Metabolomics in Hyperuricemia and Gout
Abstract
1. Introduction
2. Analytical Technology for Metabolomics
3. Metabolomics Data Analysis, Interpretation and Sharing
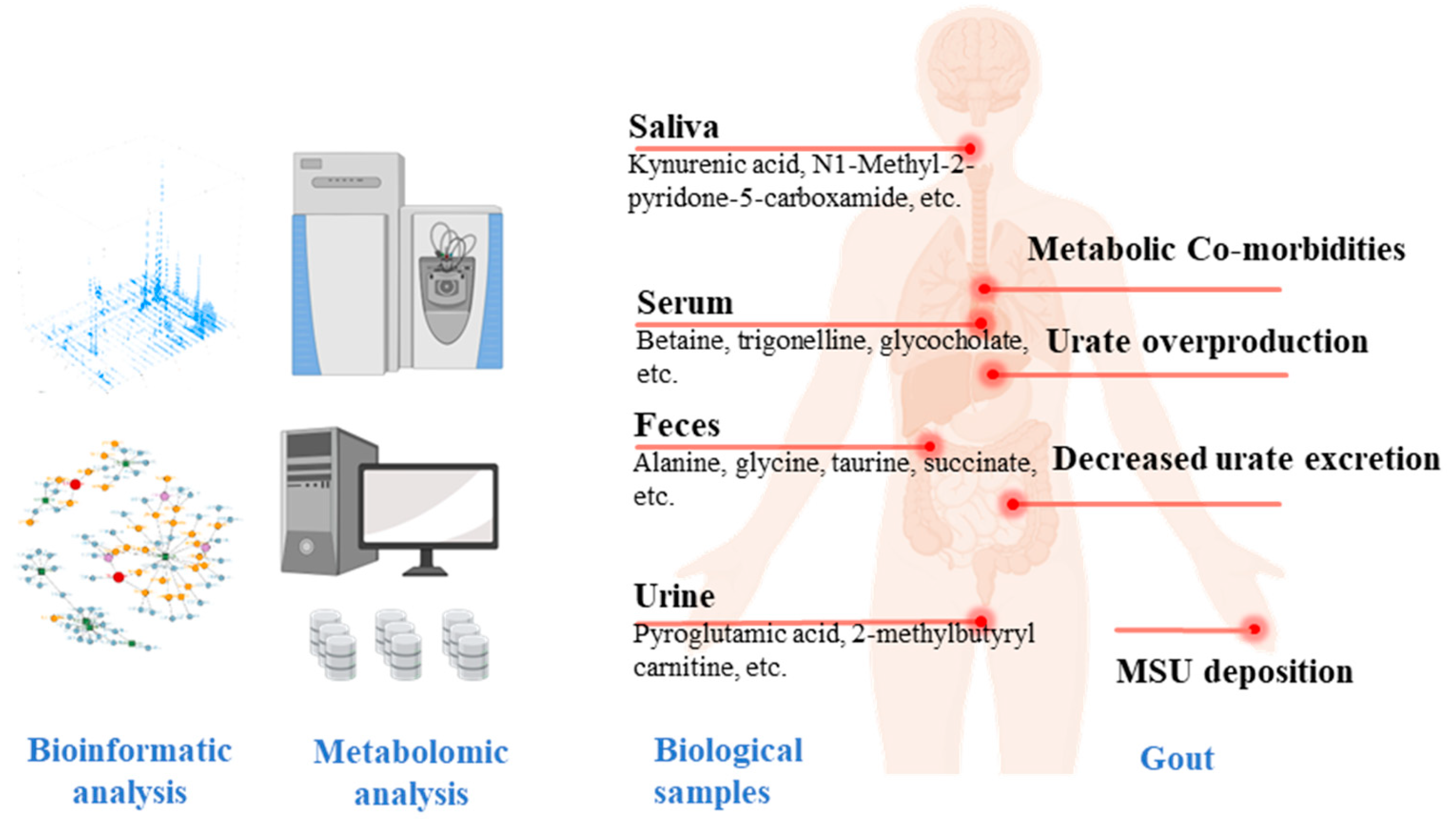
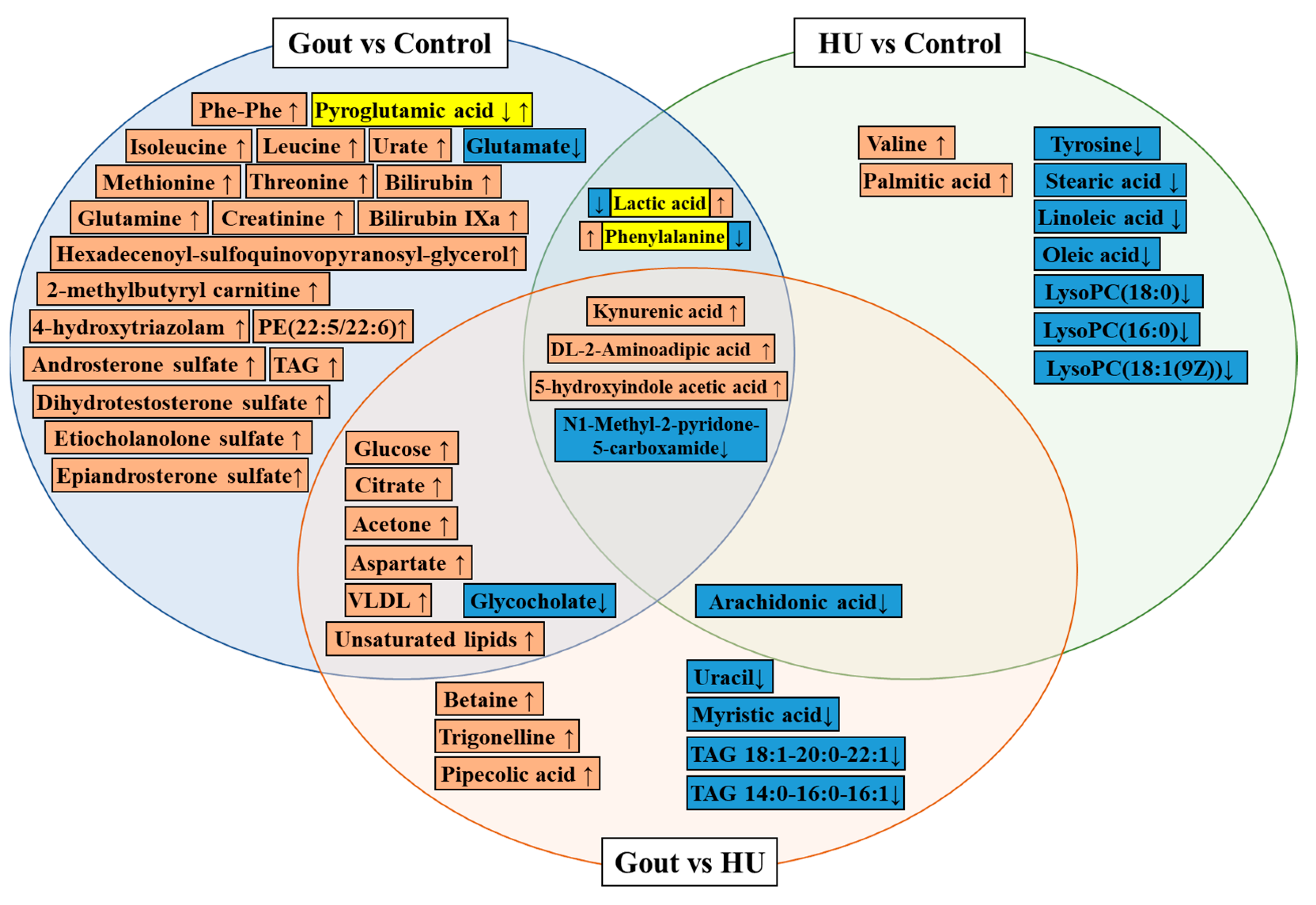
4. Metabolic Profiling and Metabolite Biomarker Discovery in Clinical Populations with HU and Gout
| Date | Sample | Discovery Cohort | Validation Cohort | Technique Platform | Upregulated Biomarkers | Downregulated Biomarkers | Conclusions |
|---|---|---|---|---|---|---|---|
| 2021 [31] | Serum | Gout (109) vs. healthy (119) | LCMS | Pyroglutamic acid, glycocholate, lactic acid, glutamate | Glycine, serine, and threonine metabolism disorder Arginine and proline metabolism disorder Ascorbate and aldarate metabolism disorder Alanine, aspartate, and glutamate metabolism disorder | ||
| Gout (109) vs. HU (102) | Betaine, trigonelline, pipecolic acid, | Glycocholate, uracil, myristic acid, arachidonate | Arginine biosynthesis↑ Glycine, serine, and threonine metabolism disorder | ||||
| 2018 [67] | Serum | Gout (49) vs. healthy (50) | NMR | VLDL, isoleucine, leucine, glutamine, methionine, acetone, citrate, aspartate, creatinine, glucose, threonine, triglycerides, unsaturated lipids and phenylalanine | Aminoacyl-tRNA biosynthesis↓ Valine, leucine and isoleucine biosynthesis↑ Nitrogen metabolism↑ Alanine, aspartate and glutamate metabolism↑ D-glutamine and D-glutamate metabolism disorder | ||
| Gout (49) vs. HU (50) | VLDL, lipid, acetone, citrate, aspartate, glucose | ||||||
| 2020 [68] | Serum | Gout (31) vs. healthy (31) | LCMS | 4-hydroxytriazolam, bilirubin, urate, 4E,15Z-Bilirubin IXa, androsterone sulfate, 5a-dihydrotestosterone sulfate, etiocholanolone sulfate, epiandrosterone sulfate, 1,2-Di-O-(8-hexadecenoyl)-3-O-(6-sulfoquinovopyranosyl)glycerol, PE(22:5(4Z,7Z,10Z,13Z,16Z)/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) | Primary bile acid biosynthesis↓ Purine metabolism↑ Glycerophospholipid metabolism disorder | ||
| 2020 [69] | Serum and Urine | Gout (30) vs. healthy (30) | Gout (50) vs. healthy (50) | LCMS | Pyroglutamic acid, Phe-Phe, 2-methylbutyryl carnitine | Purine metabolism disorder Branched-chain amino acids (BCAAs) metabolism disorder Tricarboxylic acid cycle disorder Synthesis and degradation of ketone bodies disorder Bile secretion disorder Arachidonic acid metabolism disorder | |
| 2018 [71] | Urine | Gout (35) vs. healthy (29) | GCMS | Urate, isoxanthopterin | Purine nucleotide synthesis↑ Amino acid metabolism↓ Purine metabolism↑ Lipid and carbohydrate metabolism disorder | ||
| 2017 [72] | Feces | Gout (26) vs. healthy (26) | NMR | Alanine, glycine, taurine, succinate, acetate, a-glucose, b-glucose, a-xylose | Valine, asparagine, aspartate, citrulline, phenylalanine, a-ketoisocaproate | Urate excretion disorder Purine metabolism disorder Inflammatory responses disorder | |
| 2021 [73] | Serum | Gout (50) vs. HU (50) | Gout (69) vs. HU (50) | LCMS | TAG 18:1-20:0-22:1, TAG 14:0-16:0-16:1 | Lipid disorders | |
