Age-Related Fourier-Transform Infrared Spectroscopic Changes in Protein Conformation in an Aging Model of Human Dermal Fibroblasts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. FTIR Spectroscopy
2.2.1. Sample Preparation
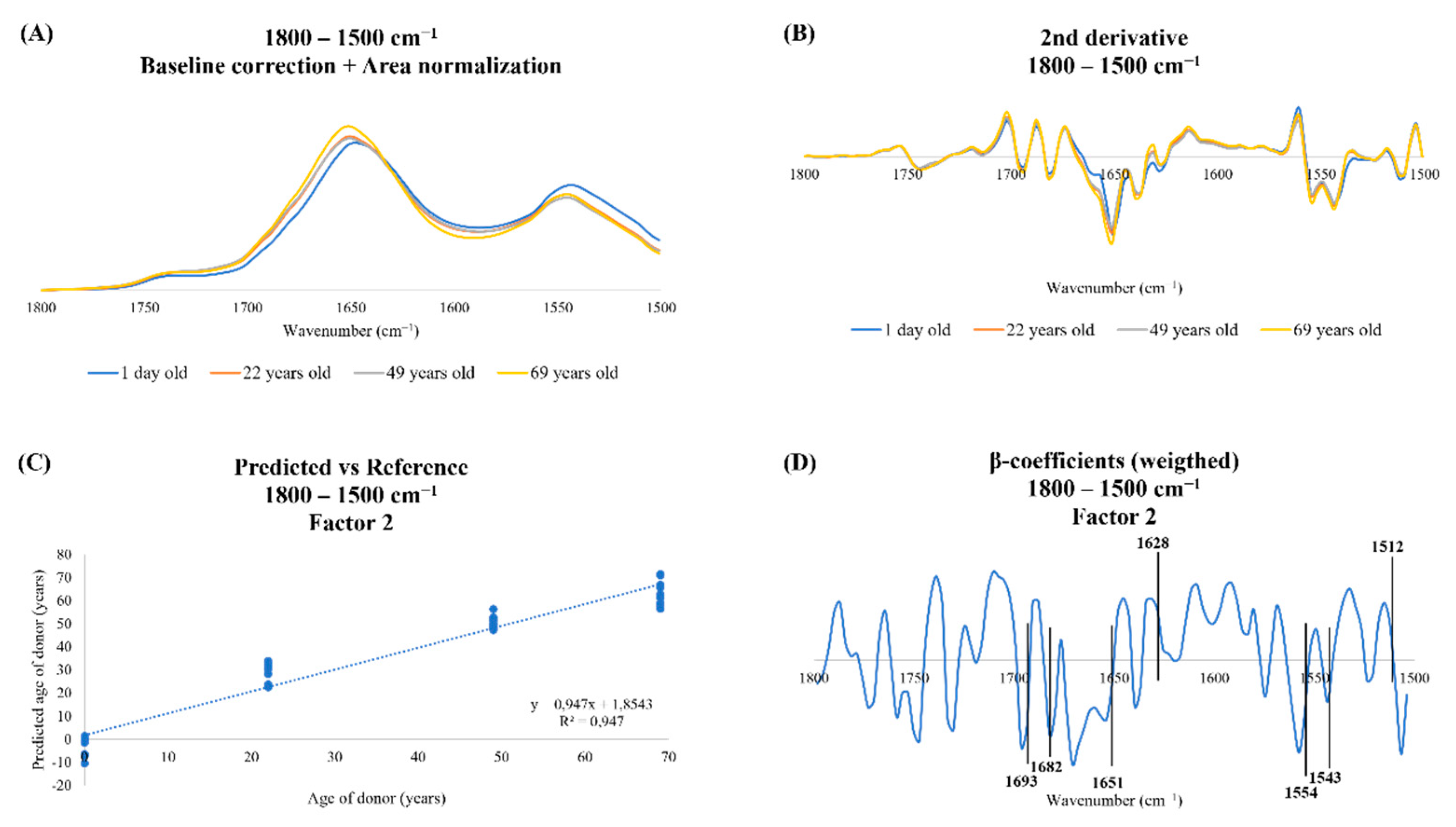
2.2.2. Spectra Processing
2.2.3. Peak Intensity Analysis
2.2.4. Partial Least Squares Regression Analysis
3. Results
3.1. Peak Intensity Analysis
3.2. Spectroscopic Profile of Human Dermal Fibroblasts
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Micó, V.; Berninches, L.; Tapia, J.; Daimiel, L. NutrimiRAging: Micromanaging Nutrient Sensing Pathways through Nutrition to Promote Healthy Aging. Int. J. Mol. Sci. 2017, 18, 915. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of Aging: An Expanding Universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef] [PubMed]
- Clausen, L.; Abildgaard, A.B.; Gersing, S.K.; Stein, A.; Lindorff-Larsen, K.; Hartmann-Petersen, R. Protein Stability and Degradation in Health and Disease. Adv. Protein Chem. Struct. Biol. 2019, 114, 61–83. [Google Scholar] [CrossRef]
- Taylor, R.C.; Dillin, A. Aging as an Event of Proteostasis Collapse. Cold Spring Harb. Perspect. Biol. 2011, 3, a004440. [Google Scholar] [CrossRef]
- Stefani, M.; Dobson, C.M.; Stefani, M.; Dobson, C.M. Protein Aggregation and Aggregate Toxicity: New Insights into Protein Folding, Misfolding Diseases and Biological Evolution. J. Mol. Med. 2003, 81, 678–699. [Google Scholar] [CrossRef]
- Chiti, F.; Stefani, M.; Taddei, N.; Ramponi, G.; Dobson, C.M. Rationalization of the Effects of Mutations on Peptide and Protein Aggregation Rates. Nature 2003, 424, 805–808. [Google Scholar] [CrossRef]
- Cuanalo-Contreras, K.; Schulz, J.; Mukherjee, A.; Park, K.W.; Armijo, E.; Soto, C. Extensive Accumulation of Misfolded Protein Aggregates during Natural Aging and Senescence. Front. Aging Neurosci. 2023, 14, 1090109. [Google Scholar] [CrossRef] [PubMed]
- Martins, F.; Sousa, J.; Pereira, C.D.; da Cruz e Silva, O.A.B.; Rebelo, S. Nuclear Envelope Dysfunction and Its Contribution to the Aging Process. Aging Cell 2020, 19, e13143. [Google Scholar] [CrossRef] [PubMed]
- Lees, H.; Walters, H.; Cox, L.S. Animal and Human Models to Understand Ageing. Maturitas 2016, 93, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Cristofalo, V.J.; Beck, J.; Allen, R.G.; Smith, J.R. Cell Senescence: An Evaluation of Replicative Senescence in Culture as a Model for Cell Aging in Situ. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2003, 58, 776–781. [Google Scholar] [CrossRef] [PubMed]
