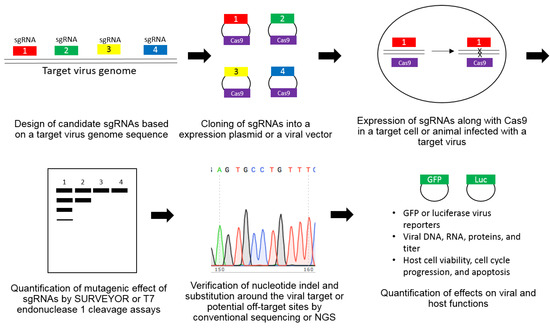
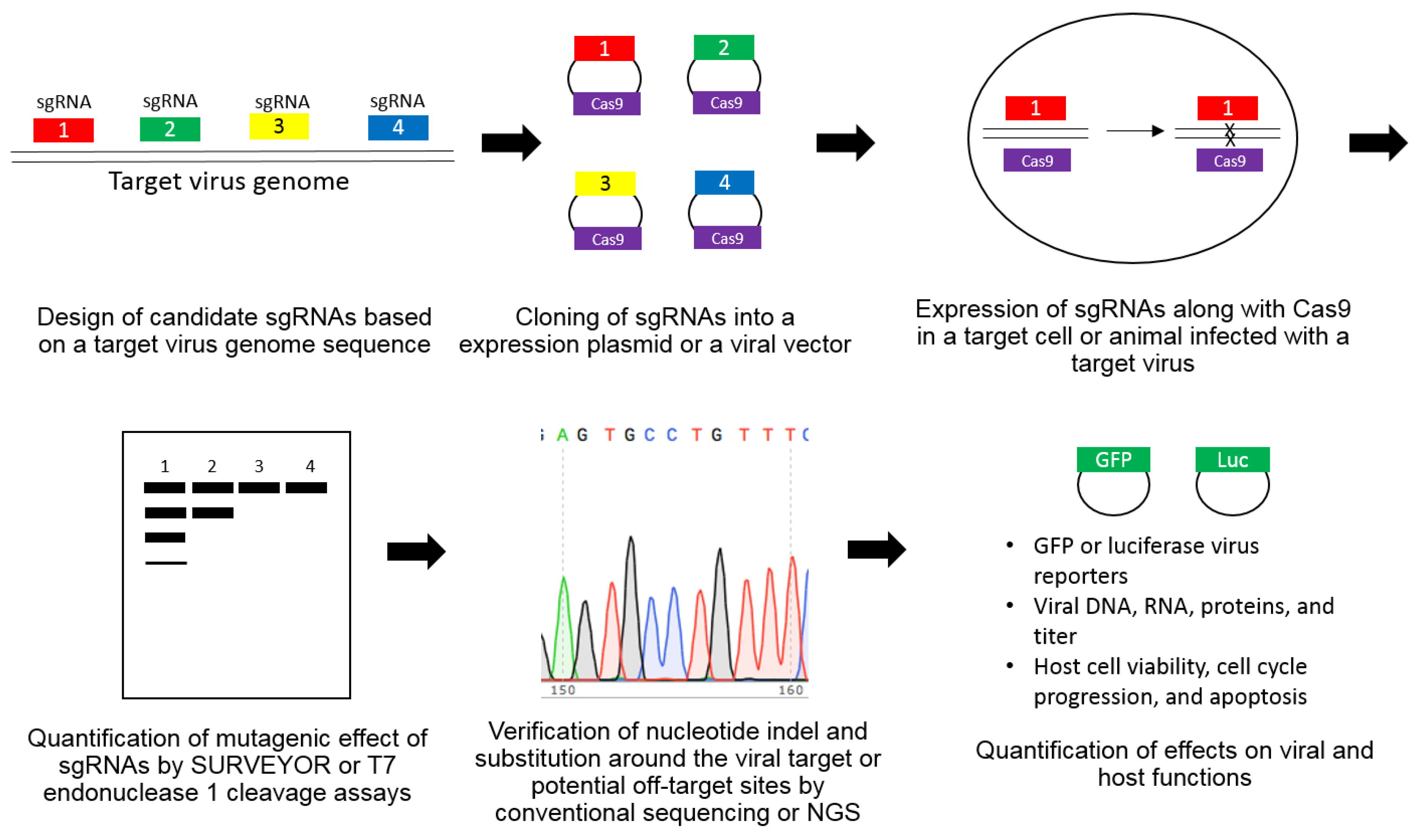
Abstract
From its unexpected discovery as a bacterial adaptive immune system to its countless applications as one of the most versatile gene-editing tools, the CRISPR/Cas9 system has revolutionized every field of life science. Virology is no exception to this ever-growing list of CRISPR/Cas9-based applications. Direct manipulation of a virus genome by CRISPR/Cas9 has enabled a systematic study of cis-elements and trans-elements encoded in a virus genome. In addition, this virus genome-specific mutagenesis by CRISPR/Cas9 was further funneled into the development of a novel class of antiviral therapy targeting many incurable chronic viral infections. In this review, a general concept on the CRISPR/Cas9-based antiviral strategy will be described first. To understand the current status of the CRISPR/Cas9-based antiviral approach, a series of recently published antiviral studies involving CRISPR/Cas9-mediated control of several clinically-relevant viruses including human immunodeficiency virus, hepatitis B virus, herpesviruses, human papillomavirus, and other viruses will be presented. Lastly, the potential challenge and future prospect for successful clinical translation of this CRISPR/Cas9-based antiviral method will be discussed.