| 2022 [75] | Serum | HU (20) vs. healthy (20) | LCMS untargeted + targeted | Lactic acid, valine, palmitic acid, | Tyrosine, phenylalanine, arachidonic acid, stearic acid, linoleic acid, oleic acid, lipids, LysoPC(18:0), LysoPC(16:0), LysoPC(18:1(9Z)) | Glycerophospholipid metabolism disorder Arachidonic acid metabolism disorder Sphingolipid metabolism disorder Linoleic acid metabolism disorder α-linolenic acid metabolism disorder Phenylalanine, tyrosine, and tryptophan biosynthesis disorder Phenylalanine metabolism disorder | |
| 2017 [74] | Saliva | Gout (8) vs. healthy (15) | Gout (30) vs. healthy (30) | CICMS + assay kits | Urate, oxalic acid, L-homocysteic acid (HCA) | ||
| Gout (8) vs. HU (15) | Gout (30) vs. HU (30) | Urate, oxalic acid, L-homocysteic acid (HCA) | |||||
| 2022 [70] | Serum | 5 sequential stages (347) | 5 sequential stages (200) | LCMS untargeted + targeted | kynurenic acid (KYNA), 5-hydroxyindole acetic acid (5-HIAA), DL-2-Aminoadipic acid (2AMIA) | N1-Methyl-2-pyridone-5-carboxamide (2PY) | KYNA and 5-HIAA are related to acute inflammation of gouty arthritis 2PY and 2AMIA are related to renal function damage caused by long-term HU |
5. Multi-Omics and Big Data
6. Metabolomics in Experiment Models
| Data | Analytical Platform | Rodent Models | Differential Metabolites/ Metabolic Pathways |
|---|---|---|---|
| 2022 [87] | UPLC-QTOF-MS/MS | MSU Crystal-Induced Gouty arthritis Rats | Arachidonic acid, sphingolipid, and glycerophospholipid metabolism |
| 2021 [88] | UPLC-QTOF/MS | High fructose combined with potassium oxonate (HFCPO)-induced hyperuricemia Rats | Acylcarnitine and amino acid related metabolites |
| 2020 [89] | 1H NMR and UHPLC/Q-Orbitrap-MS | Potassium oxonate induced hyperuricemia rats | Pyruvate, lactate, creatine, glycine, LysoPC and PC and etc |
| 2019 [91] | Capillary electrophoresis–time-of-flight mass spectrometry (CE-TOFMS) | Rat model of renal I/R | Purine/pyrimidine metabolism |
| 2022 [90] | Shimadzu Nexera XR HPLC-MS SCIEX Triple Quad™ 3500 | Acute gouty peritonitis mouse model | Glycolysis pathway |
7. Summary and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dalbeth, N.; Gosling, A.L.; Gaffo, A.; Abhishek, A. Gout. Lancet 2021, 397, 1843–1855. [Google Scholar] [CrossRef]
- Desai, J.; Steiger, S.; Anders, H.-J. Molecular Pathophysiology of Gout. Trends Mol. Med. 2017, 23, 756–768. [Google Scholar] [CrossRef] [PubMed]
- Dehlin, M.; Jacobsson, L.; Roddy, E. Global epidemiology of gout: Prevalence, incidence, treatment patterns and risk factors. Nat. Rev. Rheumatol. 2020, 16, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Dalbeth, N.; Phipps-Green, A.; Frampton, C.; Neogi, T.; Taylor, W.J.; Merriman, T.R. Relationship between serum urate concentration and clinically evident incident gout: An individual participant data analysis. Ann. Rheum. Dis. 2018, 77, 1048–1052. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, J.D.; Dalbeth, N.; Mikuls, T.; Brignardello-Petersen, R.; Guyatt, G.; Abeles, A.M.; Gelber, A.C.; Harrold, L.R.; Khanna, D.; King, C.; et al. 2020 American College of Rheumatology Guideline for the Management of Gout. Arthritis Care Res. 2020, 72, 744–760. [Google Scholar] [CrossRef]
