Taxonogenomic Analysis of Marine-Derived Streptomyces sp. N11-50 and the Profile of NRPS and PKS Gene Clusters
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Taxonomic Position of Streptomyces sp. N11-50 and Related WGS-Published Strains
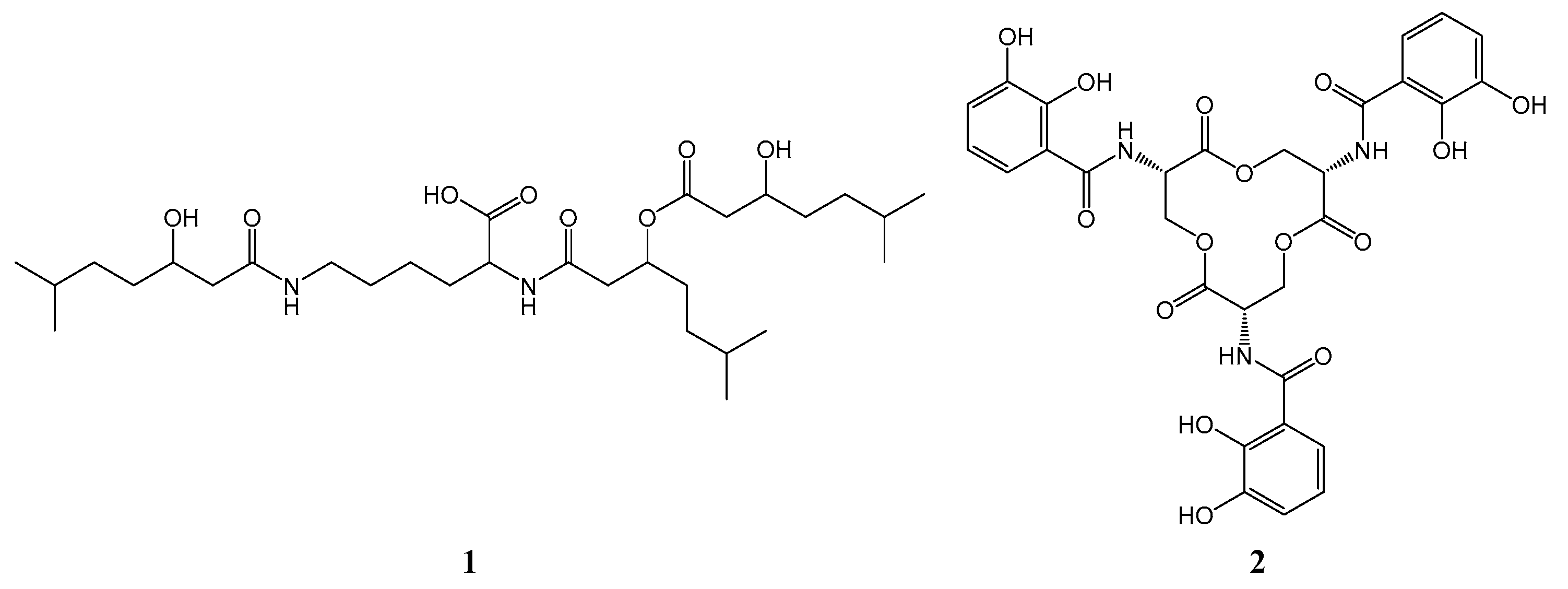
3.2. NRPS and Hybrid PKS/NRPS Gene Clusters in S. albus N11-50
3.3. Type-I and Type-II PKS Gene Clusters in S. albus N11-50
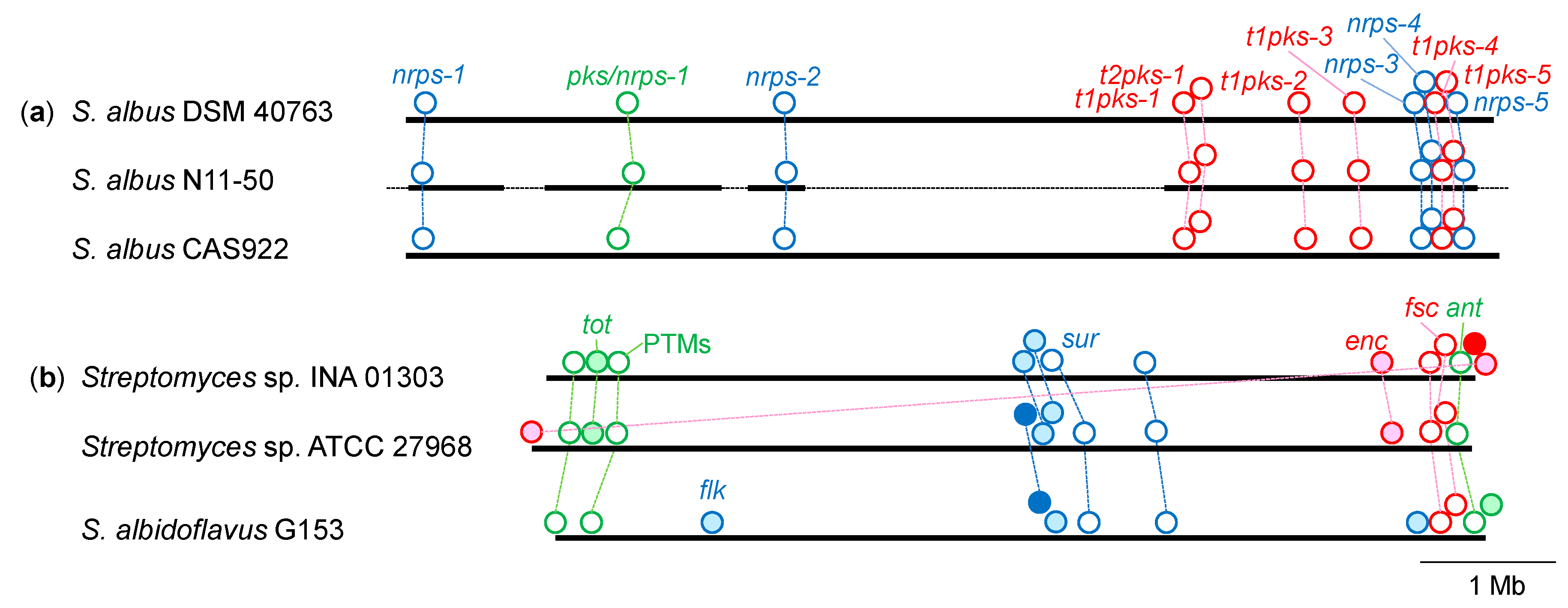
3.4. Distribution of NRPS and PKS Gene Clusters Found in S. albus N11-50
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baltz, R.H. Gifted Microbes for Genome Mining and Natural Product Discovery. J. Ind. Microbiol. Biotechnol. 2017, 44, 573–588. [Google Scholar] [CrossRef] [PubMed]
- Nett, M.; Ikeda, H.; Moore, B.S. Genomic Basis for Natural Product Biosynthetic Diversity in the Actinomycetes. Nat. Prod. Rep. 2009, 26, 1362–1384. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, M.A.; Walsh, C.T. Assembly-Line Enzymology for Polyketide and Nonribosomal Peptide Antibiotics: Logic, Machinery, and Mechanisms. Chem. Rev. 2006, 106, 3468–3496. [Google Scholar] [CrossRef] [PubMed]
- Komaki, H. Recent Progress of Reclassification of the Genus Streptomyces. Microorganisms 2023, 11, 831. [Google Scholar] [CrossRef] [PubMed]
- Manivasagan, P.; Venkatesan, J.; Sivakumar, K.; Kim, S. Pharmaceutically Active Secondary Metabolites of Marine Actinobacteria. Microbiol. Res. 2014, 169, 262–278. [Google Scholar] [CrossRef]
- Subramani, R.; Aalbersberg, W. Marine Actinomycetes: An Ongoing Source of Novel Bioactive Metabolites. Microbiol. Res. 2012, 167, 571–580. [Google Scholar] [CrossRef]
- Harunari, E.; Ogino, K.; Kanaki, S.; Kumagai, T.; Igarashi, Y. Isolation and Metabolites Analysis of Actinomycetes from Deep-Sea Water in Toyama Bay. Deep Ocean Water Res. 2021, 22, 49–57. [Google Scholar]
- Igarashi, Y.; Zhou, T.; Sato, S.; Matsumoto, T.; Yu, L.; Oku, N. Akaeolide, a Carbocyclic Polyketide from Marine-Derived Streptomyces. Org. Lett. 2013, 15, 5678–5681. [Google Scholar] [CrossRef]