1. Introduction
Along with a proteinaceous structural component, a nucleic acid is an essential building block for assembly of an infectious virus particle. Therefore, efficient viral genome replication inside a host cell is one of the most important tasks for the successful completion of a virus life cycle. In theory, the ablation of viral genetic elements has been regarded as one of the most ideal antiviral strategies. However, the lack of a virus gene-specific destruction method has been a big hurdle for the realization of this virus genome-targeting antiviral strategy. Recently, a variety of sequence-specific endonucleases have been introduced and tested for their therapeutic potentials for direct manipulation of a viral genome in preclinical studies. They include zinc finger nucleases, transcription activator-like effectors nucleases (TALENS), and clustered regulatory interspaced short palindromic repeat (CRISPR)-associated (Cas) nucleases [1,2]. Among them, the CRISPR/Cas system has been one of the most preferred choices for various antiviral applications due to its relative versatility, specificity, and ease of use [3,4,5,6,7,8]. Originally, CRISPR/Cas was discovered as one of the bacterial adaptive immune systems for defense against a foreign nucleic acid attack such as a phage infection and an exogenous plasmid uptake [9,10]. In order to achieve precise and specific digestion of these potentially harmful genetic elements from intruders, the CRISPR/Cas system was evolved to employ a foreign DNA-derived RNA as a guide molecule for sequence-specific destruction of target viral DNAs [11]. This RNA-directed sequence specificity of the CRISPR/Cas system has enabled a powerful and versatile genetic manipulation of genomes from diverse eukaryotic as well as prokaryotic organisms. Many successful applications to several human viral pathogens in cell-based and animal studies were highly encouraging, which is enough to hope for their accelerated translation in the clinical setting [3,4,5,6,7,8]. However, increasing concerns regarding the safety of CRISPR/Cas system due to its potential off-target activity and emergence of CRISPR/Cas-resistant escape mutant viruses along with the difficulty in its efficient delivery to every single virus-infected cell still seems to be a daunting task for full fruition of this promising antiviral approach [12,13]. In this review, the general concept regarding the design and efficacy validation method for CRISPR/Cas-based antiviral strategy will first be introduced and reviewed. Then, the current status of the CRISPR/Cas-based antiviral approach to control major pathogenic human viruses including human immunodeficiency virus (HIV), hepatitis B virus (HBV), herpes viruses, human papillomavirus (HPV), and other viruses will be summarized next. Lastly, this review will be concluded with thoughts regarding a potential challenge for the realization of CRISPR/Cas-based therapy and prospect for CRISPR/Cas-based antiviral strategy in the future.
5. Conclusions
Tight linkage of a viral life cycle to a host cellular metabolism has made clearance of this invader out of our body a therapeutic challenge. Pharmacological intervention of viral infection by using a small molecule inhibitor against a virus-specific enzyme has been one of the best antiviral options so far. However, this conventional antiviral approach has been inappropriate for the control of most of the latency-associated chronic viral infections. In this regard, the introduction of the CRISPR/Cas technology with an unprecedented capability for direct targeting of a viral genome would contribute to the buildup of a new antiviral armamentarium aiming for a previously unthinkable, complete cure. In spite of many challenges ahead that need to be resolved for the full transition of this CRISPR/Cas technology from a preclinical study to a practical antiviral therapy, complete curative potential of the CRISPR/Cas-based antiviral strategy should provide continuous motivation for the development of novel antiviral therapeutics. Without a doubt, this new antiviral approach should help chronically infected patients stop taking a life-long medication in the near future.
Author Contributions
C.L. collected all the necessary references and wrote the paper.
Funding
The Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education [No. 2016R1D1A1B03933100] supported this research.
Conflicts of Interest
The author declares that there are no competing interest regarding the publication of this paper.