- Schumacher, H.R.; Taylor, W.; Edwards, L.; Grainger, R.; Schlesinger, N.; Dalbeth, N.; Sivera, F.; Singh, J.; Evans, R.; Waltrip, R.W.; et al. Outcome Domains for Studies of Acute and Chronic Gout. J. Rheumatol. 2009, 36, 2342–2345. [Google Scholar] [CrossRef]
- Hirsch, J.D.; Terkeltaub, R.; Khanna, D.; Singh, J.; Sarkin, A.; Shieh, M.; Kavanaugh, A.; Lee, S.J. Gout disease-specific quality of life and the association with gout characteristics. Patient Relat. Outcome Meas. 2010, 1, 1–8. [Google Scholar] [CrossRef]
- Li, Q.; Li, X.; Wang, J.; Liu, H.; Kwong, J.S.; Chen, H.; Li, L.; Chung, S.C.; Shah, A.; Chen, Y.; et al. Diagnosis and treatment for hyperuricemia and gout: A systematic review of clinical practice guidelines and consensus statements. BMJ Open 2019, 9, e026677. [Google Scholar] [CrossRef]
- Soltani, Z.; Rasheed, K.; Kapusta, D.R.; Reisin, E. Potential Role of Uric Acid in Metabolic Syndrome, Hypertension, Kidney Injury, and Cardiovascular Diseases: Is It Time for Reappraisal? Curr. Hypertens. Rep. 2013, 15, 175–181. [Google Scholar] [CrossRef]
- Puig, J.G.; Martínez, M.A. Hyperuricemia, gout and the metabolic syndrome. Curr. Opin. Rheumatol. 2008, 20, 187–191. [Google Scholar] [CrossRef]
- Perez De Souza, L.; Alseekh, S.; Scossa, F.; Fernie, A.R. Ultra-high-performance liquid chromatography high-resolution mass spectrometry variants for metabolomics research. Nat. Methods 2021, 18, 733–746. [Google Scholar] [CrossRef] [PubMed]
- Alseekh, S.; Aharoni, A.; Brotman, Y.; Contrepois, K.; D’Auria, J.; Ewald, J.; Ewald, J.C.; Fraser, P.D.; Giavalisco, P.; Hall, R.D.; et al. Mass spectrometry-based metabolomics: A guide for annotation, quantification and best reporting practices. Nat. Methods 2021, 18, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef]
- Metallo, C.M.; Vander Heiden, M.G. Understanding metabolic regulation and its influence on cell physiology. Mol. Cell 2013, 49, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef]
- Boon, R.; Silveira, G.G.; Mostoslavsky, R. Nuclear metabolism and the regulation of the epigenome. Nat. Metab. 2020, 2, 1190–1203. [Google Scholar] [CrossRef]
- Renaudin, F.; Orliaguet, L.; Castelli, F.; Fenaille, F.; Prignon, A.; Alzaid, F.; Combes, C.; Delvaux, A.; Adimy, Y.; Cohen-Solal, M.; et al. Gout and pseudo-gout-related crystals promote GLUT1-mediated glycolysis that governs NLRP3 and interleukin-1beta activation on macrophages. Ann. Rheum. Dis. 2020, 79, 1506–1514. [Google Scholar] [CrossRef]
- Cobo, I.; Cheng, A.; Murillo-Saich, J.; Coras, R.; Torres, A.; Abe, Y.; Lana, A.J.; Schlachetzki, J.; Liu-Bryan, R.; Terkeltaub, R.; et al. Monosodium urate crystals regulate a unique JNK-dependent macrophage metabolic and inflammatory response. Cell Rep. 2022, 38, 110489. [Google Scholar] [CrossRef]