- Fukushima, K.; Yazawa, K.; Arai, T. Biological Activities of Albonoursin. J. Antibiot. 1973, 26, 175–176. [Google Scholar] [CrossRef]
- Kanzaki, H.; Imura, D.; Sashida, R.; Kobayashi, A.; Kawazu, K. Effective Production of Dehydro Cyclic Dipeptide Albonoursin Exhibiting Pronuclear Fusion Inhibitory Activity. I. Taxonomy and Fermentation. J. Antibiot. 1999, 52, 1017–1022. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Xi, L.; Liu, P.; Wang, Y.; Wang, W.; Huang, Y.; Zhu, W. Diketopiperazine Derivatives from the Marine-Derived Actinomycete Streptomyces sp. FXJ7.328. Mar. Drugs 2013, 11, 1035–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Lai, Y.; Lu, Y.; Yang, Y.; Chen, S. Analysis of the Biosynthesis of Antibacterial Cyclic Dipeptides in Nocardiopsis alba. Arch. Microbiol. 2014, 196, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Gondry, M.; Sauguet, L.; Belin, P.; Thai, R.; Amouroux, R.; Tellier, C.; Tuphile, K.; Jacquet, M.; Braud, S.; Courcon, M.; et al. Cyclodipeptide Synthases Are a Family of tRNA-Dependent Peptide Bond-Forming Enzymes. Nat. Chem. Biol. 2009, 5, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Lautru, S.; Gondry, M.; Genet, R.; Pernodet, J.L. The Albonoursin Gene Cluster of S. noursei Biosynthesis of Diketopiperazine Metabolites Independent of Nonribosomal Peptide Synthetases. Chem. Biol. 2002, 9, 1355–1364. [Google Scholar] [CrossRef] [Green Version]
- Komaki, H.; Tamura, T.; Igarashi, Y. Classification and Secondary Metabolite-Biosynthetic Gene Clusters of Marine Streptomyces Strains Including a Lobophorin- and Divergolide-Producer. Hydrobiology 2023, 2, 151–161. [Google Scholar] [CrossRef]
- Yoon, S.; Ha, S.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A Taxonomically United Database of 16S rRNA Gene Sequences and Whole-Genome Assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Rong, X.; Huang, Y. Taxonomic Evaluation of the Streptomyces hygroscopicus Clade Using Multilocus Sequence Analysis and DNA-DNA Hybridization, Validating the MLSA Scheme for Systematics of the Whole Genus. Syst. Appl. Microbiol. 2012, 35, 7–18. [Google Scholar] [CrossRef]
