The Link That Binds: The Linker of Hsp70 as a Helm of the Protein’s Function
Abstract
:1. Hsp70 Molecular Chaperones
2. Hsp70 Structure
3. The General Features of Naturally Occurring Linker Peptides
4. The Hsp70 Linker Peptides
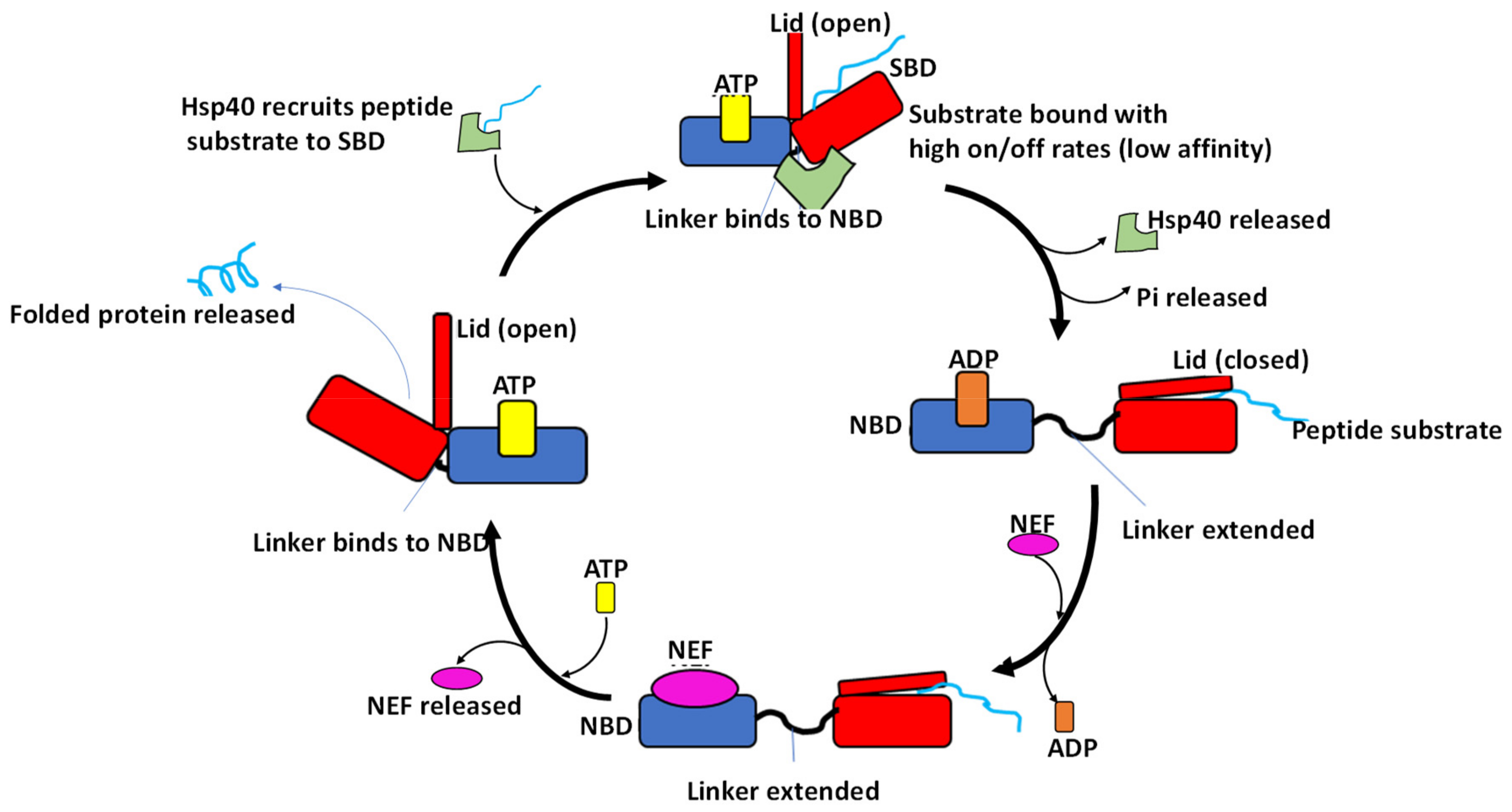
5. Hsp70 Functional Cycle
6. The Role of the Linker of Hsp70 in Facilitating ATP Binding and Hydrolysis
7. The Role of the Hsp70 Linker in Substrate Binding
8. The Role of the Linker on the Association of Hsp70 with Co-Chaperones
9. The Role of the Linker of Hsp70 in Regulating Oligomerization
10. The Role of the Linker of Hsp70 in Regulating Its Stability
11. Targeting the Linker in Drug Discovery
12. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Lindquist, S.; Craig, E.A. The Heat shock proteins. Annu. Rev. Genet. 1988, 22, 633–677. [Google Scholar] [CrossRef] [PubMed]
- Szabo, A.; Langer, T.; Schröder, H.; Flanagan, J.; Bukau, B.; Hartl, F.U. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc. Natl. Acad. Sci. USA 1994, 91, 10345–10349. [Google Scholar] [CrossRef] [PubMed]
- Bercovich, B.; Stancovski, I.; Mayer, A.; Blumenfeld, N.; Laszlo, A.; Schwartz, A.L.; Ciechanover, A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J. Biol. Chem. 1997, 272, 9002–9010. [Google Scholar] [CrossRef] [PubMed]
- Calloni, G.; Chen, T.; Schermann, S.; Chang, C.; Genevaux, P.; Agostini, F.; Tartaglia, G.; Hayer-Hartl, M.; Hartl, F.U. DnaK functions as a central hub in the E. coli chaperone network. Cell 2012, 3, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P. Gymnastics of molecular chaperones. Mol. Cell. 2010, 39, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 2013, 38, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Young, J.C. Mechanisms of the Hsp70 chaperone system. Biochem. Cell Biol. 2010, 88, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Kurt, N.; Rajagopalan, S.; Cavagnero, S. Effect of Hsp70 chaperone on the folding and misfolding of polypeptides modelling an elongating protein chain. J Mol. Biol. 2006, 355, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Shonhai, A. Plasmodial heat shock proteins: Targets for chemotherapy. FEMS Immunol. Med. Microbiol. 2010, 58, 61–74. [Google Scholar] [CrossRef]
- Zuiderweg, E.; Bertelsen, E.; Rousaki, A.; Mayer, M.P.; Gestwicki, J.E.; Ahmad, A. Allostery in the Hsp70 chaperone proteins. Top. Curr. Chem. 2013, 328, 99–153. [Google Scholar]
- Easton, D.P.; Kaneko, Y.; Subjeck, J.R. The Hsp110 and Grp170 stress proteins: Newly recognized relatives of the Hsp70s. Cell Stress Chaperones 2000, 5, 276–290. [Google Scholar] [CrossRef]
- Liu, Q.; Hendrickson, W.A. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 2007, 131, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.J.; Easton, D.; Murawski, M.; Kaneneko, Y.; Subjeck, J.R. The chaperoning activity of Hsp110: Identification of functional domains by use of targeted deletions. J. Biol. Chem. 1999, 274, 15712–15718. [Google Scholar] [CrossRef] [PubMed]
- Polier, S.; Dragovic, Z.; Hartl, F.U.; Bracher, A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 2008, 133, 1068–1079. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Sternberg, M.J. Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar] [CrossRef]