- Newgard, C.B. Metabolomics and Metabolic Diseases: Where Do We Stand? Cell Metab. 2017, 25, 43–56. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Alseekh, S.; Fernie, A.R. Metabolomics 20 years on: What have we learned and what hurdles remain? Plant J. 2018, 94, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef] [PubMed]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020, 48, D440–D444. [Google Scholar] [CrossRef]
- Shao, Y.; Le, W. Recent advances and perspectives of metabolomics-based investigations in Parkinson’s disease. Mol. Neurodegener. 2019, 14, 3. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.N.; Higashi, R.M.; Fan, T.W.M. NMR and MS-based Stable Isotope-Resolved Metabolomics and applications in cancer metabolism. TrAC Trends Anal. Chem. 2019, 120, 115322. [Google Scholar] [CrossRef]
- Griffin, J.L. Metabonomics: NMR spectroscopy and pattern recognition analysis of body fluids and tissues for characterisation of xenobiotic toxicity and disease diagnosis. Curr. Opin. Chem. Biol. 2003, 7, 648–654. [Google Scholar] [CrossRef]
- Vignoli, A.; Ghini, V.; Meoni, G.; Licari, C.; Takis, P.G.; Tenori, L.; Turano, P.; Luchinat, C. High-Throughput Metabolomics by 1D NMR. Angew. Chem. Int. Ed. 2019, 58, 968–994. [Google Scholar] [CrossRef]
- Takis, P.G.; Ghini, V.; Tenori, L.; Turano, P.; Luchinat, C. Uniqueness of the NMR approach to metabolomics. TrAC Trends Anal. Chem. 2019, 120, 115300. [Google Scholar] [CrossRef]
- Markley, J.L.; Brüschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytical techniques in metabolomics analysis. Analyst 2012, 137, 293–300. [Google Scholar] [CrossRef]
- Shen, X.; Wang, C.; Liang, N.; Liu, Z.; Li, X.; Zhu, Z.J.; Merriman, T.R.; Dalbeth, N.; Terkeltaub, R.; Li, C.; et al. Serum metabolomics identifies dysregulated pathways and potential metabolic biomarkers for hyperuricemia and gout. Arthritis Rheumatol. 2021, 73, 1738–1748. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; TeSlaa, T.; Xu, X.; Zeng, X.; Yang, L.; Xing, G.; Tesz, G.J.; Clasquin, M.F.; Rabinowitz, J.D. Serine catabolism generates liver NADPH and supports hepatic lipogenesis. Nat. Metab. 2021, 3, 1608–1620. [Google Scholar] [CrossRef] [PubMed]
- Hui, S.; Ghergurovich, J.M.; Morscher, R.J.; Jang, C.; Teng, X.; Lu, W.; Esparza, L.A.; Reya, T.; Le, Z.; Yanxiang Guo, J.; et al. Glucose feeds the TCA cycle via circulating lactate. Nature 2017, 551, 115–118. [Google Scholar] [CrossRef]
- Li, M.; He, X.; Guo, W.; Yu, H.; Zhang, S.; Wang, N.; Liu, G.; Sa, R.; Shen, X.; Jiang, Y.; et al. Aldolase B suppresses hepatocellular carcinogenesis by inhibiting G6PD and pentose phosphate pathways. Nat. Cancer 2020, 1, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Shi, X.; Wang, S.; Xu, G. Multidimensional liquid chromatography-mass spectrometry for metabolomic and lipidomic analyses. TrAC Trends Anal. Chem. 2019, 120, 115302. [Google Scholar] [CrossRef]