- Saito, H.; Miura, K. Preparation of Transforming Deoxyribonucleic Acid by Phenol Treatment. Biochim. Biophys. Acta 1963, 72, 619–629. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A Database Tandem for Fast and Reliable Genome-Based Classification and Nomenclature of Prokaryotes. Nucleic Acids Res. 2021, 50, D801–D807. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Goker, M.; Sproer, C.; Klenk, H.P. When Should a DDH Experiment Be Mandatory in Microbial Taxonomy? Arch. Microbiol. 2013, 195, 413–418. [Google Scholar] [CrossRef]
- Vela Gurovic, M.S.; Díaz, M.L.; Gallo, C.A.; Dietrich, J. Phylogenomics, CAZyome and Core Secondary Metabolome of Streptomyces albus Species. Mol. Genet. Genom. 2021, 296, 1299–1311. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.-W.; De Meyer, S.; et al. Proposed Minimal Standards for the Use of Genome Data for the Taxonomy of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Glaeser, S.P.; Kämpfer, P. Multilocus Sequence Analysis (MLSA) in Prokaryotic Taxonomy. Syst. Appl. Microbiol. 2015, 38, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lasch, C.; Stierhof, M.; Estevez, M.R.; Myronovskyi, M.; Zapp, J.; Luzhetskyy, A. Dudomycins: New Secondary Metabolites Produced after Heterologous Expression of an NRPS Cluster from Streptomyces albus ssp. chlorinus NRRL B-24108. Microorganisms 2020, 8, 1800. [Google Scholar] [CrossRef] [PubMed]
- Patzer, S.I.; Braun, V. Gene Cluster Involved in the Biosynthesis of Griseobactin, a Catechol-Peptide Siderophore of Streptomyces sp. ATCC 700974. J. Bacteriol. 2010, 192, 426–435. [Google Scholar] [CrossRef] [Green Version]
