Insights into the Phylogeny and Evolution of Cold Shock Proteins: From Enteropathogenic Yersinia and Escherichia coli to Eubacteria
Abstract
1. Introduction
2. Results
2.1. Phylogeny and Consistency of Csps in Enteropathogenic Yersinia Enterocolitica and Yersinia Pseudotuberculosis
2.2. Phylogeny and Consistency of Csps in E. coli
2.3. Comparison of Csps in Enteropathogenic Yersinia and E. coli
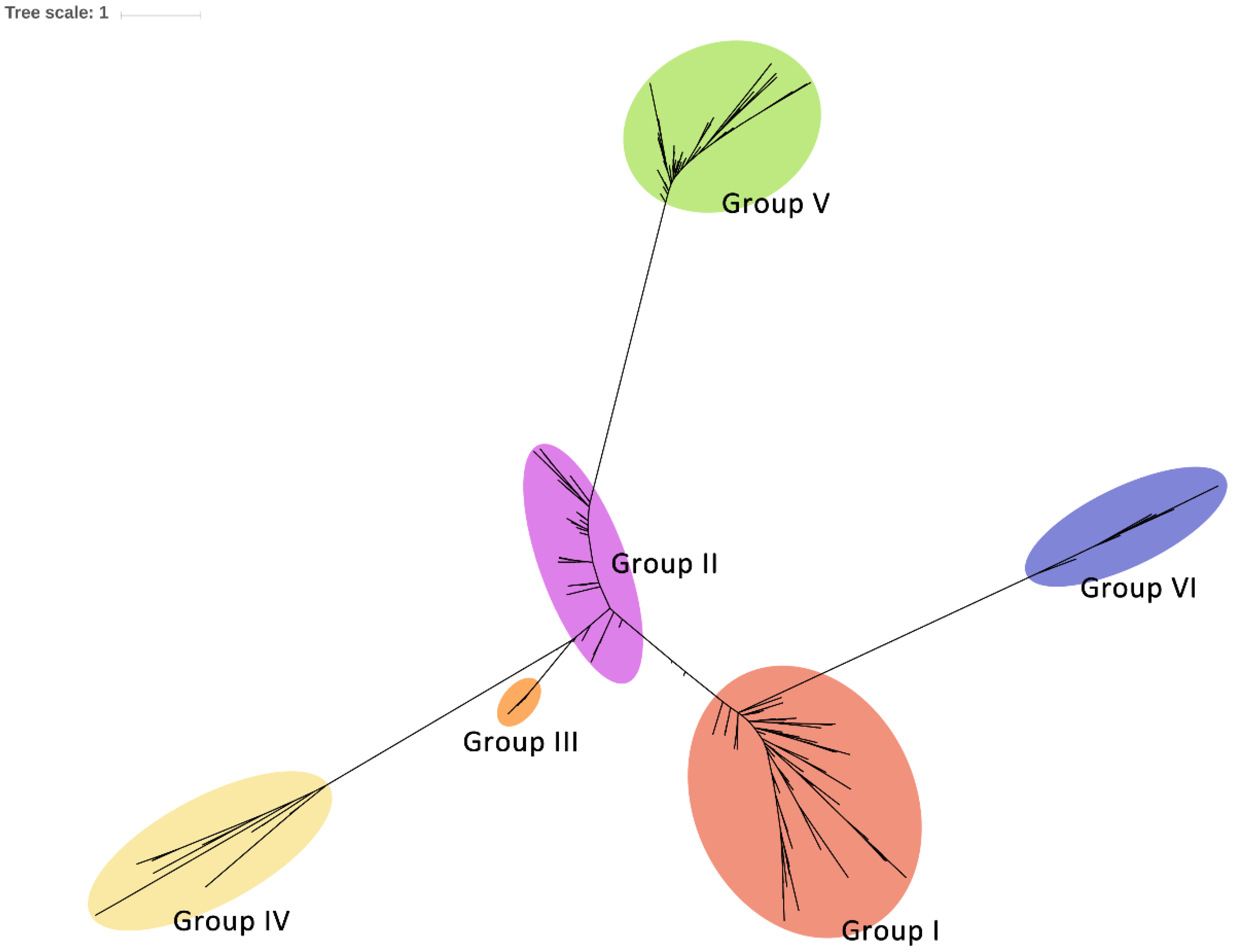
2.4. Phylogenetic Diversity of Csps in Enterobacteriales
2.5. Clarification of Phylogeny and Origin Time of Csp Genes in Eubacteria
3. Discussion
4. Materials and Methods
4.1. Selection of Csp Sequences from Databases
4.2. Protein Clustering, Alignment and Phylogeny
4.3. Estimation of Substitution Rate and Origin Time of Csp and 16S rRNA
4.4. Molecular Clock Calibration
4.5. Data Availability
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 5′-UTR | 5′ untranslated region |
| aa | amino acids |
| BEAST | Bayesian Evolutionary Analysis Sampling Trees |
| Cips | Cold-induced proteins |
| CSD | Cold shock domain |
| CSPs | Cold shock proteins |
| HPD | Highest posterior density |
| MAFFT | Multiple Alignment using Fast Fourier Transform |
| MCMC | Markov chain Monte Carlo |
| MCC | Maximum clade credibility |
| MYA | Million years ago |
| NCBI | National Center for Biotechnology Information |
| PATRIC | Pathosystems Resource Integration Center |
| RAxML | Randomized Axelerated Maximum Likelihood |
| s.d. | standard deviation |
| tMRCA | time to the most recent common ancestor |
References
- Laukkanen-Ninios, R.; Fredriksson-Ahomaa, M.; Korkeala, H. Enteropathogenic Yersinia in the pork production chain: Challenges for control. Compr. Rev. Food Sci. Food Saf. 2014, 13, 1165–1191. [Google Scholar] [CrossRef]
- Fredriksson-Ahomaa, M.; Lindström, M.; Korkeala, H. Yersinia enterocolitica and Yersinia pseudotuberculosis. In Pathogens and Toxins in Foods; Juneja, V.K., Sofos, J.N., Eds.; ASM Press: Washington, DC, USA, 2010; Volume 11, pp. 164–180. [Google Scholar]
- Keto-Timonen, R.; Pöntinen, A.; Aalto-Araneda, M.; Korkeala, H. Growth of Yersinia pseudotuberculosis strains at different temperatures, pH values, and NaCl and ethanol concentrations. J. Food Prot. 2017, 81, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Barria, C.; Malecki, M.; Arraiano, C.M. Bacterial adaptation to cold. Microbiology 2013, 159, 2437–2443. [Google Scholar] [CrossRef] [PubMed]
- Keto-Timonen, R.; Hietala, N.; Palonen, E.; Hakakorpi, A.; Lindström, M.; Korkeala, H. Cold shock proteins: A minireview with special emphasis on Csp-family of enteropathogenic Yersinia. Front. Microbiol. 2016, 7, 1–7. [Google Scholar] [CrossRef]