- Zhou, Z.; Shen, X.; Chen, X.; Tu, J.; Xiong, X.; Zhu, Z.-J. LipidIMMS Analyzer: Integrating multi-dimensional information to support lipid identification in ion mobility—Mass spectrometry based lipidomics. Bioinformatics 2019, 35, 698–700. [Google Scholar] [CrossRef]
- Haag, A.M. Mass Analyzers and Mass Spectrometers. In Modern Proteomics—Sample Preparation, Analysis and Practical Applications; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 157–169. [Google Scholar]
- Doerr, A. Global metabolomics. Nat. Methods 2017, 14, 32. [Google Scholar] [CrossRef]
- Zhou, J.; Yin, Y. Strategies for large-scale targeted metabolomics quantification by liquid chromatography-mass spectrometry. Analyst 2016, 141, 6362–6373. [Google Scholar] [CrossRef]
- Zheng, F.; Zhao, X.; Zeng, Z.; Wang, L.; Lv, W.; Wang, Q.; Xu, G. Development of a plasma pseudotargeted metabolomics method based on ultra-high-performance liquid chromatography–mass spectrometry. Nat. Protoc. 2020, 15, 2519–2537. [Google Scholar] [CrossRef]
- Bonner, R.; Hopfgartner, G. SWATH data independent acquisition mass spectrometry for metabolomics. TrAC Trends Anal. Chem. 2019, 120, 115278. [Google Scholar] [CrossRef]
- Van Der Greef, J.; Van Wietmarschen, H.; Van Ommen, B.; Verheij, E. Looking back into the future: 30 years of metabolomics at TNO. Mass Spectrom. Rev. 2013, 32, 399–415. [Google Scholar] [CrossRef]
- Fernie, A.R.; Trethewey, R.N.; Krotzky, A.J.; Willmitzer, L. Metabolite profiling: From diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 2004, 5, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; Vandergheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Kirkwood, K.I.; Pratt, B.S.; Shulman, N.; Tamura, K.; Maccoss, M.J.; Maclean, B.X.; Baker, E.S. Utilizing Skyline to analyze lipidomics data containing liquid chromatography, ion mobility spectrometry and mass spectrometry dimensions. Nat. Protoc. 2022, 17, 2415–2430. [Google Scholar] [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Kind, T.; Liu, K.-H.; Lee, D.Y.; Defelice, B.; Meissen, J.K.; Fiehn, O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 2013, 10, 755–758. [Google Scholar] [CrossRef]
- Montenegro-Burke, J.R.; Guijas, C.; Siuzdak, G. METLIN: A Tandem Mass Spectral Library of Standards. In Computational Methods and Data Analysis for Metabolomics; Springer: New York, NY, USA, 2020; pp. 149–163. [Google Scholar] [CrossRef]
- Schmid, R.; Petras, D.; Nothias, L.-F.; Wang, M.; Aron, A.T.; Jagels, A.; Tsugawa, H.; Rainer, J.; Garcia-Aloy, M.; Dührkop, K.; et al. Ion identity molecular networking for mass spectrometry-based metabolomics in the GNPS environment. Nat. Commun. 2021, 12, 3832. [Google Scholar] [CrossRef]
- Shen, X.; Wang, R.; Xiong, X.; Yin, Y.; Cai, Y.; Ma, Z.; Liu, N.; Zhu, Z.-J. Metabolic reaction network-based recursive metabolite annotation for untargeted metabolomics. Nat. Commun. 2019, 10, 1516. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Lu, W.; Wang, L.; Xing, X.; Chen, Z.; Teng, X.; Zeng, X.; Muscarella, A.D.; Shen, Y.; Cowan, A.; et al. Metabolite discovery through global annotation of untargeted metabolomics data. Nat. Methods 2021, 18, 1377–1385. [Google Scholar] [CrossRef] [PubMed]