- Robbins, N.; Spitzer, M.; Wang, W.; Waglechner, N.; Patel, D.J.; O’Brien, J.S.; Ejim, L.; Ejim, O.; Tyers, M.; Wright, G.D. Discovery of Ibomycin, a Complex Macrolactone that Exerts Antifungal Activity by Impeding Endocytic Trafficking and Membrane Function. Cell Chem. Biol. 2016, 23, 1383–1394. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Carandang, R.R.; Harada, Y.; Okada, S.; Yoshitake, K.; Asakawa, S.; Nogi, Y.; Matsunaga, S.; Takada, K. Lactomycins A–C, Dephosphorylated Phoslactomycin Derivatives that Inhibit Cathepsin B, from the Marine-derived Streptomyces sp. ACT232. Mar. Drugs 2018, 16, 70. [Google Scholar] [CrossRef] [Green Version]
- Geyer, K.; Sundaram, S.; Sušnik, P.; Koert, U.; Erb, T.J. Understanding Substrate Selectivity of Phoslactomycin Polyketide Synthase by Using Reconstituted In Vitro Systems. ChemBioChem 2020, 21, 2080–2085. [Google Scholar] [CrossRef]
- Horsman, G.P.; Van Lanen, S.G.; Shen, B. Iterative Type I Polyketide Synthases for Enediyne Core Biosynthesis. Methods Enzymol. 2009, 459, 97–112. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, L.; Kong, L.; Wang, T.; Chu, Y.; Deng, Z.; You, D. Unveiling the Post-PKS Redox Tailoring Steps in Biosynthesis of the Type II Polyketide Antitumor Antibiotic Xantholipin. Chem. Biol. 2012, 19, 422–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.; Zhang, L.; Ito, T.; Qu, X.; Asakawa, Y.; Awakawa, T.; Abe, I.; Liu, W. Biosynthetic Pathway for High Structural Diversity of a Common Dilactone Core in Antimycin Production. Org. Lett. 2012, 14, 4142–4145. [Google Scholar] [CrossRef] [PubMed]
- Piel, J.; Hertweck, C.; Shipley, P.R.; Hunt, D.M.; Newman, M.S.; Moore, B.S. Cloning, Sequencing and Analysis of the Enterocin Biosynthesis Gene Cluster from the Marine Isolate ‘Streptomyces maritimus’: Evidence for the Derailment of an Aromatic Polyketide Synthase. Chem. Biol. 2000, 7, 943–955. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez Estévez, M.; Myronovskyi, M.; Rosenkränzer, B.; Paululat, T.; Petzke, L.; Ristau, J.; Luzhetskyy, A. Novel Fredericamycin Variant Overproduced by a Streptomycin-Resistant Streptomyces albus subsp. chlorinus Strain. Mar. Drugs 2020, 18, 284. [Google Scholar] [CrossRef]