- Lidzbarsky, G.; Gutman, D.; Shekhidem, H.A.; Sharvit, L.; Atzmon, G. Genomic Instabilities, Cellular Senescence, and Aging: In Vitro, In Vivo and Aging-Like Human Syndromes. Front. Med. 2018, 5, 104. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, S.; Yan, Z.; Pei, M. A Prospect of Cell Immortalization Combined with Matrix Microenvironmental Optimization Strategy for Tissue Engineering and Regeneration. Cell Biosci. 2019, 9, 7. [Google Scholar] [CrossRef]
- Brunet, A. Old and New Models for the Study of Human Ageing. Nat. Rev. Mol. Cell Biol. 2020, 21, 491–493. [Google Scholar] [CrossRef] [PubMed]
- Phipps, S.M.O.; Berletch, J.B.; Andrews, L.G.; Tollefsbol, T.O. Aging Cell Culture: Methods and Observations. Methods Mol. Biol. 2007, 371, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Tigges, J.; Krutmann, J.; Fritsche, E.; Haendeler, J.; Schaal, H.; Fischer, J.W.; Kalfalah, F.; Reinke, H.; Reifenberger, G.; Stühler, K.; et al. The Hallmarks of Fibroblast Ageing. Mech. Ageing Dev. 2014, 138, 26–44. [Google Scholar] [CrossRef] [PubMed]
- Fleischer, J.G.; Schulte, R.; Tsai, H.H.; Tyagi, S.; Ibarra, A.; Shokhirev, M.N.; Huang, L.; Hetzer, M.W.; Navlakha, S. Predicting Age from the Transcriptome of Human Dermal Fibroblasts. Genome Biol. 2018, 19, 221. [Google Scholar] [CrossRef] [PubMed]
- Lagoid, J.C.; Puzzi, M.B. The Effect of Aging in Primary Human Dermal Fibroblasts. PLoS ONE 2019, 14, e0219165. [Google Scholar] [CrossRef]
- Gey, C.; Seeger, K. Metabolic Changes during Cellular Senescence Investigated by Proton NMR-Spectroscopy. Mech. Ageing Dev. 2013, 134, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, K.; Beleites, C.; Marthandan, S.; Matthäus, C.; Diekmann, S.; Popp, J. Raman and Infrared Spectroscopy Distinguishing Replicative Senescent from Proliferating Primary Human Fibroblast Cells by Detecting Spectral Differences Mainly Due to Biomolecular Alterations. Anal. Chem. 2017, 89, 2937–2947. [Google Scholar] [CrossRef]
- Eberhardt, K.; Matthäus, C.; Marthandan, S.; Diekmann, S.; Popp, J. Raman and Infrared Spectroscopy Reveal That Proliferating and Quiescent Human Fibroblast Cells Age by Biochemically Similar but Not Identical Processes. PLoS ONE 2018, 13, e0207380. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.; Yuan, H.; Soifer, I.; Maile, T.M.; Wang, R.Y.; Ireland, A.; O’Brien, J.; Goudeau, J.; Chan, L.; Vijay, T.; et al. Novel Insights from a Multiomics Dissection of the Hayflick Limit. eLife 2022, 11, e70283. [Google Scholar] [CrossRef]
- Magalhães, S.; Almeida, I.; Pereira, C.D.; Rebelo, S.; Goodfellow, B.J.; Nunes, A. The Long-Term Culture of Human Fibroblasts Reveals a Spectroscopic Signature of Senescence. Int. J. Mol. Sci. 2022, 23, 5830. [Google Scholar] [CrossRef] [PubMed]