- Ruiz, D.M.; Turowski, V.R.; Murakamib, M.R. Effects of the linker region on the structure and function of modular GH5 cellulase. Sci. Rep. 2016, 6, 28504. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, R.S.; Khosla, C. Role of linkers in communication between protein modules. Curr. Opin. Chem. Biol. 2000, 4, 22–27. [Google Scholar] [CrossRef]
- George, R.A.; Heringa, J. An analysis of protein domain linkers: Their classification and role in protein folding. Protein Eng. 2002, 15, 871–879. [Google Scholar] [CrossRef]
- Van Leeuwen, H.C.; Strating, M.J.; Rensen, M.; de Laat, W.; van der Vliet, P.C. Linker length and composition influence the flexibility of Oct-1 DNA binding. EMBO J. 1997, 16, 2043–2053. [Google Scholar] [CrossRef] [Green Version]
- Robinson, C.R.; Sauer, R.T. Optimizing the stability of single-chain proteins by linker length and composition mutagenesis. Proc. Natl, Acad. Sci. USA 1998, 95, 5929–5934. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Zaro, J.; Shen, W.C. Fusion protein linkers: Property, design and functionality. Adv. Drug. Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef] [PubMed]
- Chichili, V.P.R.; Kumar, V.; Sivaraman, J. Linkers in the structural biology of protein-protein interactions. Protein Sci. 2012, 22, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Wriggers, W.; Chakravarty, S.; Jennings, P.A. Control of protein functional dynamics by peptide linkers. Biopolymers 2005, 80, 736–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argos, P. An investigation of oligopeptides linking domains in protein tertiary structures and possible candidates for general gene fusion. J. Mol. Biol. 1990, 211, 943–958. [Google Scholar] [CrossRef]
- Williamson, M.P. The structure and function of proline-rich regions in proteins. Biochem. J. 1994, 297, 249–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, S.L.; Russell, G.C.; Williamson, M.P.; Guest, J.R. Restructuring an interdomain linker in the dihydrolipoamide acetyltransferase component of the pyruvate dehydrogenase complex of Escherichia coli. Protein Eng. 1993, 6, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Aurora, R.; Creamer, T.P.; Srinivasan, R.; Rose, G.D. Local interactions in protein folding: Lessons from the alpha-helix. J. Biol. Chem. 1997, 272, 1413–1416. [Google Scholar] [CrossRef] [PubMed]
- Vogel, M.; Mayer, M.P.; Bukau, B. Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J. Biol. Chem. 2006, 281, 38705–38711. [Google Scholar] [CrossRef]
- Swain, J.F.; Dinler, G.; Sivendran, R.; Montgomery, D.; Stotz, M.; Gierasch, L.M. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell. 2007, 26, 27–39. [Google Scholar] [CrossRef]
- English, C.A.; Sherman, W.; Meng, W.; Gierasch, L.M. The Hsp70 interdomain linker is a dynamic switch that enables allosteric communication between two structured domains. J. Biol. Chem. 2017, 292, 14765–14774. [Google Scholar] [CrossRef] [Green Version]
- General, I.J.; Liu, Y.; Blackburn, M.E.; Mao, W.; Gierasch, L.M.; Bahar, I. ATPase subdomain IA is a mediator of interdomain allostery in Hsp70 molecular chaperones. PLoS Comput. Biol. 2014, 10, e1003624. [Google Scholar] [CrossRef] [PubMed]