- Horbal, L.; Stierhof, M.; Palusczak, A.; Eckert, N.; Zapp, J.; Luzhetskyy, A. Cyclofaulknamycin with the Rare Amino Acid D-capreomycidine Isolated from a Well-Characterized Streptomyces albus Strain. Microorganisms 2021, 9, 1609. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Huang, X.; Zhou, X.; Bai, L.; He, J.; Jeong, K.J.; Lee, S.Y.; Deng, Z. Organizational and Mutational Analysis of a Complete FR-008/Candicidin Gene Cluster Encoding a Structurally Related Polyene Complex. Chem. Biol. 2003, 10, 1065–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Huang, H.; Liang, J.; Wang, M.; Lu, L.; Shao, Z.; Cobb, R.E.; Zhao, H. Activation and Characterization of a Cryptic Polycyclic Tetramate Macrolactam Biosynthetic Gene Cluster. Nat. Commun. 2013, 4, 2894. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Nazari, B.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. Discovery of a Cryptic Antifungal Compound from Streptomyces albus J1074 Using High-Throughput Elicitor Screens. J. Am. Chem. Soc. 2017, 139, 9203–9212. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Zhang, Q.; Tan, B.; Zheng, L.; Li, H.; Zhu, Y.; Zhang, C. Genome Mining and Activation of a Silent PKS/NRPS Gene Cluster Direct the Production of Totopotensamides. Org. Lett. 2017, 19, 5697–5700. [Google Scholar] [CrossRef] [PubMed]
- Seipke, R.F. Strain-Level Diversity of Secondary Metabolism in Streptomyces albus. PLoS ONE 2015, 10, e0116457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komaki, H.; Sakurai, K.; Hosoyama, A.; Kimura, A.; Igarashi, Y.; Tamura, T. Diversity of Nonribosomal Peptide Synthetase and Polyketide Synthase Gene Clusters among Taxonomically Close Streptomyces Strains. Sci. Rep. 2018, 8, 6888. [Google Scholar] [CrossRef] [PubMed] [Green Version]

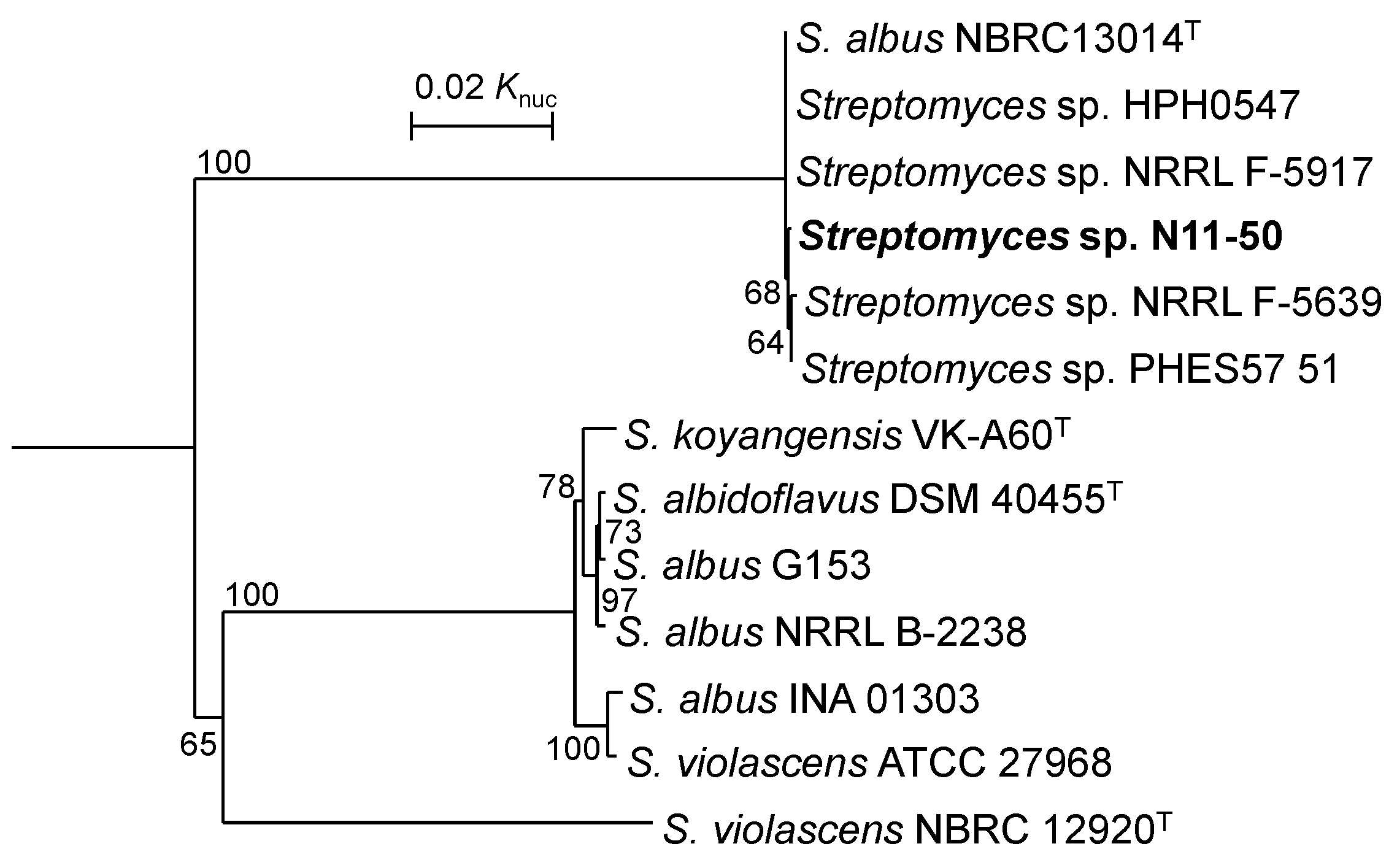
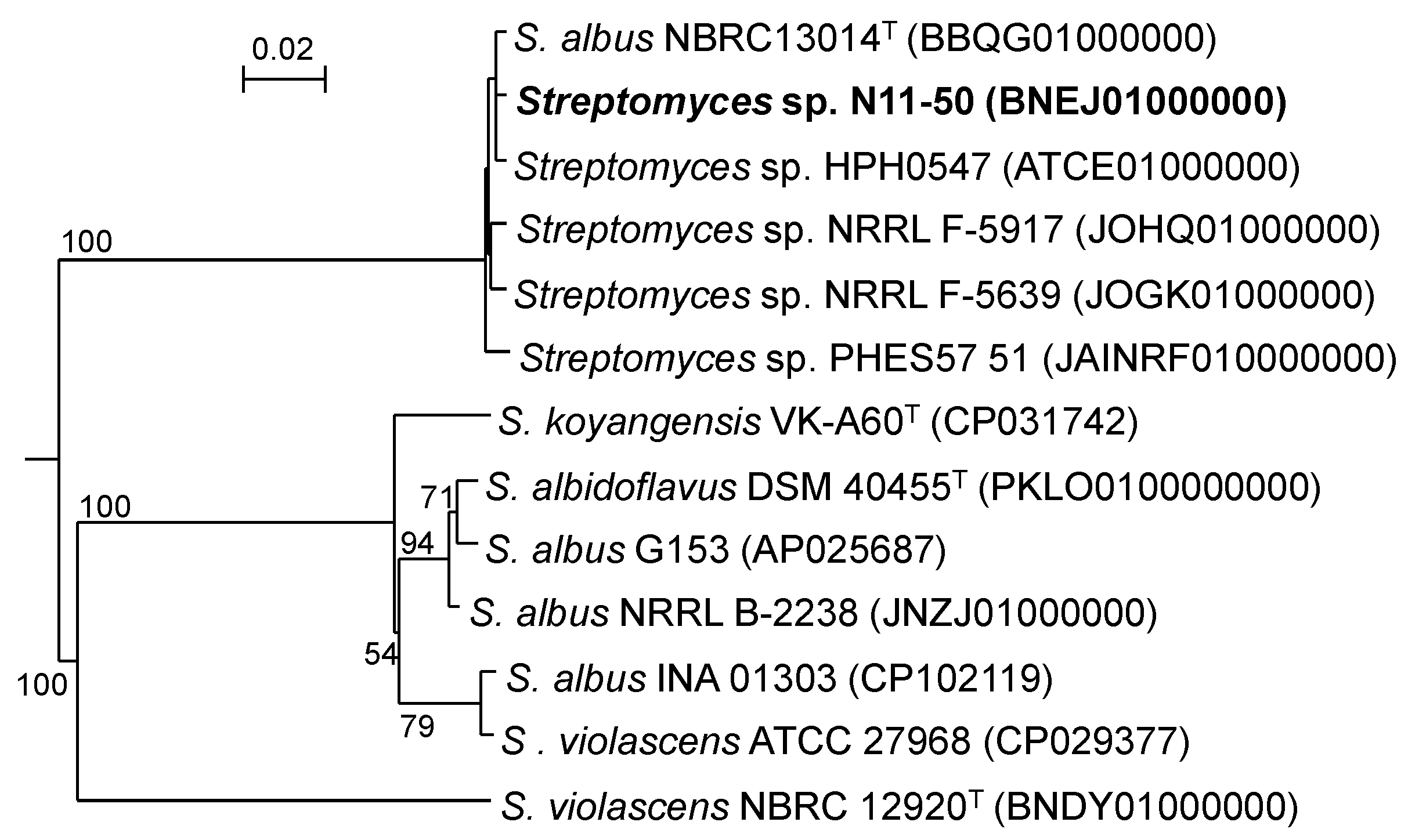

| Strain | Evolutionary Distance in MLSA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