References
- Carr, A. Toxicity of antiretroviral therapy and implications for drug development. Nat. Rev. Drug Discov. 2003, 2, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Bella, R.; Kaminski, R.; Mancuso, P.; Young, W.B.; Chen, C.; Sariyer, R.; Fischer, T.; Amini, S.; Ferrante, P.; Jacobson, J.M.; et al. Removal of HIV DNA by CRISPR from patient blood engrafts in humanized mice. Mol. Ther. Nucleic Acids 2018, 12, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Ebina, H.; Misawa, N.; Kanemura, Y.; Koyanagi, Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013, 3, 2510. [Google Scholar] [CrossRef]
- Hu, W.; Kaminski, R.; Yang, F.; Zhang, Y.; Cosentino, L.; Li, F.; Luo, B.; Alvarez-Carbonell, D.; Garcia-Mesa, Y.; Karn, J.; et al. Rna-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc. Natl. Acad. Sci. USA 2014, 111, 11461–11466. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, R.; Bella, R.; Yin, C.; Otte, J.; Ferrante, P.; Gendelman, H.E.; Li, H.; Booze, R.; Gordon, J.; Hu, W.; et al. Excision of HIV-1 DNA by gene editing: A proof-of-concept in vivo study. Gene Ther. 2016, 23, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, R.; Chen, Y.; Fischer, T.; Tedaldi, E.; Napoli, A.; Zhang, Y.; Karn, J.; Hu, W.; Khalili, K. Elimination of HIV-1 genomes from human t-lymphoid cells by CRISPR/Cas9 gene editing. Sci. Rep. 2016, 6, 22555. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, R.; Chen, Y.; Salkind, J.; Bella, R.; Young, W.B.; Ferrante, P.; Karn, J.; Malcolm, T.; Hu, W.; Khalili, K. Negative feedback regulation of HIV-1 by gene editing strategy. Sci. Rep. 2016, 6, 31527. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.K.; Gu, Y.; Diaz, A.; Marlett, J.; Takahashi, Y.; Li, M.; Suzuki, K.; Xu, R.; Hishida, T.; Chang, C.J.; et al. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat. Commun. 2015, 6, 6413. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, S.; Liu, Z.; Ke, Z.; Li, C.; Yu, X.; Chen, S.; Guo, D. Genome-scale screening identification of saCas9/grnas for targeting HIV-1 provirus and suppression of HIV-1 infection. Virus Res. 2018, 250, 21–30. [Google Scholar] [CrossRef]
- Yin, C.; Zhang, T.; Li, F.; Yang, F.; Putatunda, R.; Young, W.B.; Khalili, K.; Hu, W.; Zhang, Y. Functional screening of guide RNAs targeting the regulatory and structural HIV-1 viral genome for a cure of aids. AIDS 2016, 30, 1163–1174. [Google Scholar] [CrossRef]
- Yin, C.; Zhang, T.; Qu, X.; Zhang, Y.; Putatunda, R.; Xiao, X.; Li, F.; Xiao, W.; Zhao, H.; Dai, S.; et al. In vivo excision of HIV-1 provirus by saCas9 and multiplex single-guide RNAs in animal models. Mol. Ther. 2017, 25, 1168–1186. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Lei, R.; Le Duff, Y.; Li, J.; Guo, F.; Wainberg, M.A.; Liang, C. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology 2015, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; Kim, S.; Kim, J.M.; Kim, J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; Kim, S.; Kim, Y.; Kweon, J.; Kim, H.S.; Bae, S.; Kim, J.S. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014, 24, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Cradick, T.J.; Fine, E.J.; Antico, C.J.; Bao, G. CRISPR/Cas9 systems targeting beta-globin and ccr5 genes have substantial off-target activity. Nucleic Acids Res. 2013, 41, 9584–9592. [Google Scholar] [CrossRef]
- Hou, P.; Chen, S.; Wang, S.; Yu, X.; Chen, Y.; Jiang, M.; Zhuang, K.; Ho, W.; Hou, W.; Huang, J.; et al. Genome editing of cxcr4 by CRISPR/Cas9 confers cells resistant to HIV-1 infection. Sci. Rep. 2015, 5, 15577. [Google Scholar] [CrossRef] [PubMed]
- Hultquist, J.F.; Schumann, K.; Woo, J.M.; Manganaro, L.; McGregor, M.J.; Doudna, J.; Simon, V.; Krogan, N.J.; Marson, A. A Cas9 ribonucleoprotein platform for functional genetic studies of HIV-host interactions in primary human t cells. Cell Rep. 2016, 17, 1438–1452. [Google Scholar] [CrossRef]