- Jones, P.G.; VanBogelen, R.A.; Neidhardt, F.C. Induction of proteins in response to low temperature in Escherichia coli. J. Bacteriol. 1987, 169, 2092–2095. [Google Scholar] [CrossRef]
- Schmid, B.; Klumpp, J.; Raimann, E.; Loessner, M.J.; Stephan, R.; Tasara, T. Role of cold shock proteins in growth of Listeria monocytogenes under cold and osmotic stress conditions. Appl. Environ. Microbiol. 2009, 75, 1621–1627. [Google Scholar] [CrossRef]
- Graumann, P.; Schröder, K.; Schmid, R.; Marahiel, M.A. Cold shock stress-induced proteins in Bacillus subtilis. J. Bacteriol. 1996, 178, 4611–4619. [Google Scholar] [CrossRef]
- Wemekamp-Kamphuis, H.H.; Karatzas, A.K.; Wouters, J.A.; Abee, T. Enhanced levels of cold shock proteins in Listeria monocytogenes LO28 upon exposure to low temperature and high hydrostatic pressure. Appl. Environ. Microbiol. 2002, 68, 456–463. [Google Scholar] [CrossRef]
- Wang, N.; Yamanaka, K.; Inouye, M. CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J. Bacteriol. 1999, 181, 1603–1609. [Google Scholar]
- Goldstein, J.; Pollitt, N.S.; Inouye, M. Major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 1990, 87, 283–287. [Google Scholar] [CrossRef]
- Yamanaka, K.; Fang, L.; Inouye, M. The CspA family in Escherichia coli: Multiple gene duplication for stress adaptation. Mol. Microbiol. 1998, 27, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Kanamaru, K.; Mizuno, T.; Horikoshi, K. A novel member of the cspA family of genes that is induced by cold shock in Escherichai coli. J. Bacteriol. 1996, 178, 2994–2997. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Fang, L.; Inouye, M. The role of the 5′-end untranslated region of the mRNA for CspA, the major cold-shock protein of Escherichia coli, in cold-shock adaptation. J. Bacteriol. 1996, 178, 4919–4925. [Google Scholar] [CrossRef] [PubMed]
- Eshwar, A.K.; Guldimann, C.; Oevermann, A.; Tasara, T. Cold-shock domain family proteins (Csps) are involved in regulation of virulence, cellular aggregation, and flagella-based motility in Listeria monocytogenes. Front. Cell. Infect. Microbiol. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Uppal, S.; Rao Akkipeddi, V.S.N.; Jawali, N. Posttranscriptional regulation of cspE in Escherichia coli: Involvement of the short 5′-untranslated region. FEMS Microbiol. Lett. 2008, 279, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Lathem, W.W.; Crosby, S.D.; Miller, V.L.; Goldman, W.E. Progression of primary pneumonic plague: A mouse model of infection, pathology, and bacterial transcriptional activity. Proc. Natl. Acad. Sci. USA 2005, 102, 17786–17791. [Google Scholar] [CrossRef] [PubMed]
- Phadtare, S. Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 2004, 6, 125–136. [Google Scholar] [PubMed]