| 1. S. albidoflavus DSM 40455T | - | 0.001 | 0.002 | 0.011 | 0.010 | 0.122 | 0.008 | 0.153 | 0.154 | 0.153 | 0.154 | 0.155 | 0.153 |
| 2. S. albus G153 | 91.7 | - | 0.003 | 0.011 | 0.010 | 0.121 | 0.008 | 0.154 | 0.155 | 0.154 | 0.155 | 0.156 | 0.154 |
| 3. S. albus NRRL B-2238 | 92.1 | 91.8 | - | 0.010 | 0.010 | 0.124 | 0.008 | 0.154 | 0.154 | 0.154 | 0.154 | 0.156 | 0.154 |
| 4. S. albus INA 01303 | 64.6 | 64.8 | 65.7 | - | 0.003 | 0.125 | 0.013 | 0.156 | 0.156 | 0.156 | 0.156 | 0.157 | 0.156 |
| 5. S. violascens ATCC 27968 | 65.7 | 65.8 | 66.7 | 93.5 | - | 0.126 | 0.012 | 0.156 | 0.156 | 0.156 | 0.156 | 0.157 | 0.156 |
| 6. S. violascens NBRC 12920T | 22.7 | 22.8 | 24.4 | 22.6 | 22.6 | - | 0.124 | 0.165 | 0.166 | 0.165 | 0.166 | 0.167 | 0.165 |
| 7. S. koyangensis VK-A60T | 64.6 | 64.5 | 64.8 | 61.1 | 61.9 | 22.8 | - | 0.156 | 0.156 | 0.156 | 0.156 | 0.157 | 0.156 |
| 8. S. albus NBRC 13014T | 21.4 | 21.6 | 23.3 | 21.3 | 21.4 | 21.8 | 21.5 | - | 0.000 | 0.000 | 0.001 | 0.001 | 0.000 |
| 9. Streptomyces sp. N11-50 | 21.4 | 21.6 | 23.4 | 21.3 | 21.5 | 21.7 | 21.5 | 95.5 | - | 0.000 | 0.001 | 0.001 | 0.000 |
| 10. Streptomyces sp. HPH0547 | 21.5 | 21.7 | 23.4 | 21.5 | 21.5 | 21.8 | 21.5 | 96.0 | 95.4 | - | 0.001 | 0.001 | 0.000 |
| 11. Streptomyces sp. PHES57 51 | 21.3 | 21.6 | 23.2 | 21.3 | 21.5 | 21.8 | 21.4 | 90.1 | 89.6 | 89.9 | - | 0.001 | 0.001 |
| 12. Streptomyces sp. NRRL F-5639 | 21.4 | 21.6 | 23.3 | 21.4 | 21.5 | 21.7 | 21.5 | 92.4 | 91.5 | 92.0 | 89.6 | - | 0.001 |
| 13. Streptomyces sp. NRRL F-5917 | 21.4 | 21.6 | 23.2 | 21.4 | 21.5 | 21.7 | 21.6 | 91.9 | 91.0 | 91.3 | 89.4 | 92.8 | - |
| DNA-DNA Relatedness | |||||||||||||
| Gene Cluster | ORF | Domain Organization | Putative Product |
|---|---|---|---|
| nrps-1 | TPA0909_06090 | Aval/PCP-C | Thr-Val-Ser-Y |
| TPA0909_06100 | Aser/PCP-C/PCP-TE | ||
| TPA0909_06130 | Athr/PCP | ||
| nrps-2 | TPA0909_10490 | C/A/PCP-TE | dudomycin (1) |
| nrps-3 | TPA0909_28110 | AdiOH-Bz | enterobactin (2) |
| TPA0909_28090 | PCP | ||
| TPA0909_28080 | C/Aser/PCP-TE | ||