- Pain, S.; Dezutter, C.; Reymermier, C.; Vogelgesang, B.; Delay, E.; André, V. Age-Related Changes in pro-Opiomelanocortin (POMC) and Related Receptors in Human Epidermis. Int. J. Cosmet. Sci. 2010, 32, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Crowley, L.C.; Marfell, B.J.; Christensen, M.E.; Waterhouse, N.J. Measuring Cell Death by Trypan Blue Uptake and Light Microscopy. Cold Spring Harb. Protoc. 2016, 2016, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, S.; Goodfellow, B.J.; Nunes, A. FTIR Spectroscopy in Biomedical Research: How to Get the Most out of Its Potential. Appl. Spectrosc. Rev. 2021, 56, 869–907. [Google Scholar] [CrossRef]
- Agahian, F.; Funt, B. Outlier Modeling for Spectral Data Reduction. J. Opt. Soc. Am. 2014, 31, 1445. [Google Scholar] [CrossRef]
- Nieuwoudt, H.H.; Prior, B.A.; Pretorius, I.S.; Manley, M.; Bauer, F.F. Principal Component Analysis Applied to Fourier Transform Infrared Spectroscopy for the Design of Calibration Sets for Glycerol Prediction Models in Wine and for the Detection and Classification of Outlier Samples. J. Agric. Food Chem. 2004, 52, 3726–3735. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, S.; Almeida, I.; Martins, F.; Camões, F.; Soares, A.R.; Goodfellow, B.J.; Rebelo, S.; Nunes, A. FTIR Spectroscopy as a Tool to Study Age-Related Changes in Cardiac and Skeletal Muscle of Female C57BL/6J Mice. Molecules 2021, 26, 6410. [Google Scholar] [CrossRef]
- Juszczyk, P.; Kolodziejczyk, A.S.; Grzonka, Z. FTIR Spectroscopic Studies on Aggregation Process of the Beta-Amyloid 11-28 Fragment and Its Variants. J. Pept. Sci. 2009, 15, 23–29. [Google Scholar] [CrossRef]
- Mateus, T.; Almeida, I.; Costa, A.; Viegas, D.; Magalhães, S.; Martins, F.; Herdeiro, M.T.; da Cruz e Silva, O.A.B.; Fraga, C.; Alves, I.; et al. Fourier-Transform Infrared Spectroscopy as a Discriminatory Tool for Myotonic Dystrophy Type 1 Metabolism: A Pilot Study. Int. J. Environ. Res. Public Health 2021, 18, 3800. [Google Scholar] [CrossRef]
- Kong, J.; Yu, S. Fourier Transform Infrared Spectroscopic Analysis of Protein Secondary Structures. Acta Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Yang, S.; Kong, J.; Dong, A.; Yu, S. Obtaining Information about Protein Secondary Structures in Aqueous Solution Using Fourier Transform IR Spectroscopy. Nat. Protoc. 2015, 10, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Usoltsev, D.; Sitnikova, V.; Kajava, A.; Uspenskaya, M. FTIR Spectroscopy Study of the Secondary Structure Changes in Human Serum Albumin and Trypsin under Neutral Salts. Biomolecules 2020, 10, 606. [Google Scholar] [CrossRef]
- Sadat, A.; Joye, I.J. Peak Fitting Applied to Fourier Transform Infrared and Raman Spectroscopic Analysis of Proteins. Appl. Sci. 2020, 10, 5918. [Google Scholar] [CrossRef]
- Wang, H.; Ju, A.; Wang, L. Ultraviolet Spectroscopic Detection of Nitrate and Nitrite in Seawater Simultaneously Based on Partial Least Squares. Molecules 2021, 26, 3685. [Google Scholar] [CrossRef] [PubMed]