- Sen, P.; Lamichhane, S.; Mathema, V.B.; McGlinchey, A.; Dickens, A.M.; Khoomrung, S.; Orešič, M. Deep learning meets metabolomics: A methodological perspective. Brief. Bioinform. 2021, 22, 1531–1542. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Jewison, T.; Su, Y.; Disfany, F.M.; Liang, Y.; Knox, C.; Maciejewski, A.; Poelzer, J.; Huynh, J.; Zhou, Y.; Arndt, D.; et al. SMPDB 2.0: Big Improvements to the Small Molecule Pathway Database. Nucleic Acids Res. 2013, 42, D478–D484. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010, 38, W71–W77. [Google Scholar] [CrossRef]
- Picart-Armada, S.; Fernández-Albert, F.; Vinaixa, M.; Yanes, O.; Perera-Lluna, A. FELLA: An R package to enrich metabolomics data. BMC Bioinform. 2018, 19, 538. [Google Scholar] [CrossRef]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2019, 48, D498–D503. [Google Scholar] [CrossRef]
- Brunk, E.; Sahoo, S.; Zielinski, D.C.; Altunkaya, A.; Dräger, A.; Mih, N.; Gatto, F.; Nilsson, A.; Preciat Gonzalez, G.A.; Aurich, M.K.; et al. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat. Biotechnol. 2018, 36, 272–281. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Fiehn, O.; Robertson, D.; Griffin, J.; Van Der Werf, M.; Nikolau, B.; Morrison, N.; Sumner, L.W.; Goodacre, R.; Hardy, N.W.; Taylor, C.; et al. The metabolomics standards initiative (MSI). Metabolomics 2007, 3, 175–178. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Sud, M.; Fahy, E.; Cotter, D.; Azam, K.; Vadivelu, I.; Burant, C.; Edison, A.; Fiehn, O.; Higashi, R.; Nair, K.S.; et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2015, 44, D463–D470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, H.; Chang, D.; Guo, F.; Pan, H.; Yang, Y. Metabolomics approach by 1H NMR spectroscopy of serum reveals progression axes for asymptomatic hyperuricemia and gout. Arthritis Res. Ther. 2018, 20, 111. [Google Scholar] [CrossRef]
- Zhong, Z.; Huang, Y.; Huang, Q.; Zheng, S.; Huang, Z.; Deng, W.; Li, T. Serum metabolic profiling analysis of gout patients based on UPLC-Q-TOF/MS. Clin. Chim. Acta 2021, 515, 52–60. [Google Scholar] [CrossRef]
- Huang, Y.; Xiao, M.; Ou, J.; Lv, Q.; Wei, Q.; Chen, Z.; Wu, J.; Tu, L.; Jiang, Y.; Zhang, X.; et al. Identification of the urine and serum metabolomics signature of gout. Rheumatology 2020, 59, 2960–2969. [Google Scholar] [CrossRef] [PubMed]
- Lyu, S.; Rao, Y.; Liu, P.; Yang, S.; Chen, W.; Yang, H.; Ke, S.; OuYang, H.; He, M.; Feng, Y. Metabolomics analysis reveals four biomarkers associated with the gouty arthritis progression in patients with sequential stages. Semin. Arthritis Rheum. 2022, 55, 152022. [Google Scholar] [CrossRef]
- Li, Q.; Wei, S.; Wu, D.; Wen, C.; Zhou, J. Urinary Metabolomics Study of Patients with Gout Using Gas Chromatography-Mass Spectrometry. Biomed. Res. Int. 2018, 2018, 3461572. [Google Scholar] [CrossRef] [PubMed]