- Valdes, H.M.; Burgos-Bravo, F.; Quiroga-Roger, D.; Casanova-Morales, N.; Wilson, C.A. Mechanical properties of chaperone BIP, the master regulator of the endoplasmic reticulum. In Endoplasmic Reticulum; Catala, A., Ed.; Intech Open: London, UK, 2019; ISBN 978-1-83880-088-8. [Google Scholar]
- Zininga, T.; Achilonu, I.; Hoppe, H.; Prinsloo, E.; Dirr, H.W.; Shonhai, A. Plasmodium falciparum Hsp70-z, an Hsp110 homologue, exhibits independent chaperone activity and interacts with Hsp70-1 in a nucleotide-dependent fashion. Cell Stress Chaperones 2016, 21, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Shonhai, A.; Botha, M.; de Beer, T.A.; Boshoff, A.; Blatch, G.L. Structure function study of a Plasmodium falciparum Hsp70 using three-dimensional modelling and in vitro analyses. Protein Pept. Lett. 2008, 15, 1117–1125. [Google Scholar] [CrossRef] [PubMed]
- Pallarès, I.; de Groot, N.S.; Iglesias, V.; Sant’Anna, R.; Biosca, A.; Fernàndez-Busquets, X.; Ventura, S. Discovering putative prion-like proteins in Plasmodium falciparum: A computational and experimental analysis. Front. Microbiol. 2018, 9, 1737. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.P.; Vorvis, C.; Sarbeng, E.; Ledesma, V.C.C.; Willis, J.E.; Liu, Q. The four hydrophobic residues on the Hsp70 interdomain linker have two distinct roles. J. Mol. Bio. 2011, 4, 1099–1113. [Google Scholar] [CrossRef] [PubMed]
- Kityk, R.; Kopp, J.; Sinning, I.; Mayer, M.P. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell. 2012, 48, 863–874. [Google Scholar] [CrossRef]
- Clerico, E.M.; Tilitsky, J.M.; Meng, W.; Gierasch, L.M. How Hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 2015, 427, 1575–1588. [Google Scholar] [CrossRef]
- Zhuravleva, A.; Gierasch, L. Allosteric signal transmission in the nucleotide binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones. Proc. Natl. Acad. Sci. USA 2011, 108, 6987–6992. [Google Scholar] [CrossRef]
- Qi, R.; Boateng Sarbeng, E.; Liu, Q.; Quynh Le, K.; Xu, X.; Xu, H.; Yang, J.; Li, W.; Vorvis, C.; Hendrickson, W.; et al. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat. Struct. Mol. Biol. 2013, 20, 900–907. [Google Scholar] [CrossRef] [Green Version]
- Buchberger, A.; Schröder, H.; Büttner, M.; Valencia, A.; Bukau, B. A conserved loop in the ATPase domain of the DnaK chaperone is essential for stable binding of GrpE. Nat. Struct. Biol. 1994, 1, 95–101. [Google Scholar] [CrossRef]
- Jiang, J.; Prasad, K.; Lafer, E.M.; Sousa, R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol. Cell. 2005, 20, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P.; Schröder, H.; Rüdiger, S.; Paal, K.; Laufen, T.; Bukau, B. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat. Struct. Mol. Biol. 2000, 7, 586–593. [Google Scholar]
- Brehme, M.; Voisine, C. Model systems of protein-misfolding diseases reveal chaperone modifiers of proteotoxicity. Dis. Mod. Mech. 2016, 9, 823–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moro, F.; Fernandez-Saiz, V.; Muga, A. The lid Subdomain of DnaK is required for the stabilization of the substrate-binding Site. J. Biol. Chem. 2004, 279, 19600–19606. [Google Scholar] [CrossRef] [PubMed]
- Liberek, K.; Marszalek, J.; Ang, D.; Georgopoulos, C.; Zylicz, M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. Natl. Acad. Sci. USA 1991, 88, 2874–2878. [Google Scholar] [CrossRef] [PubMed]