- Li, C.; Guan, X.; Du, T.; Jin, W.; Wu, B.; Liu, Y.; Wang, P.; Hu, B.; Griffin, G.E.; Shattock, R.J.; et al. Inhibition of HIV-1 infection of primary cd4+ t-cells by gene editing of ccr5 using adenovirus-delivered CRISPR/Cas9. J. Gen. Virol. 2015, 96, 2381–2393. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, Q.; Yu, X.; Li, Y.; Guo, Y.; Liu, Z.; Sun, F.; Hou, W.; Li, C.; Wu, L.; et al. Hiv-1 inhibition in cells with cxcr4 mutant genome created by CRISPR-Cas9 and piggybac recombinant technologies. Sci. Rep. 2018, 8, 8573. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, S.; Jin, X.; Wang, Q.; Yang, K.; Li, C.; Xiao, Q.; Hou, P.; Liu, S.; Wu, S.; et al. Genome editing of the HIV co-receptors ccr5 and cxcr4 by CRISPR-Cas9 protects cd4(+) t cells from HIV-1 infection. Cell Biosci. 2017, 7, 47. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, S.; Xiao, Q.; Liu, Z.; Liu, S.; Hou, P.; Zhou, L.; Hou, W.; Ho, W.; Li, C.; et al. Genome modification of cxcr4 by Staphylococcus aureus Cas9 renders cells resistance to HIV-1 infection. Retrovirology 2017, 14, 51. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ye, C.; Liu, J.; Zhang, D.; Kimata, J.T.; Zhou, P. Ccr5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS ONE 2014, 9, e115987. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yang, H.; Gao, Y.; Chen, Z.; Xie, L.; Liu, Y.; Liu, Y.; Wang, X.; Li, H.; Lai, W.; et al. CRISPR/Cas9-mediated ccr5 ablation in human hematopoietic stem/progenitor cells confers HIV-1 resistance in vivo. Mol. Ther. 2017, 25, 1782–1789. [Google Scholar] [CrossRef]
- Ye, L.; Wang, J.; Beyer, A.I.; Teque, F.; Cradick, T.J.; Qi, Z.; Chang, J.C.; Bao, G.; Muench, M.O.; Yu, J.; et al. Seamless modification of wild-type induced pluripotent stem cells to the natural ccr5delta32 mutation confers resistance to HIV infection. Proc. Natl. Acad. Sci. USA 2014, 111, 9591–9596. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Yao, Y.; Xiao, H.; Li, J.; Liu, Q.; Yang, Y.; Adah, D.; Lu, J.; Zhao, S.; Qin, L.; et al. Simultaneous knockout of cxcr4 and ccr5 genes in CD4+ T cells via CRISPR/Cas9 confers resistance to both x4- and r5-tropic human immunodeficiency virus type 1 infection. Hum. Gene Ther. 2018, 29, 51–67. [Google Scholar] [CrossRef]
- Khalili, K.; Kaminski, R.; Gordon, J.; Cosentino, L.; Hu, W. Genome editing strategies: Potential tools for eradicating HIV-1/aids. J. Neurovirol. 2015, 21, 310–321. [Google Scholar] [CrossRef]
- Darcis, G.; Das, A.T.; Berkhout, B. Tackling HIV persistence: Pharmacological versus CRISPR-based shock strategies. Viruses 2018, 10, 157. [Google Scholar] [CrossRef]
- Bialek, J.K.; Dunay, G.A.; Voges, M.; Schafer, C.; Spohn, M.; Stucka, R.; Hauber, J.; Lange, U.C. Targeted HIV-1 latency reversal using CRISPR/Cas9-derived transcriptional activator systems. PLoS ONE 2016, 11, e0158294. [Google Scholar] [CrossRef] [PubMed]
- Limsirichai, P.; Gaj, T.; Schaffer, D.V. CRISPR-mediated activation of latent HIV-1 expression. Mol. Ther. 2016, 24, 499–507. [Google Scholar] [CrossRef]
- Zhang, Y.; Yin, C.; Zhang, T.; Li, F.; Yang, W.; Kaminski, R.; Fagan, P.R.; Putatunda, R.; Young, W.B.; Khalili, K.; et al. CRISPR/gRNA-directed synergistic activation mediator (sam) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci. Rep. 2015, 5, 16277. [Google Scholar] [CrossRef]
- Ji, H.; Jiang, Z.; Lu, P.; Ma, L.; Li, C.; Pan, H.; Fu, Z.; Qu, X.; Wang, P.; Deng, J.; et al. Specific reactivation of latent HIV-1 by dCas9-Suntag-vp64-mediated guide RNA targeting the HIV-1 promoter. Mol. Ther. 2016, 24, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Saayman, S.M.; Lazar, D.C.; Scott, T.A.; Hart, J.R.; Takahashi, M.; Burnett, J.C.; Planelles, V.; Morris, K.V.; Weinberg, M.S. Potent and targeted activation of latent HIV-1 using the CRISPR/dCas9 activator complex. Mol. Ther. 2016, 24, 488–498. [Google Scholar] [CrossRef]
- Bogerd, H.P.; Kornepati, A.V.; Marshall, J.B.; Kennedy, E.M.; Cullen, B.R. Specific induction of endogenous viral restriction factors using CRISPR/Cas-derived transcriptional activators. Proc. Natl. Acad. Sci. USA 2015, 112, E7249–E7256. [Google Scholar] [CrossRef] [PubMed]
- Dufour, C.; Claudel, A.; Joubarne, N.; Merindol, N.; Maisonnet, T.; Masroori, N.; Plourde, M.B.; Berthoux, L. Editing of the human trim5 gene to introduce mutations with the potential to inhibit HIV-1. PLoS ONE 2018, 13, e0191709. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Luo, M.; Yu, T.; Chen, L.; Huang, Q.; Chen, S.; Xie, L.; Zeng, Y.; Luo, F.; Xiong, H.; et al. CRISPR/Cas9-mediated deletion of mir-146a enhances antiviral response in HIV-1 infected cells. Genes Immun. 2018. [Google Scholar] [CrossRef]