- Shao, T.; Shao, L.; Li, H.; Xie, Z.; He, Z.; Wen, C. Combined Signature of the Fecal Microbiome and Metabolome in Patients with Gout. Front. Microbiol. 2017, 8, 268. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Y.; Liu, H.; Xu, T.; Wang, M.-J.; Lu, J.; Guo, Y.; Chen, W.; Ke, M.; Zhou, G.; et al. Serum lipidomics reveals distinct metabolic profiles for asymptomatic hyperuricemic and gout patients. Rheumatology 2021, 61, 2644–2651. [Google Scholar] [CrossRef]
- Cui, L.; Liu, J.; Yan, X.; Hu, S. Identification of Metabolite Biomarkers for Gout Using Capillary Ion Chromatography with Mass Spectrometry. Anal. Chem. 2017, 89, 11737–11743. [Google Scholar] [CrossRef] [PubMed]
- Qin, N.; Qin, M.; Shi, W.; Kong, L.; Wang, L.; Xu, G.; Guo, Y.; Zhang, J.; Ma, Q. Investigation of pathogenesis of hyperuricemia based on untargeted and targeted metabolomics. Sci. Rep. 2022, 12, 13980. [Google Scholar] [CrossRef] [PubMed]
- Suhre, K.; Shin, S.-Y.; Petersen, A.-K.; Mohney, R.P.; Meredith, D.; Wägele, B.; Altmaier, E.; Deloukas, P.; Erdmann, J.; Grundberg, E.; et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature 2011, 477, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Döring, A.; Gieger, C.; Mehta, D.; Gohlke, H.; Prokisch, H.; Coassin, S.; Fischer, G.; Henke, K.; Klopp, N.; Kronenberg, F.; et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat. Genet. 2008, 40, 430–436. [Google Scholar] [CrossRef]
- Sulem, P.; Gudbjartsson, D.F.; Walters, G.B.; Helgadottir, H.T.; Helgason, A.; Gudjonsson, S.A.; Zanon, C.; Besenbacher, S.; Bjornsdottir, G.; Magnusson, O.T.; et al. Identification of low-frequency variants associated with gout and serum uric acid levels. Nat. Genet. 2011, 43, 1127–1130. [Google Scholar] [CrossRef]
- Vasiliou, V.; Sandoval, M.; Backos, D.S.; Jackson, B.C.; Chen, Y.; Reigan, P.; Lanaspa, M.A.; Johnson, R.J.; Koppaka, V.; Thompson, D.C. ALDH16A1 is a novel non-catalytic enzyme that may be involved in the etiology of gout via protein–protein interactions with HPRT1. Chem.-Biol. Interact. 2013, 202, 22–31. [Google Scholar] [CrossRef]
- Charkoftaki, G.; Chen, Y.; Han, M.; Sandoval, M.; Yu, X.; Zhao, H.; Orlicky, D.J.; Thompson, D.C.; Vasiliou, V. Transcriptomic analysis and plasma metabolomics in Aldh16a1 -null mice reveals a potential role of ALDH16A1 in renal function. Chem.-Biol. Interact. 2017, 276, 15–22. [Google Scholar] [CrossRef]
- Crane, J.K. Role of host xanthine oxidase in infection due to enteropathogenic and Shiga-toxigenicEscherichia coli. Gut Microbes 2013, 4, 388–391. [Google Scholar] [CrossRef]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions Between the Microbiota and the Immune System. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef]
- Ichida, K.; Matsuo, H.; Takada, T.; Nakayama, A.; Murakami, K.; Shimizu, T.; Yamanashi, Y.; Kasuga, H.; Nakashima, H.; Nakamura, T.; et al. Decreased extra-renal urate excretion is a common cause of hyperuricemia. Nat. Commun. 2012, 3, 764. [Google Scholar] [CrossRef]