- Annamalai, T.; Venkitanarayanan, K. Expression of major cold shock proteins and genes by Yersinia enterocolitica in synthetic medium and foods. J. Food Prot. 2005, 68, 2454–2458. [Google Scholar] [CrossRef]
- Neuhaus, K.; Francis, K.P.; Rapposch, S.; Görg, A.; Scherer, S. Pathogenic Yersinia species carry a novel, cold-inducible major cold shock protein tandem gene duplication producing both bicistronic and monocistronic mRNA. J. Bacteriol. 1999, 181, 6449–6455. [Google Scholar]
- Phadtare, S.; Inouye, M. Sequence-selective interactions with RNA by CspB, CspC and CspE, members of the CspA family of Escherichia coli. Mol. Microbiol. 1999, 33, 1004–1014. [Google Scholar] [CrossRef]
- Lee, S.J.; Xie, A.; Jiang, W.; Etchegaray, J.-P.; Jones, P.G.; Inouye, M. Family of the major cold-shock protein, CspA (CS7.4), of Escherichia coli, whose members show a high sequence similarity with the eukaryotic Y-box binding proteins. Mol. Microbiol. 1994, 11, 833–839. [Google Scholar] [CrossRef]
- Phadtare, S.; Inouye, M. Role of CspC and CspE in regulation of expression of RpoS and UspA the stress response proteins in Escherichia coli. J. Bacteriol. 2001, 183, 1205–1214. [Google Scholar] [CrossRef]
- Bae, W.; Phadtare, S.; Severinov, K.; Inouye, M. Characterization of Escherichia coli cspE, whose product negatively regulates transcription of cspA, the gene for the major cold shock protein. Mol. Microbiol. 1999, 31, 1429–1441. [Google Scholar] [CrossRef]
- Yamanaka, K.; Inouye, M. Growth-phase-dependent expression of cspD, encoding a member of the CspA family in Escherichia coli. J. Bacteriol. 1997, 179, 5126–5130. [Google Scholar] [CrossRef]
- Hébraud, M.; Potier, P. Cold shock response and low temperature adaptation in psychrotrophic bacteria. J. Mol. Microbiol. Biotechnol. 1999, 1, 211–219. [Google Scholar]
- Landsman, D. RNP-1, an RNA-binding motif is conserved in the DNA-binding cold shock domain. Nucleic Acids Res. 1992, 20, 2861–2864. [Google Scholar] [CrossRef]
- Schindelin, H.; Marahiel, M.A.; Heinemann, U. Universal nucleic acid-binding domain revealed by crystal structure of the B. subtilis major cold-shock protein. Nature 1993, 364, 164–168. [Google Scholar] [CrossRef]
- Phadtare, S.; Inouye, M.; Severinov, K. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J. Biol. Chem. 2002, 277, 7239–7245. [Google Scholar] [CrossRef]
- Rath, D.; Jawali, N. Loss of expression of cspC, a cold shock family gene, confers a gain of fitness in Escherichia coli K-12 strains. J. Bacteriol. 2006, 188, 6780–6785. [Google Scholar] [CrossRef]
- Graumann, P.; Marahiel, M.A. Some like it cold: Response of microorganisms to cold shock. Arch. Microbiol. 1996, 166, 293–300. [Google Scholar] [CrossRef]