| nrps-4 | TPA0909_28620 | A/PCP-TD | dipeptide |
| TPA0909_28750 | A | ||
| nrps-5 | TPA0909_30010 | C/Acys/MT/PCP-TE | X-Ser-Cys-mCys |
| TPA0909_30080 | A | ||
| TPA0909_30100 | PCP-C/A/PCP-C/Acys/PCP | ||
| pks/nrps-1 | TPA0909_66390 | Aasn/PCP | tetraketide with Asn |
| TPA0909_66410 | C/PCP | ||
| TPA0909_66430 | ATmm/ACP-KS/AT/KR/ACP | ||
| TPA0909_66440 | KS/ATm/KR/ACP-KS/ATm/ACP |
| Gene Cluster | ORF (TPA0909) 1 | Domain Organization | Putative Product |
|---|---|---|---|
| t1pks-1 | _14380 | ACP | unknown |
| _14390 | KS/AT/ACP | ||
| _14420 | ACP | ||
| t1pks-2 | _21420 | ACP | tambjamine BE-18591 (3) |
| _21430 | KS | ||
| _21460 2 | KS/KS | ||
| _21470 | ACP | ||
| _21480 | ACP/ACP/AmT | ||
| _21500 | TE | ||
| t1pks-3 | _24630 | KS/ATmm/ACP-KS/AT/KR/ACP-KS/ATmm/KR/ACP | ibomycin congener derived from polyketide chain shown as 4b |
| _24640 | KS/ATm/KR/ACP-KS/ATmm/KR/ACP-KS/ATmm/KR/ACP | ||
| -KS/ATm/DH/KR/ACP | |||
| _24650 | KS/ATm/DH/KR/ACP-KS/ATmm/KR/ACP | ||
| _24660 | KS/ATmm/KR/ACP-KS/ATm/KR/ACP-KS/ATmm/KR/ACP | ||
| _24670 | KS/ATm/KR/ACP-KS/ATm/KR/ACP-KS/ATm/DH/ACP | ||
| -KS/ATmm/KR/ACP-KS/ATm/DH/ER/KR/ACP | |||
| _24680 | KS/ATm/KR/ACP-KS/ATm/KR/ACP | ||
| _24690 | KS/ATmm/DH/KR/ACP-KS/ATm/DH/KR/ACP-TE | ||
| t1pks-4 | _28890 | AT/ACP-KS/ATm/DH/KR/ACP | congener of lactomycins and phoslactomycin derived from 5c |
| _28880 | KS/ATm/DH/KR/ACP-KS/AT/KR/ACP | ||
| _28870 | KS/ATm/KR/ACP | ||
| _28860 | KS/ATm/KR/ACP-TE | ||
| _28850 | KS/ATmm/DH/ER/KR/ACP | ||
| _28840 2 | KS/ATem/KR/ACP | ||
| t1pks-5 | _29480 | KS/ATm/KR/DH/ACP | enediyne |
| t2pks-1 | _14620 | ACP | aromatic compound like xantholipin (6) |
| _14680 | KSα | ||
| _14690 | KSβ (CLF) |
| Strain | nrps- | pks/nrps -1 | t1pks- | t2pks -1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |||
| S. albus N11-50 1 | + | + | + | + | + | + | + | + | + | + | + | + |
| S. albus NBRC 13014T 2 | + | + | - | + | + | + | + | + | + | + | + | + |
| S. albus CAS922 3 | + | + | + | + | + | + | + | + | + | + | + | + |