- Ueda, S.; Ebina, H.; Kanemura, Y.; Misawa, N.; Koyanagi, Y. Anti-HIV-1 potency of the CRISPR/Cas9 system insufficient to fully inhibit viral replication. Microbiol. Immunol. 2016, 60, 483–496. [Google Scholar] [CrossRef]
- Wang, G.; Zhao, N.; Berkhout, B.; Das, A.T. CRISPR-Cas9 can inhibit HIV-1 replication but NHEJ repair facilitates virus escape. Mol. Ther. 2016, 24, 522–526. [Google Scholar] [CrossRef]
- Wang, G.; Zhao, N.; Berkhout, B.; Das, A.T. A combinatorial CRISPR-Cas9 attack on HIV-1 DNA extinguishes all infectious provirus in infected t cell cultures. Cell Rep. 2016, 17, 2819–2826. [Google Scholar] [CrossRef]
- Wang, Z.; Pan, Q.; Gendron, P.; Zhu, W.; Guo, F.; Cen, S.; Wainberg, M.A.; Liang, C. CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep. 2016, 15, 481–489. [Google Scholar] [CrossRef]
- Yoder, K.E.; Bundschuh, R. Host double-strand break repair generates HIV-1 strains resistant to CRISPR/Cas9. Sci. Rep. 2016, 6, 29530. [Google Scholar] [CrossRef]
- Lebbink, R.J.; de Jong, D.C.; Wolters, F.; Kruse, E.M.; van Ham, P.M.; Wiertz, E.J.; Nijhuis, M. A combinational CRISPR/cas9 gene-editing approach can halt HIV replication and prevent viral escape. Sci. Rep. 2017, 7, 41968. [Google Scholar] [CrossRef] [PubMed]
- Mefferd, A.L.; Bogerd, H.P.; Irwan, I.D.; Cullen, B.R. Insights into the mechanisms underlying the inactivation of HIV-1 proviruses by CRISPR/Cas. Virology 2018, 520, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Cullen, B.R. Role and mechanism of action of the apobec3 family of antiretroviral resistance factors. J. Virol. 2006, 80, 1067–1076. [Google Scholar] [CrossRef] [PubMed]
- Doehle, B.P.; Schafer, A.; Cullen, B.R. Human apobec3b is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 vif. Virology 2005, 339, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, X.; Stremlau, M.; Lee, M.; Sodroski, J. Removal of arginine 332 allows human trim5alpha to bind human immunodeficiency virus capsids and to restrict infection. J. Virol. 2006, 80, 6738–6744. [Google Scholar] [CrossRef] [PubMed]
- Sawyer, S.L.; Wu, L.I.; Emerman, M.; Malik, H.S. Positive selection of primate trim5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 2005, 102, 2832–2837. [Google Scholar] [CrossRef]
- Jung, U.; Urak, K.; Veillette, M.; Nepveu-Traversy, M.E.; Pham, Q.T.; Hamel, S.; Rossi, J.J.; Berthoux, L. Preclinical assessment of mutant human trim5alpha as an anti-HIV-1 transgene. Hum. Gene Ther. 2015, 26, 664–679. [Google Scholar] [CrossRef]
- Lee, W.M. Hepatitis b virus infection. N. Engl. J. Med. 1997, 337, 1733–1745. [Google Scholar] [CrossRef]
- Ott, J.J.; Stevens, G.A.; Groeger, J.; Wiersma, S.T. Global epidemiology of hepatitis b virus infection: New estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine 2012, 30, 2212–2219. [Google Scholar] [CrossRef]
- Block, T.M.; Rawat, S.; Brosgart, C.L. Chronic hepatitis b: A wave of new therapies on the horizon. Antiviral Res. 2015, 121, 69–81. [Google Scholar] [CrossRef]
- Shlomai, A.; Rice, C.M. Virology. Getting rid of a persistent troublemaker to cure hepatitis. Science 2014, 343, 1212–1213. [Google Scholar] [CrossRef] [PubMed]
- Schinazi, R.F.; Ehteshami, M.; Bassit, L.; Asselah, T. Towards HBV curative therapies. Liver Int. 2018, 38 (Suppl. 1), 102–114. [Google Scholar] [CrossRef]
- Bloom, K.; Maepa, M.B.; Ely, A.; Arbuthnot, P. Gene therapy for chronic HBV-can we eliminate cccDNA? Genes (Basel) 2018, 9, 207. [Google Scholar] [CrossRef] [PubMed]
- Pankowicz, F.P.; Jarrett, K.E.; Lagor, W.R.; Bissig, K.D. CRISPR/Cas9: At the cutting edge of hepatology. Gut 2017, 66, 1329–1340. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Lu, M.; Yang, D. CRISPR/Cas9-based tools for targeted genome editing and replication control of HBV. Virol. Sin. 2015, 30, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.; Zhang, K.; Li, J. Application of CRISPR/Cas9 technology to HBV. Int. J. Mol. Sci. 2015, 16, 26077–26086. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.C.; Chen, P.J. The potential and challenges of CRISPR-Cas in eradication of hepatitis b virus covalently closed circular DNA. Virus Res. 2018, 244, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Moyo, B.; Bloom, K.; Scott, T.; Ely, A.; Arbuthnot, P. Advances with using CRISPR/Cas-mediated gene editing to treat infections with hepatitis b virus and hepatitis c virus. Virus Res. 2018, 244, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Ely, A.; Moyo, B.; Arbuthnot, P. Progress with developing use of gene editing to cure chronic infection with hepatitis b virus. Mol. Ther. 2016, 24, 671–677. [Google Scholar] [CrossRef]