- Chapot-Chartier, M.P.; Schouler, C.; Lepeuple, A.S.; Gripon, J.C.; Chopin, C. Characterization of cspB, a cold-shock-inducible gene from Lactococcus lactis, and evidence for a family of genes homologous to the Escherichia coli cspA major cold shock gene. J. Bacteriol. 1997, 179, 5589–5593. [Google Scholar] [CrossRef]
- Mayo, B.; Derzelle, S.; Fernandez, M.; Leonard, C.; Ferain, T.; Hols, P.; Suárez, J.E.; Delcour, J. Cloning and characterization of cspL and cspP, two cold-inducible genes from Lactobacillus plantarum. J. Bacteriol. 1997, 179, 3039–3042. [Google Scholar] [CrossRef]
- Ochman, H.; Elwyn, S.; Moran, N.A. Calibrating bacterial evolution. Proc. Natl. Acad. Sci. USA 1999, 96, 12638–12643. [Google Scholar] [CrossRef]
- Battistuzzi, F.U.; Feijao, A.; Hedges, S.B. A genomic timescale of prokaryote evolution: Insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 2004, 4, 44. [Google Scholar] [CrossRef]
- Shenhar, Y.; Biran, D.; Ron, E.Z. Resistance to environmental stress requires the RNA chaperones CspC and CspE. Environ. Microbiol. Rep. 2012, 4, 532–539. [Google Scholar] [CrossRef]
- Graumann, P.L.; Marahiel, M.A. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 1998, 23, 286–290. [Google Scholar] [CrossRef]
- Virtanen, J.-P.; Keto-Timonen, R.; Jaakkola, K.; Salin, N.; Korkeala, H. Changes in transcriptome of Yersinia pseudotuberculosis IP32953 grown at 3 °C and 28 °C detected by RNA sequencing shed light on cold adaptation. Front. Cell. Infect. Microbiol. 2018, 8, 416. [Google Scholar] [CrossRef]
- Cavicchioli, R. Cold-adapted archaea. Nat. Rev. Microbiol. 2006, 4, 331. [Google Scholar] [CrossRef]
- Giaquinto, L.; Curmi, P.M.G.; Siddiqui, K.S.; Poljak, A.; DeLong, E.; DasSarma, S.; Cavicchioli, R. Structure and function of cold shock proteins in archaea. J. Bacteriol. 2007, 189, 5738–5748. [Google Scholar] [CrossRef]
- Mihailovich, M.; Militti, C.; Gabaldón, T.; Gebauer, F. Eukaryotic cold shock domain proteins: Highly versatile regulators of gene expression. BioEssays 2010, 32, 109–118. [Google Scholar] [CrossRef]
- Deckert, G.; Warren, P.V.; Gaasterland, T.; Young, W.G.; Lenox, A.L.; Graham, D.E.; Overbeek, R.; Snead, M.A.; Keller, M.; Aujay, M.; et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 1998, 392, 353. [Google Scholar] [CrossRef]
- Perl, D.; Welker, C.; Schindler, T.; Schröder, K.; Marahiel, M.A.; Jaenicke, R.; Schmid, F.X. Conservation of rapid two-state folding in mesophilic, thermophilic and hyperthermophilic cold shock proteins. Nat. Struct. Biol. 1998, 5, 229. [Google Scholar] [CrossRef]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, 1–6. [Google Scholar] [CrossRef]




| Clade 1 | Subclade 1 | Representative csp Gene(s) | Currently Used Name(s) 2 | RNP1 3 | RNP2 3 | Known Function of Csps and Occurrence in Bacteria |
|---|---|---|---|---|---|---|
| I | Ia | aq_1303a | cspC | KGYGFITX | FVH | Exists in the hyperthermophilic bacterium. |