- Kampinga, H.H.; Craig, E.A. The Hsp70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P.; Gierasch, L.M. Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J. Biol. Chem. 2019, 294, 2085–2097. [Google Scholar] [CrossRef] [Green Version]
- Mayer, M.P.; Kityk, R. Insights into the molecular mechanism of allostery in Hsp70s. Front. Mol. Biosci. 2015, 2, 58. [Google Scholar] [CrossRef] [Green Version]
- Mayer, M.P. Intra-molecular pathways of allosteric control in Hsp70s. Philos. Trans. R. Soc. Lond. Biol. Sci. 2018, 373, 2017. [Google Scholar] [CrossRef]
- Kityk, R.; Vogel, M.; Schlecht, R.; Bukau, B.; Mayer, P.M. Pathways of allosteric regulation in Hsp70 chaperones. Nat. Commun. 2015, 6, 8308. [Google Scholar] [CrossRef] [Green Version]
- Zhuravleva, A.; Gierasch, L.M. Substrate-binding domain conformational dynamics mediate Hsp70 allostery. Proc. Natl. Acad. Sci. USA 2015, 112, 2865–2873. [Google Scholar] [CrossRef] [PubMed]
- Chiappori, F.; Merelli, I.; Milanesi, L.; Colombo, G.; Morra, G. An atomistic view of Hsp70 allosteric crosstalk: From the nucleotide to the substrate binding domain and back. Sci. Rep. 2016, 6, 23474. [Google Scholar] [CrossRef] [PubMed]
- Zhuravleva, A.; Clerico, M.E.; Gierasch, L.M. An interdomain energetic tug of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell 2012, 151, 1296–1307. [Google Scholar] [CrossRef] [PubMed]
- Alderson, T.R.; Kim, J.H.; Cai, K.; Frederick, R.O.; Tonelli, M.; John, L. Markley. The specialized Hsp70 (HscA) interdomain linker binds to its nucleotide-binding domain and stimulates ATP hydrolysis in both cis and trans configurations. Biochemistry 2014, 53, 7148–7159. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Kurochkin, A.V.; Yip, G.N.; Zhang, Y.; Bertelsen, E.B.; Zuiderweg, E.R. Allostery in Hsp70 chaperones is transduced by subdomain rotations. J. Mol. Biol. 2009, 388, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Bertelsen, E.B.; Chang, L.; Gestwicki, J.E.; Zuiderweg, E.R. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. USA 2009, 106, 8471–8476. [Google Scholar] [CrossRef]
- Leidig, C.; Bange, G.; Kopp, J.; Amlacher, S.; Aravind, A.; Wickles, S.; Witte, G.; Hurt, E.; Beckmann, R.; Sinning, I. Structural characterization of a eukaryotic chaperone—The ribosome-associated complex. Nat. Struct. Mol. Biol. 2013, 20, 23–28. [Google Scholar] [CrossRef]
- Wisniewska, M.; Karlberg, T.; Lehtiö, L.; Johansson, I.; Kotenyova, T.; Moche, M.; Schüler, H. Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B’, and HSPA5/BiP/GRP78. PLoS ONE 2010, 5, e8625. [Google Scholar] [CrossRef]
- Amick, J.; Schlanger, S.E.; Wachnowsky, C.; Moseng, M.A.; Emerson, C.C.; Dare, M.; Luo, W.I.; Ithychanda, S.S.; Nix, J.C.; Cowan, J.A.; et al. Crystal structure of the nucleotide-binding domain of mortalin, the mitochondrial Hsp70 chaperone. Protein Sci. 2014, 23, 833–842. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Leu, J.; Murphy, M.E.; George, D.L.; Marmorstein, R. Crystal structure of the stress-inducible human heat shock protein 70 substrate-binding domain in complex with peptide substrate. PLoS ONE 2014, 9, e103518. [Google Scholar] [CrossRef]