- Sharaf, Y.A.; Ibrahim, A.E.; El Deeb, S.; Sayed, R.A. Green Chemometric Determination of Cefotaxime Sodium in the Presence of Its Degradation Impurities Using Different Multivariate Data Processing Tools; GAPI and AGREE Greenness Evaluation. Molecules 2023, 28, 2187. [Google Scholar] [CrossRef]
- Glassford, S.E.; Byrne, B.; Kazarian, S.G. Recent Applications of ATR FTIR Spectroscopy and Imaging to Proteins. Biochim. Biophys. Acta 2013, 1834, 2849–2858. [Google Scholar] [CrossRef]
- Asuero, A.G.; Sayago, A.; González, A.G. The Correlation Coefficient: An Overview. Crit. Rev. Anal. Chem. 2007, 36, 41–59. [Google Scholar] [CrossRef]
- Mortality and Life Expectancy Statistics—Statistics Explained. Available online: https://ec.europa.eu/eurostat/statistics-explained/index.php?title=Mortality_and_life_expectancy_statistics#Life_expectancy_at_birth (accessed on 20 June 2022).
- Mitchell, S.J.; Scheibye-Knudsen, M.; Longo, D.L.; de Cabo, R. Animal Models of Aging Research: Implications for Human Aging and Age-Related Diseases. Annu. Rev. Anim. Biosci. 2015, 3, 283–303. [Google Scholar] [CrossRef] [PubMed]
- Ferrucci, L.; Kuchel, G.A. Heterogeneity of Aging: Individual Risk Factors, Mechanisms, Patient Priorities, and Outcomes. J. Am. Geriatr. Soc. 2021, 69, 610–612. [Google Scholar] [CrossRef]
- Anisimova, A.S.; Alexandrov, A.I.; Makarova, N.E.; Gladyshev, V.N.; Dmitriev, S.E. Protein Synthesis and Quality Control in Aging. Aging 2018, 10, 4269. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, A.S.; Meerson, M.B.; Gerashchenko, M.V.; Kulakovskiy, I.V.; Dmitriev, S.E.; Gladyshev, V.N. Multifaceted Deregulation of Gene Expression and Protein Synthesis with Age. Proc. Natl. Acad. Sci. USA 2020, 117, 15581–15590. [Google Scholar] [CrossRef] [PubMed]
- Vilchez, D.; Saez, I.; Dillin, A. The Role of Protein Clearance Mechanisms in Organismal Ageing and Age-Related Diseases. Nat. Commun. 2014, 5, 5659. [Google Scholar] [CrossRef] [PubMed]
- Frankowska, N.; Lisowska, K.; Witkowski, J.M. Proteolysis Dysfunction in the Process of Aging and Age-Related Diseases. Front. Aging 2022, 3, 85. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef] [PubMed]
- Cerf, E.; Sarroukh, R.; Tamamizu-Kato, S.; Breydo, L.; Derclayes, S.; Dufrênes, Y.F.; Narayanaswami, V.; Goormaghtigh, E.; Ruysschaert, J.M.; Raussens, V. Antiparallel Beta-Sheet: A Signature Structure of the Oligomeric Amyloid Beta-Peptide. Biochem. J. 2009, 421, 415–423. [Google Scholar] [CrossRef]
- Kayed, R.; Sokolov, Y.; Edmonds, B.; McIntire, T.M.; Milton, S.C.; Hall, J.E.; Glabe, C.G. Permeabilization of Lipid Bilayers Is a Common Conformation-Dependent Activity of Soluble Amyloid Oligomers in Protein Misfolding Diseases. J. Biol. Chem. 2004, 279, 46363–46366. [Google Scholar] [CrossRef]
- Cheng, P.N.; Pham, J.D.; Nowick, J.S. The Supramolecular Chemistry of β-Sheets. J. Am. Chem. Soc. 2013, 135, 5477–5492. [Google Scholar] [CrossRef] [PubMed]