| Ib | Lmo2016, BSU05120, lmo1879, L172505, CPE1242 | cspB, cspC, cspD, cspE, cspL | KGGFIXX | XFH | Involves in regulation of cold and osmotic stress tolerance, virulence, cellular aggregation and flagella-based motility [7,15]; mainly exists in gram-positive bacteria. | |
| II | IIa | b3556, b1557, b0990, b1552 | cspA, cspB, cspG, cspI | KGFGFIP | VFVHF | Cold-inducible [6,11,18]; exists in Gammaproteobacteria. |
| IIb | b1823, b0623 | cspC, cspE | KGFGFITP | VFVHF | Involves in cold adaptation (CspE) [29], transcriptional regulation and/or chromosome condensation [12,23,36]; exists in Gammaproteobacteria. | |
| IIc | YPTB1423, YE1546 | cspB, cspE2 | GGFI | VVX | Involves in stress response in vivo [17]; mainly exists in Yersiniaceae. | |
| IId | VCA0166 | - | KGFGFQ | VFVHF | Exists in Vibrionaceae. | |
| IIe | PA0456 | - | KGGFIX | FH | Exists in Gammaproteobacteria (not including Enterobacteriales). | |
| III | IIIa | b0880 | cspD | KGFGFI | IFAHY | Plays a role in the nutrient-stress response [25,37]; exists in Gammaproteobacteria. |
| IIIb | RSp1053, RSc3156 | cspD2, cspC | KGGFIX | LFAH | Mainly exists in Betaproteobacteria. | |
| IV | IVa | SMc04318, SMc04234 | csp1, csp4 | KFGFIXP | XFVHX | Exists in Alphaproteobacteria. |
| IVb | Rv3648c | cspA | KGFGFIAP | XFVH | Mainly exists in Actinobacteria. | |
| V | V | b0989, b1558 | cspF, cspH | SGKGIP | VQH | Function unknown [10]; only exists in Enterobacteriaceae. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, T.; Keto-Timonen, R.; Jiang, X.; Virtanen, J.-P.; Korkeala, H. Insights into the Phylogeny and Evolution of Cold Shock Proteins: From Enteropathogenic Yersinia and Escherichia coli to Eubacteria. Int. J. Mol. Sci. 2019, 20, 4059. https://doi.org/10.3390/ijms20164059
Yu T, Keto-Timonen R, Jiang X, Virtanen J-P, Korkeala H. Insights into the Phylogeny and Evolution of Cold Shock Proteins: From Enteropathogenic Yersinia and Escherichia coli to Eubacteria. International Journal of Molecular Sciences. 2019; 20(16):4059. https://doi.org/10.3390/ijms20164059
Chicago/Turabian StyleYu, Tao, Riikka Keto-Timonen, Xiaojie Jiang, Jussa-Pekka Virtanen, and Hannu Korkeala. 2019. "Insights into the Phylogeny and Evolution of Cold Shock Proteins: From Enteropathogenic Yersinia and Escherichia coli to Eubacteria" International Journal of Molecular Sciences 20, no. 16: 4059. https://doi.org/10.3390/ijms20164059
APA StyleYu, T., Keto-Timonen, R., Jiang, X., Virtanen, J.-P., & Korkeala, H. (2019). Insights into the Phylogeny and Evolution of Cold Shock Proteins: From Enteropathogenic Yersinia and Escherichia coli to Eubacteria. International Journal of Molecular Sciences, 20(16), 4059. https://doi.org/10.3390/ijms20164059