- Flaherty, K.M.; DeLuca-Flaherty, C.; McKay, D.B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 1990, 346, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhao, X.; Burkholder, W.F.; Gragerov, A.; Ogata, C.M.; Gottesman, M.E.; Hendrickson, W.A. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 1996, 272, 1606–1614. [Google Scholar] [CrossRef] [PubMed]
- Rüdiger, S.; Germeroth, L.; Schneider-Mergener, J.; Bukau, B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997, 16, 1501–1507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liebscher, M.; Roujeinikova, A. Allosteric coupling between the lid and interdomain linker in DnaK revealed by inhibitor binding studies. J. Bacteriol. 2009, 191, 1456–1462. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.E.; Voisine, C.; Craig, E.A. Intragenic suppressors of Hsp70 mutants: Interplay between the ATPase- and peptide-binding domains. Proc. Natl. Acad. Sci. USA 1999, 96, 9269–9276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.; Maes, E.G.; Alex, B.; Taylor, B.; Wang, L.; Hinck, A.P.; Lafer, E.M.; Sousa, R. Structural basis of J cochaperone binding and regulation of Hsp70. Mol. Cell. 2007, 28, 422–433. [Google Scholar] [CrossRef]
- Sharma, D.; Masison, D.C. Hsp70 structure, function, regulation and influence on yeast prions. Protein Pept. Lett. 2009, 16, 571–581. [Google Scholar] [CrossRef]
- Alderson, T.R.; Kim, J.H.; Markley, J.L. Dynamical structures of Hsp70 and Hsp70-Hsp40 complexes. Structure 2016, 24, 1014–1030. [Google Scholar] [CrossRef]
- Young, J.C.; Agashe, V.R.; Siegers, K.; Hartl, F.U. Pathways of chaperone mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 2004, 5, 781–791. [Google Scholar] [CrossRef]
- Han, W.; Christen, P. Mutations in the interdomain linker region of DnaK abolish the chaperone action of the DnaK/DnaJ/GrpE system. FEBS Lett. 2001, 497, 55–58. [Google Scholar] [CrossRef]
- Genest, O.; Wickner, S.; Doyle, S.M. Hsp90 and Hsp70 chaperones: Collaborators in protein remodeling. J. Biol. Chem. 2018, 294, 2109–2120. [Google Scholar] [CrossRef] [PubMed]
- Lässle, M.; Blatch, G.L.; Kundra, V.; Takatori, T.; Zetter, B.R. Stress-inducible, murine protein mSTI1. Characterization of binding domains for heat shock proteins and in vitro phosphorylation by different kinases. J. Biol. Chem. 1997, 272, 1876–1884. [Google Scholar] [CrossRef] [PubMed]
- Rohl, A.; Wengler, D.; Madl, T.; Lagleder, S.; Tippel, F.; Herrmann, M. Hsp90 regulates the dynamics of its cochaperone Sti1 and the transfer of Hsp70 between modules. Nat. Commun. 2015, 6, 6655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zininga, T.; Makumire, S.; Gitau, G.W.; Njunge, J.M.; Pooe, O.J.; Klimek, H.; Scheurr, R.; Raifer, H.; Prinsloo, E.; Przyborski, J.M.; et al. Plasmodium falciparum Hop (PfHop) Interacts with the Hsp70 chaperone in a nucleotide-dependent fashion and exhibits ligand selectivity. PLoS ONE 2015, 10, e0135326. [Google Scholar] [CrossRef] [PubMed]
- Zininga, T.; Pooe, O.J.; Makhado, P.B.; Ramatsui, L.; Prinsloo, E.; Achilonu, I.; Dirr, H.; Shonhai, A. Polymyxin B inhibits the chaperone activity of Plasmodium falciparum Hsp70. Cell Stress Chaperones 2017, 22, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Zininga, T.; Ramatsui, L.; Makhado, P.B.; Makumire, S.; Achilinou, I.; Hoppe, H.; Dirr, H.W.; Shonhai, A. (-)-Epigallocatechin-3-Gallate inhibits the chaperone activity of Plasmodium falciparum Hsp70 chaperones and abrogates their association with functional partners. Molecules 2017, 22, 2139. [Google Scholar] [CrossRef] [PubMed]