| S. albus DSM 40763 3 | + | + | + | + | + | + | + | + | + | + | + | + |
| S. albus NRRL B-1335 3 | + | + | + | + | + | + | + | + | + | + | + | + |
| S. albus NRRL B-1685 3 | + | + | + | + | + | + | + | + | + | + | + | + |
| S. albus NRRL B-2362 3 | + | + | + | + | + | + | + | + | + | + | + | + |
| S. albus NRRL B-2465 3 | + | + | + | + | + | + | + | + | + | + | + | + |
| S. albus NRRL B-12378 3 | + | + | + | + | + | + | + | + | + | + | + | + |
| S. albidoflavus G153 4 | - | - | - | - | - | - | - | - | - | - | - | - |
| S. albidoflavus NRRL B-2238 4 | - | - | - | - | - | - | - | - | - | - | - | - |
| Streptomyces sp. INA 01303 5 | - | - | - | - | - | - | - | - | - | - | - | - |
| S. albus NRRL F-5639 1 | + | + | + | + | + | + | + | + | + | + | + | + |
| S. albus NRRL F-5917 1 | + | + | + | + | + | + | + | + | + | + | + | + |
| S. albus HPH0547 1 | + | + | + | + | + | + | + | + | + | + | + | + |
| S. albus PHES57 51 1 | + | + | + | + | + | + | + | + | + | + | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komaki, H.; Igarashi, Y.; Tamura, T. Taxonogenomic Analysis of Marine-Derived Streptomyces sp. N11-50 and the Profile of NRPS and PKS Gene Clusters. Hydrobiology 2023, 2, 382-394. https://doi.org/10.3390/hydrobiology2020025
Komaki H, Igarashi Y, Tamura T. Taxonogenomic Analysis of Marine-Derived Streptomyces sp. N11-50 and the Profile of NRPS and PKS Gene Clusters. Hydrobiology. 2023; 2(2):382-394. https://doi.org/10.3390/hydrobiology2020025
Chicago/Turabian StyleKomaki, Hisayuki, Yasuhiro Igarashi, and Tomohiko Tamura. 2023. "Taxonogenomic Analysis of Marine-Derived Streptomyces sp. N11-50 and the Profile of NRPS and PKS Gene Clusters" Hydrobiology 2, no. 2: 382-394. https://doi.org/10.3390/hydrobiology2020025
APA StyleKomaki, H., Igarashi, Y., & Tamura, T. (2023). Taxonogenomic Analysis of Marine-Derived Streptomyces sp. N11-50 and the Profile of NRPS and PKS Gene Clusters. Hydrobiology, 2(2), 382-394. https://doi.org/10.3390/hydrobiology2020025