- Tycko, R.; Wickner, R.B. Molecular Structures of Amyloid and Prion Fibrils: Consensus versus Controversy. Acc. Chem. Res. 2013, 46, 1487–1496. [Google Scholar] [CrossRef] [PubMed]
- Ke, P.C.; Zhou, R.; Serpell, L.C.; Riek, R.; Knowles, T.P.J.; Lashuel, H.A.; Gazit, E.; Hamley, I.W.; Davis, T.P.; Fändrich, M.; et al. Half a Century of Amyloids: Past, Present and Future. Chem. Soc. Rev. 2020, 49, 5473–5509. [Google Scholar] [CrossRef] [PubMed]
- Riek, R. The Three-Dimensional Structures of Amyloids. Cold Spring Harb. Perspect. Biol. 2017, 9, a023572. [Google Scholar] [CrossRef]
- Diaz-Espinoza, R. Catalytically Active Amyloids as Future Bionanomaterials. Nanomaterials 2022, 12, 3802. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.Y.; Chi, J.T.; Dudoit, S.; Bondre, C.; Van De Rijn, M.; Botstein, D.; Brown, P.O. Diversity, Topographic Differentiation, and Positional Memory in Human Fibroblasts. Proc. Natl. Acad. Sci. USA 2002, 99, 12877–12882. [Google Scholar] [CrossRef]
- Sacco, A.M.; Belviso, I.; Romano, V.; Carfora, A.; Schonauer, F.; Nurzynska, D.; Montagnani, S.; Di Meglio, F.; Castaldo, C. Diversity of Dermal Fibroblasts as Major Determinant of Variability in Cell Reprogramming. J. Cell Mol. Med. 2019, 23, 4256–4268. [Google Scholar] [CrossRef] [PubMed]
- Sriram, G.; Bigliardi, P.L.; Bigliardi-Qi, M. Fibroblast Heterogeneity and Its Implications for Engineering Organotypic Skin Models in Vitro. Eur. J. Cell Biol. 2015, 94, 483–512. [Google Scholar] [CrossRef] [PubMed]
- Driskell, R.R.; Watt, F.M. Understanding Fibroblast Heterogeneity in the Skin. Trends Cell Biol. 2015, 25, 92–99. [Google Scholar] [CrossRef]
- Ahuja, A.K.; Pontiggia, L.; Moehrlen, U.; Biedermann, T. The Dynamic Nature of Human Dermal Fibroblasts Is Defined by Marked Variation in the Gene Expression of Specific Cytoskeletal Markers. Life 2022, 12, 935. [Google Scholar] [CrossRef] [PubMed]
- Rittié, L.; Fisher, G.J. Natural and Sun-Induced Aging of Human Skin. Cold Spring Harb. Perspect. Med. 2015, 5, a015370. [Google Scholar] [CrossRef] [PubMed]
- Bulbiankova, D.; Díaz-Puertas, R.; Álvarez-Martínez, F.J.; Herranz-López, M.; Barrajón-Catalán, E.; Micol, V. Hallmarks and Biomarkers of Skin Senescence: An Updated Review of Skin Senotherapeutics. Antioxidants 2023, 12, 444. [Google Scholar] [CrossRef]
- Foo, H.; Mather, K.A.; Thalamuthu, A.; Sachdev, P.S. The Many Ages of Man: Diverse Approaches to Assessing Ageing-Related Biological and Psychological Measures and Their Relationship to Chronological Age. Curr. Opin. Psychiatry 2019, 32, 130–137. [Google Scholar] [CrossRef]
- Wu, L.; Xie, X.; Liang, T.; Ma, J.; Yang, L.; Yang, J.; Li, L.; Xi, Y.; Li, H.; Zhang, J.; et al. Integrated Multi-Omics for Novel Aging Biomarkers and Antiaging Targets. Biomolecules 2021, 12, 39. [Google Scholar] [CrossRef] [PubMed]