- Benaroudj, N.; Triniolles, F.; Ladjimi, M.M. Effect of nucleotides, peptides, and unfolded proteins on the self-association of the molecular chaperone HSc70. J. Biol. Chem. 1996, 271, 18471–18476. [Google Scholar] [CrossRef]
- Benaroudj, N.; Fouchaq, B.; Ladjimi, M.M. The COOH terminal peptide binding domain is essential for self-association of the molecular chaperone HSc70. J. Biol. Chem. 1997, 272, 8744–8751. [Google Scholar] [CrossRef] [PubMed]
- Fouchaq, B.; Benaroudj, N.; Ebel, C.; Ladjimi, M.M. Oligomerization of the 17-kDa peptide-binding domain of the molecular chaperone HSC70. Eur. J. Biochem. 1999, 259, 379–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohno, M.; Kitabatake, N.; Tani, F. Role of the C-terminal region of mouse inducible Hsp72 in the recognition of peptide substrate for chaperone activity. FEBS Lett. 2004, 576, 381–386. [Google Scholar] [CrossRef] [Green Version]
- Aprile, F.A.; Dhulesia, A.; Stengel, F.; Roodveldt, C.; Benesch, J.L.P.; Tortora, P.; Robinson, C.V.; Salvatella, X.; Dobson, C.M.; Cremades, N. Hsp70 oligomerization is mediated by an interaction between the interdomain linker and the substrate-binding domain. PLoS ONE 2013, 8, e67961. [Google Scholar] [CrossRef] [PubMed]
- Morgner, N.; Schmidt, C.; Beilsten-Edmands, V.; Ebong, I.; Patel, N.A.; Clerico, E.M.; Kirschke, E.; Daturpalli, S.; Jackson, S.E.; Agard, D.; et al. Hsp70 forms antiparallel dimers stabilized by post-translational modifications to position clients for transfer to Hsp90. Cell Rep. 2015, 11, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Sarbeng, E.B.; Liu, Q.; Tian, X.; Yang, J.; Li, H.; Wong, J.L.; Zhou, L.; Liu, Q. A functional DnaK dimer is essential for the efficient interaction with heat shock protein 40 kDa (Hsp40). J. Biol. Chem. 2015, 290, 8849–8862. [Google Scholar] [CrossRef] [PubMed]
- Trcka, F.; Durech, M.; Vankova, P.; Chmelik, P.; Martinkova, V.; Hausner, J.; Kadek, A.; Marcoux, J.; Klumpler, T.; Vojtesek, B.; et al. Human stress inducible Hsp70 has a high propensity to form ATP-dependent antiparallel dimers that are differentially regulated by cochaperone binding. Mol. Cell Proteomics 2019, 18, 320–337. [Google Scholar] [PubMed]
- Schönfeld, H.J.; Schmidt, D.; Schröder, H.; Bukau, B. The DnaK chaperone system of Escherichia coli: Quaternary structures and interactions of the DnaK and GrpE components. J. Biol. Chem. 1995, 270, 2183–2189. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.C.; Forouhar, F.; Yeh, Y.H.; Shr, H.L.; Wang, C.; Hsiao, C.D. Crystal structure of the C-terminal 10-kDa subdomain of Hsc70. J. Biol. Chem. 2003, 278, 30311–30316. [Google Scholar] [CrossRef] [PubMed]
- Marcion, G.; Seigneuric, R.; Chavanne, E.; Artur, Y.; Briand, L.; Hadi, T.; Gobbo, J.; Garrido, C.; Neiers, F. C-terminal amino acids are essential for human heat shock protein 70 dimerization. Cell Stress Chaperones 2015, 20, 61–72. [Google Scholar] [CrossRef]
- Nemoto, T.K.; Fukuma, Y.; Itoh, H.; Takagi, T.; Ono, T. A disulfide bridge mediated by cysteine 574 is formed in the dimer of the 70-kDa heat shock protein. J. Biochem. 2006, 139, 677–687. [Google Scholar] [CrossRef]