- Kennedy, E.M.; Kornepati, A.V.; Cullen, B.R. Targeting hepatitis b virus cccDNA using CRISPR/Cas9. Antiviral Res. 2015, 123, 188–192. [Google Scholar] [CrossRef]
- Lin, S.R.; Yang, H.C.; Kuo, Y.T.; Liu, C.J.; Yang, T.Y.; Sung, K.C.; Lin, Y.Y.; Wang, H.Y.; Wang, C.C.; Shen, Y.C.; et al. The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo. Mol. Ther. Nucleic Acids 2014, 3, e186. [Google Scholar] [CrossRef] [PubMed]
- Seeger, C.; Sohn, J.A. Targeting hepatitis b virus with CRISPR/Cas9. Mol. Ther. Nucleic Acids 2014, 3, e216. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Qu, L.; Wang, H.; Wei, L.; Dong, Y.; Xiong, S. Targeting hepatitis b virus cccDNA by CRISPR/Cas9 nuclease efficiently inhibits viral replication. Antiviral Res. 2015, 118, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Karimova, M.; Beschorner, N.; Dammermann, W.; Chemnitz, J.; Indenbirken, D.; Bockmann, J.H.; Grundhoff, A.; Luth, S.; Buchholz, F.; Schulze zur Wiesch, J.; et al. CRISPR/Cas9 nickase-mediated disruption of hepatitis b virus open reading frame S and X. Sci. Rep. 2015, 5, 13734. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, E.M.; Bassit, L.C.; Mueller, H.; Kornepati, A.V.R.; Bogerd, H.P.; Nie, T.; Chatterjee, P.; Javanbakht, H.; Schinazi, R.F.; Cullen, B.R. Suppression of hepatitis b virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology 2015, 476, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hao, R.; Chen, S.; Guo, D.; Chen, Y. Inhibition of hepatitis b virus by the CRISPR/Cas9 system via targeting the conserved regions of the viral genome. J. Gen. Virol. 2015, 96, 2252–2261. [Google Scholar] [CrossRef] [PubMed]
- Ramanan, V.; Shlomai, A.; Cox, D.B.; Schwartz, R.E.; Michailidis, E.; Bhatta, A.; Scott, D.A.; Zhang, F.; Rice, C.M.; Bhatia, S.N. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis b virus. Sci. Rep. 2015, 5, 10833. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, Z.W.; Liu, S.; Zhang, R.Y.; Ding, S.L.; Xie, X.M.; Long, L.; Chen, X.M.; Zhuang, H.; Lu, F.M. Dual gRNAs guided CRISPR/Cas9 system inhibits hepatitis b virus replication. World J. Gastroenterol. 2015, 21, 9554–9565. [Google Scholar] [CrossRef]
- Zhen, S.; Hua, L.; Liu, Y.H.; Gao, L.C.; Fu, J.; Wan, D.Y.; Dong, L.H.; Song, H.F.; Gao, X. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/crispr-associated Cas9 system to disrupt the hepatitis b virus. Gene Ther. 2015, 22, 404–412. [Google Scholar] [CrossRef]
- Sakuma, T.; Masaki, K.; Abe-Chayama, H.; Mochida, K.; Yamamoto, T.; Chayama, K. Highly multiplexed CRISPR-Cas9-nuclease and Cas9-nickase vectors for inactivation of hepatitis b virus. Genes Cells 2016, 21, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Seeger, C.; Sohn, J.A. Complete spectrum of CRISPR/Cas9-induced mutations on HBV cccDNA. Mol. Ther. 2016, 24, 1258–1266. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Xie, K.; Xu, Y.; Wang, L.; Chen, K.; Zhang, L.; Fang, J. CRISPR/Cas9 produces anti-hepatitis b virus effect in hepatoma cells and transgenic mouse. Virus Res. 2016, 217, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Mei, M.; Li, B.; Zhu, X.; Zu, W.; Tian, Y.; Wang, Q.; Guo, Y.; Dong, Y.; Tan, X. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 2017, 27, 440–443. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sheng, C.; Wang, S.; Yang, L.; Liang, Y.; Huang, Y.; Liu, H.; Li, P.; Yang, C.; Yang, X.; et al. Removal of integrated hepatitis b virus DNA using CRISPR-Cas9. Front. Cell. Infect. Microbiol. 2017, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Scott, T.; Moyo, B.; Nicholson, S.; Maepa, M.B.; Watashi, K.; Ely, A.; Weinberg, M.S.; Arbuthnot, P. Ssaavs containing cassettes encoding saCas9 and guides targeting hepatitis b virus inactivate replication of the virus in cultured cells. Sci. Rep. 2017, 7, 7401. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, R.; Zhang, R.; Ding, S.; Zhang, T.; Yuan, Q.; Guan, G.; Chen, X.; Zhang, T.; Zhuang, H.; et al. The gRNA-miRNA-gRNA ternary cassette combining CRISPR/Cas9 with RNAi approach strongly inhibits hepatitis b virus replication. Theranostics 2017, 7, 3090–3105. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, M.; Gong, M.; Xu, Y.; Xie, C.; Deng, H.; Li, X.; Wu, H.; Wang, Z. Inhibition of hepatitis b virus replication via HBV DNA cleavage by Cas9 from Staphylococcus aureus. Antiviral Res. 2018, 152, 58–67. [Google Scholar] [CrossRef]
