100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guedes, A.M.; Santos Alves, T.F.; Salústio, P.J.; Cabral-Marques, H.M.; Ribeiro, M.H.L. Design of a Cyclodextrin Bioproduction Process Using Bacillus pseudofirmus and Paenibacillus macerans. Future Pharmacol. 2023, 3, 568-584. https://doi.org/10.3390/futurepharmacol3030035
Guedes AM, Santos Alves TF, Salústio PJ, Cabral-Marques HM, Ribeiro MHL. Design of a Cyclodextrin Bioproduction Process Using Bacillus pseudofirmus and Paenibacillus macerans. Future Pharmacology. 2023; 3(3):568-584. https://doi.org/10.3390/futurepharmacol3030035
Chicago/Turabian StyleGuedes, Alexandre Miguel, Tiago Filipe Santos Alves, Paulo J. Salústio, Helena M. Cabral-Marques, and Maria H. L. Ribeiro. 2023. "Design of a Cyclodextrin Bioproduction Process Using Bacillus pseudofirmus and Paenibacillus macerans" Future Pharmacology 3, no. 3: 568-584. https://doi.org/10.3390/futurepharmacol3030035
APA StyleGuedes, A. M., Santos Alves, T. F., Salústio, P. J., Cabral-Marques, H. M., & Ribeiro, M. H. L. (2023). Design of a Cyclodextrin Bioproduction Process Using Bacillus pseudofirmus and Paenibacillus macerans. Future Pharmacology, 3(3), 568-584. https://doi.org/10.3390/futurepharmacol3030035