- Liu, Q.; Li, H.; Yang, Y.; Tian, X.; Su, J.; Zhou, L.; Liu, Q. A disulfide bonded DnaK dimer is maintained in an ATP-bound state. Cell Stress Chaperones 2017, 22, 201–212. [Google Scholar] [CrossRef]
- Blamowska, M.; Neupert, W.; Hell, K. Biogenesis of the mitochondrial Hsp70 chaperone. J. Cell Biol. 2012, 199, 125–135. [Google Scholar] [CrossRef] [Green Version]
- Bauer, D.; Merz, D.R.; Pelz, B.; Theisen, K.E.; Yacyshyn, G.; Mokranjac, D.; Dima, R.I.; Rief, M.; Žoldák, G. Nucleotides regulate the mechanical hierarchy between subdomains of the nucleotide binding domain of the Hsp70 chaperone DnaK. Proc. Natl. Acad. Sci. USA 2015, 112, 10389–10394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klucken, J.; Shin, Y.; Masliah, E.; Hyman, B.T.; McLean, P.J. Hsp70 reduces alpha-synuclein aggregation and toxicity. J Biol Chem. 2004, 279, 25497–25502. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.G.; Chang, L.; Gestwicki, J.E. Heat shock protein 70 (hsp70) as an emerging drug target. J. Med. Chem. 2010, 53, 4585–4602. [Google Scholar] [CrossRef] [PubMed]
- Shonhai, A. The role of Hsp70s in the development and pathogenicity of Plasmodium species. In Heat Shock Proteins of Malaria; Shonhai, A., Blatch, G., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 47–69. [Google Scholar]
- Zininga, T.; Shonhai, A. Are heat shock proteins druggable candidates? Am. J. Biochem. Biotechnol. 2014, 10, 208–210. [Google Scholar] [CrossRef]
- Hendriks, L.E.; Dingemans, A.M.C. Heat shock protein antagonists in early stage clinical trials for NSCLC. Expert Opin. Inv. Drugs. 2017, 26, 541–550. [Google Scholar] [CrossRef]
- Chatterjee, S.; Burns, T. Targeting heat shock proteins in cancer: A promising therapeutic approach. Int. J. Mol. Sci. 2017, 18, 1978. [Google Scholar] [CrossRef]
- Daniyan, M.; Pryzyboski, J.; Shonhai, A. Partners in mischief: Functional networks of Heat shock proteins of Plasmodium falciparum and their influence on parasite virulence. Biomolecules 2019, 9, 295. [Google Scholar] [CrossRef]
- Chakafana, G.; Zininga, T.; Shonhai, A. Comparative structure function features of Hsp70s of Plasmodium falciparum and human origins. Biophys. Rev. 2019, 11, 591–602. [Google Scholar] [CrossRef]






| Cytosolic Hsp70s * | E.R. Localized Grp78 | Cytosolic Hsp110s | Grp170s | |
|---|---|---|---|---|
| First Section | G384 (non-polar, hydrophobic) | G (non-polar, hydrophobic) | P (non-polar, hydrophobic) | K (polar, positively charged, hydrophilic) |
| D385 (polar, negatively charged, hydrophilic) | D (polar, negatively charged, hydrophilic) | K (polar, positively charged, hydrophilic) | S (polar, uncharged, hydrophilic) | |
| V386 (non-polar, hydrophobic) | G (non-polar, hydrophobic) | V (non-polar, hydrophobic) | E (polar, negatively charged, hydrophilic) | |
| Eukaryotic Insertion | K (polar, positively charged, hydrophilic) | - | - | - |
| S (polar, uncharged, hydrophilic) | E (polar, negatively charged, hydrophilic) | - | - | |
| E (polar, negatively charged, hydrophilic) | D (polar, negatively charged, hydrophilic) | A (non-polar, hydrophobic) | E (polar, negatively charged, hydrophilic) | |
| N (polar, uncharged, hydrophilic) | T (polar, uncharged, hydrophilic) | F (non-polar, hydrophobic) | V (non-polar, hydrophobic) | |