- Méndez-Salazar, E.O.; Martínez-Nava, G.A. Uric acid extrarenal excretion: The gut microbiome as an evident yet understated factor in gout development. Rheumatol. Int. 2022, 42, 403–412. [Google Scholar] [CrossRef]
- Joshi, A.; McCormick, N.; Yokose, C.; Lu, N.; Choi, H. OP0164 A Population-Based, Prospective Metabolomics Study in the UK Biobank Identifies Glycoprotein Acetyls as a Novel Biomarker of Incident Gout. Ann. Rheum. Dis. 2022, 81 (Suppl. S1), 108. [Google Scholar] [CrossRef]
- Lu, J.; Dalbeth, N.; Yin, H.; Li, C.; Merriman, T.R.; Wei, W.H. Mouse models for human hyperuricaemia: A critical review. Nat. Rev. Rheumatol. 2019, 15, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Lyu, S.; Ding, R.; Liu, P.; Ouyang, H.; Feng, Y.; Rao, Y.; Yang, S. LC-MS Analysis of Serum for the Metabolomic Investigation of the Effects of Pulchinenoside b4 Administration in Monosodium Urate Crystal-Induced Gouty Arthritis Rat Model. Molecules 2019, 24, 3161. [Google Scholar] [CrossRef]
- Shan, B.; Chen, T.; Huang, B.; Liu, Y.; Chen, J. Untargeted metabolomics reveal the therapeutic effects of Ermiao wan categorized formulas on rats with hyperuricemia. J. Ethnopharmacol. 2021, 281, 114545. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-M.; Wang, P.; Teka, T.; Zhang, Y.-C.; Yang, W.-Z.; Zhang, Y.; Wang, T.; Liu, L.-X.; Han, L.-F.; Liu, C.-X. 1H NMR and UHPLC/Q-Orbitrap-MS-Based Metabolomics Combined with 16S rRNA Gut Microbiota Analysis Revealed the Potential Regulation Mechanism of Nuciferine in Hyperuricemia Rats. J. Agric. Food Chem. 2020, 68, 14059–14070. [Google Scholar] [CrossRef]
- Zhao, L.; Zhao, T.; Yang, X.; Cao, L.; Xu, R.; Liu, J.; Lin, C.; Yu, Y.; Xuan, D.; Zhu, X.; et al. IL-37 blocks gouty inflammation by shaping macrophages into a non-inflammatory phagocytic phenotype. Rheumatology 2022, 61, 3841–3853. [Google Scholar] [CrossRef] [PubMed]
- Tani, T.; Okamoto, K.; Fujiwara, M.; Katayama, A.; Tsuruoka, S. Metabolomics analysis elucidates unique influences on purine/pyrimidine metabolism by xanthine oxidoreductase inhibitors in a rat model of renal ischemia-reperfusion injury. Mol. Med. 2019, 25, 40. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Liang, N.; Tao, Y.; Yin, H. Metabolomics in Hyperuricemia and Gout. Gout Urate Cryst. Depos. Dis. 2023, 1, 49-61. https://doi.org/10.3390/gucdd1010006
Li R, Liang N, Tao Y, Yin H. Metabolomics in Hyperuricemia and Gout. Gout, Urate, and Crystal Deposition Disease. 2023; 1(1):49-61. https://doi.org/10.3390/gucdd1010006
Chicago/Turabian StyleLi, Rui, Ningning Liang, Yongzhen Tao, and Huiyong Yin. 2023. "Metabolomics in Hyperuricemia and Gout" Gout, Urate, and Crystal Deposition Disease 1, no. 1: 49-61. https://doi.org/10.3390/gucdd1010006
APA StyleLi, R., Liang, N., Tao, Y., & Yin, H. (2023). Metabolomics in Hyperuricemia and Gout. Gout, Urate, and Crystal Deposition Disease, 1(1), 49-61. https://doi.org/10.3390/gucdd1010006