- Henney, A.M. The Promise and Challenge of Personalized Medicine: Aging Populations, Complex Diseases, and Unmet Medical Need. Croat. Med. J. 2012, 53, 207. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D.; Dunn, W.; Schmidt, M.A.; Gross, S.S.; Kirwan, J.A.; Cascante, M.; Brennan, L.; Wishart, D.S.; Oresic, M.; Hankemeier, T.; et al. Metabolomics Enables Precision Medicine: “A White Paper, Community Perspective. Metabolomics 2016, 12, 149. [Google Scholar] [CrossRef] [PubMed]
- Poonprasartporn, A.; Chan, K.L.A. Live-Cell ATR-FTIR Spectroscopy as a Novel Bioanalytical Tool for Cell Glucose Metabolism Research. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 119024. [Google Scholar] [CrossRef] [PubMed]


| Characteristics | AG22153 | AG10803 | AG02222 | AG16102 |
|---|---|---|---|---|
| Age of the donor | 1 day old | 22 years old | 49 years old | 69 years old |
| Passage cells were received | P1 | P4 | P6 | P8 |
| Passage cells were used | P12 | P12 | P12 | P12 |
| Biopsy source | Foreskin | Abdomen | Abdomen | Arm |
| Gender of the donor | Male | Male | Male | Male |
| Ethnicity of the donor | White/East Indian | White | White | White |
| Samples | Peak | Biological Meaning | References |
|---|---|---|---|
| younger | 1693 | antiparallel β-sheets | [22,29,30] |
| 1682 | β-sheets | [22,31,32] | |
| 1651 | α-helices | [22,30,32] | |
| 1554 | amide II | [38] | |
| 1543 | |||
| older | 1628 | intermolecular β-sheets | [22,33,34] |
| 1512 | amide II | [38] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martins, C.; Almeida, I.; Rebelo, S.; Magalhães, S.; Nunes, A. Age-Related Fourier-Transform Infrared Spectroscopic Changes in Protein Conformation in an Aging Model of Human Dermal Fibroblasts. Spectrosc. J. 2023, 1, 37-48. https://doi.org/10.3390/spectroscj1010004
Martins C, Almeida I, Rebelo S, Magalhães S, Nunes A. Age-Related Fourier-Transform Infrared Spectroscopic Changes in Protein Conformation in an Aging Model of Human Dermal Fibroblasts. Spectroscopy Journal. 2023; 1(1):37-48. https://doi.org/10.3390/spectroscj1010004
Chicago/Turabian StyleMartins, Cláudia, Idália Almeida, Sandra Rebelo, Sandra Magalhães, and Alexandra Nunes. 2023. "Age-Related Fourier-Transform Infrared Spectroscopic Changes in Protein Conformation in an Aging Model of Human Dermal Fibroblasts" Spectroscopy Journal 1, no. 1: 37-48. https://doi.org/10.3390/spectroscj1010004
APA StyleMartins, C., Almeida, I., Rebelo, S., Magalhães, S., & Nunes, A. (2023). Age-Related Fourier-Transform Infrared Spectroscopic Changes in Protein Conformation in an Aging Model of Human Dermal Fibroblasts. Spectroscopy Journal, 1(1), 37-48. https://doi.org/10.3390/spectroscj1010004