| Hinge | Q387 (polar, uncharged, hydrophilic) | G (non-polar, hydrophobic) | K (polar, positively charged, hydrophilic) | K (polar, positively charged, hydrophilic) |
| Second Section | D388 (polar, negatively charged, hydrophilic) | D (polar, negatively charged, hydrophilic) | E (polar, negatively charged, hydrophilic) | D (polar, negatively charged, hydrophilic) |
| L389 (non-polar, hydrophobic) | L (non-polar, hydrophobic) | F (non-polar, hydrophobic) | F (non-polar, hydrophobic) | |
| L390 (non-polar, hydrophobic) | V (non-polar, hydrophobic) | S (polar, uncharged, hydrophilic) | L (non-polar, hydrophobic) | |
| Hinge | L391 (non-polar, hydrophobic) | L (non-polar, hydrophobic) | V (non-polar, hydrophobic) | V (non-polar, hydrophobic) |
| Third Section | L392 (non-polar, hydrophobic) | L (non-polar, hydrophobic) | T (polar, uncharged, hydrophilic) | L (non-polar, hydrophobic) |
| D393 (polar, negatively charged, hydrophilic) | D (polar, negatively charged, hydrophilic) | D (polar, negatively charged, hydrophilic) | D (polar, negatively charged, hydrophilic) | |
| V394 (non-polar, hydrophobic) | V (non-polar, hydrophobic) | G (non-polar, hydrophobic) | V (non-polar, hydrophobic) |
| Nucleotide | Linker Residues | Contact Residues [Domain] | H Bond Length/Å |
|---|---|---|---|
| ATP | D388 (O) | K214 (N) [NBD] | 3.078 |
| L390 (N) | K214 (O) [NBD] | 2.761 | |
| L390 (O) | F216 (N) [NBD] | 2.797 | |
| L392 (N) | F216 (O) [NBD] | 2.894 | |
| L392 (O) | V218 (N) [NBD] | 2.980 | |
| D393 (O) | I418 (N) [SBD] | 2.913 | |
| D393 (O.D) | V394(N) [linker] | 2.794 | |
| ADP | L392 (O) | T417 (H) [SBD] | 1.771 |
| V394 (N) | D415 (O) [SBD] | 2.593 |
| Bound Nucleotide/Substrate | Subdomain Conformation | Overall Subdomain Dynamics | ||||||
|---|---|---|---|---|---|---|---|---|
| NBD | SBD | Nucleotide binding cleft | Linker binding cleft | Linker | SBD | Lid | NBD | SBD |
| ATP | - | closed | open | docked | docked | released | rigid | flexible(a) |
| ADP | - | open | closed | mobile | undocked | docked | flexible | rigid(b) |
| - | peptide | open | closed | mobile | undocked | docked | flexible | rigid(c) |
| ADP | peptide | open | closed | mobile | undocked | docked | flexible | flexible(d) |
| ATP | peptide | closed | open | docked | docked | released | rigid | flexible(e) |
| - | - | open | closed | mobile | undocked | released | flexible | flexible(f) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakafana, G.; Zininga, T.; Shonhai, A. The Link That Binds: The Linker of Hsp70 as a Helm of the Protein’s Function. Biomolecules 2019, 9, 543. https://doi.org/10.3390/biom9100543
Chakafana G, Zininga T, Shonhai A. The Link That Binds: The Linker of Hsp70 as a Helm of the Protein’s Function. Biomolecules. 2019; 9(10):543. https://doi.org/10.3390/biom9100543
Chicago/Turabian StyleChakafana, Graham, Tawanda Zininga, and Addmore Shonhai. 2019. "The Link That Binds: The Linker of Hsp70 as a Helm of the Protein’s Function" Biomolecules 9, no. 10: 543. https://doi.org/10.3390/biom9100543
APA StyleChakafana, G., Zininga, T., & Shonhai, A. (2019). The Link That Binds: The Linker of Hsp70 as a Helm of the Protein’s Function. Biomolecules, 9(10), 543. https://doi.org/10.3390/biom9100543