- Schiwon, M.; Ehrke-Schulz, E.; Oswald, A.; Bergmann, T.; Michler, T.; Protzer, U.; Ehrhardt, A. One-vector system for multiplexed CRISPR/Cas9 against hepatitis b virus cccDNA utilizing high-capacity adenoviral vectors. Mol. Ther. Nucleic Acids 2018, 12, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhang, X.; Ge, Q.; Yuan, C.; Chu, L.; Liang, H.F.; Liao, Z.; Liu, Q.; Zhang, Z.; Zhang, B. CRISPR/Cas9-mediated knockout of HBsAg inhibits proliferation and tumorigenicity of HBV-positive hepatocellular carcinoma cells. J. Cell. Biochem. 2018, 119, 8419–8431. [Google Scholar] [CrossRef]
- Mali, P.; Aach, J.; Stranges, P.B.; Esvelt, K.M.; Moosburner, M.; Kosuri, S.; Yang, L.; Church, G.M. Cas9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013, 31, 833–838. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Schillinger, J.A.; Sternberg, M.R.; Johnson, R.E.; Lee, F.K.; Nahmias, A.J.; Markowitz, L.E. Seroprevalence and coinfection with herpes simplex virus type 1 and type 2 in the united states, 1988-1994. J. Infect. Dis. 2002, 185, 1019–1024. [Google Scholar] [CrossRef] [PubMed]
- Griffin, B.D.; Verweij, M.C.; Wiertz, E.J. Herpesviruses and immunity: The art of evasion. Vet. Microbiol. 2010, 143, 89–100. [Google Scholar] [CrossRef] [PubMed]
- van Diemen, F.R.; Lebbink, R.J. CRISPR/Cas9, a powerful tool to target human herpesviruses. Cell. Microbiol. 2017, 19. [Google Scholar] [CrossRef] [PubMed]
- Coen, D.M.; Schaffer, P.A. Antiherpesvirus drugs: A promising spectrum of new drugs and drug targets. Nat. Rev. Drug Discov. 2003, 2, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Westerberg, B.D.; Atashband, S.; Kozak, F.K. A systematic review of the incidence of sensorineural hearing loss in neonates exposed to herpes simplex virus (HSV). Int. J. Pediatr. Otorhinolaryngol. 2008, 72, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Whitley, R.J.; Roizman, B. Herpes simplex virus infections. Lancet 2001, 357, 1513–1518. [Google Scholar] [CrossRef]
- Xu, X.; Fan, S.; Zhou, J.; Zhang, Y.; Che, Y.; Cai, H.; Wang, L.; Guo, L.; Liu, L.; Li, Q. The mutated tegument protein ul7 attenuates the virulence of herpes simplex virus 1 by reducing the modulation of alpha-4 gene transcription. Virol. J. 2016, 13, 152. [Google Scholar] [CrossRef]
- Roehm, P.C.; Shekarabi, M.; Wollebo, H.S.; Bellizzi, A.; He, L.; Salkind, J.; Khalili, K. Inhibition of HSV-1 replication by gene editing strategy. Sci. Rep. 2016, 6, 23146. [Google Scholar] [CrossRef]
- Raab-Traub, N. Novel mechanisms of EBV-induced oncogenesis. Curr. Opin. Virol. 2012, 2, 453–458. [Google Scholar] [CrossRef]
- Wang, J.; Quake, S.R. Rna-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc. Natl. Acad. Sci. USA 2014, 111, 13157–13162. [Google Scholar] [CrossRef] [PubMed]
- Yuen, K.S.; Chan, C.P.; Wong, N.H.; Ho, C.H.; Ho, T.H.; Lei, T.; Deng, W.; Tsao, S.W.; Chen, H.; Kok, K.H.; et al. CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human cells. J. Gen. Virol. 2015, 96, 626–636. [Google Scholar] [CrossRef] [PubMed]
- Yuen, K.S.; Wang, Z.M.; Wong, N.M.; Zhang, Z.Q.; Cheng, T.F.; Lui, W.Y.; Chan, C.P.; Jin, D.Y. Suppression of Epstein-Barr virus DNA load in latently infected nasopharyngeal carcinoma cells by CRISPR/Cas9. Virus Res. 2018, 244, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Boeckh, M.; Geballe, A.P. Cytomegalovirus: Pathogen, paradigm, and puzzle. J. Clin. Investig. 2011, 121, 1673–1680. [Google Scholar] [CrossRef] [PubMed]
- Gergen, J.; Coulon, F.; Creneguy, A.; Elain-Duret, N.; Gutierrez, A.; Pinkenburg, O.; Verhoeyen, E.; Anegon, I.; Nguyen, T.H.; Halary, F.A.; et al. Multiplex CRISPR/Cas9 system impairs HCV replication by excising an essential viral gene. PLoS ONE 2018, 13, e0192602. [Google Scholar] [CrossRef] [PubMed]
- van Diemen, F.R.; Kruse, E.M.; Hooykaas, M.J.; Bruggeling, C.E.; Schurch, A.C.; van Ham, P.M.; Imhof, S.M.; Nijhuis, M.; Wiertz, E.J.; Lebbink, R.J. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog. 2016, 12, e1005701. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Yu, L.; Zhu, D.; Ding, W.; Wang, X.; Zhang, C.; Wang, L.; Jiang, X.; Shen, H.; He, D.; et al. Disruption of hpv16-e7 by CRISPR/Cas system induces apoptosis and growth inhibition in hpv16 positive human cervical cancer cells. Biomed. Res. Int. 2014, 2014, 612823. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, E.M.; Kornepati, A.V.; Goldstein, M.; Bogerd, H.P.; Poling, B.C.; Whisnant, A.W.; Kastan, M.B.; Cullen, B.R. Inactivation of the human papillomavirus e6 or e7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J. Virol. 2014, 88, 11965–11972. [Google Scholar] [CrossRef]
- Zhen, S.; Hua, L.; Takahashi, Y.; Narita, S.; Liu, Y.H.; Li, Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem. Biophys. Res. Commun. 2014, 450, 1422–1426. [Google Scholar] [CrossRef]
- Liu, Y.C.; Cai, Z.M.; Zhang, X.J. Reprogrammed CRISPR-Cas9 targeting the conserved regions of hpv6/11 e7 genes inhibits proliferation and induces apoptosis in e7-transformed keratinocytes. Asian J. Androl. 2016, 18, 475–479. [Google Scholar]
- Zhen, S.; Lu, J.J.; Wang, L.J.; Sun, X.M.; Zhang, J.Q.; Li, X.; Luo, W.J.; Zhao, L. In vitro and in vivo synergistic therapeutic effect of cisplatin with human papillomavirus16 e6/e7 CRISPR/Cas9 on cervical cancer cell line. Transl. Oncol. 2016, 9, 498–504. [Google Scholar] [CrossRef]
- Yu, L.; Hu, Z.; Gao, C.; Feng, B.; Wang, L.; Tian, X.; Ding, W.; Jin, X.; Ma, D.; Wang, H. Deletion of hpv18 e6 and e7 genes using dual sgRNA-directed CRISPRCas9 inhibits growth of cervical cancer cells. Int. J. Clin. Exp. Med. 2017, 10, 9206. [Google Scholar]
- Hsu, D.S.; Kornepati, A.V.; Glover, W.; Kennedy, E.M.; Cullen, B.R. Targeting hpv16 DNA using CRISPR/Cas inhibits anal cancer growth in vivo. Future Virol. 2018, 13, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.R. The clinical features of PML. Cleve Clin. J. Med. 2011, 78 (Suppl. 2), S8–12. [Google Scholar] [CrossRef]
- Wollebo, H.S.; Bellizzi, A.; Kaminski, R.; Hu, W.; White, M.K.; Khalili, K. CRISPR/Cas9 system as an agent for eliminating polyomavirus JC infection. PLoS ONE 2015, 10, e0136046. [Google Scholar] [CrossRef] [PubMed]
- Guinat, C.; Gogin, A.; Blome, S.; Keil, G.; Pollin, R.; Pfeiffer, D.U.; Dixon, L. Transmission routes of African swine fever virus to domestic pigs: Current knowledge and future research directions. Vet. Rec. 2016, 178, 262–267. [Google Scholar] [CrossRef]
- Hubner, A.; Petersen, B.; Keil, G.M.; Niemann, H.; Mettenleiter, T.C.; Fuchs, W. Efficient inhibition of African swine fever virus replication by CRISPR/Cas9 targeting of the viral p30 gene (cp204l). Sci. Rep. 2018, 8, 1449. [Google Scholar] [CrossRef] [PubMed]
- Mettenleiter, T.C. Molecular biology of pseudorabies (aujeszky’s disease) virus. Comp. Immunol. Microbiol. Infect. Dis. 1991, 14, 151–163. [Google Scholar] [CrossRef]
- Peng, Z.; Ouyang, T.; Pang, D.; Ma, T.; Chen, X.; Guo, N.; Chen, F.; Yuan, L.; Ouyang, H.; Ren, L. Pseudorabies virus can escape from CRISPR-Cas9-mediated inhibition. Virus Res. 2016, 223, 197–205. [Google Scholar] [CrossRef]
- Tang, Y.D.; Liu, J.T.; Wang, T.Y.; Sun, M.X.; Tian, Z.J.; Cai, X.H. CRISPR/Cas9-mediated multiple single guide RNAs potently abrogate pseudorabies virus replication. Arch. Virol. 2017, 162, 3881–3886. [Google Scholar] [CrossRef]
- Shepard, C.W.; Finelli, L.; Alter, M.J. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 2005, 5, 558–567. [Google Scholar] [CrossRef]
- Sampson, T.R.; Saroj, S.D.; Llewellyn, A.C.; Tzeng, Y.L.; Weiss, D.S. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 2013, 497, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Price, A.A.; Sampson, T.R.; Ratner, H.K.; Grakoui, A.; Weiss, D.S. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc. Natl. Acad. Sci. USA 2015, 112, 6164–6169. [Google Scholar] [CrossRef] [PubMed]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR-cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef]
- Kotterman, M.A.; Schaffer, D.V. Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 2014, 15, 445–451. [Google Scholar] [CrossRef]
- Li, L.; Hu, S.; Chen, X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities. Biomaterials 2018, 171, 207–218. [Google Scholar] [CrossRef]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).