SUMOylation Protects FASN Against Proteasomal Degradation in Breast Cancer Cells Treated with Grape Leaf Extract
Abstract
:1. Background
2. Methods
2.1. Cell Culture and Treatments
2.2. Human Breast Tissue Specimens
2.3. Plant Material and Extraction Procedure
2.4. Phenolic Characterization of Grapevine Extract
2.4.1. HPLC-UV Analysis
2.4.2. LC-HRMS Analysis
2.5. Western Blot and Co-Immunoprecipitation (Co-IP)
2.6. Real-Time PCR (RT-PCR) Analysis
2.7. Cell Viability
2.8. Flow Cytometry Assay
2.9. Clonogenicity Assay Analysis
2.10. Proteomic Sample Preparation
2.11. LC–MS/MS Settings
2.12. Preparative Data Analysis and Peptide Identification Search
2.13. Protein Stability Assay and Half-Life Determination
2.14. Proteasomal Activity Assays
2.15. RNA Interference
2.16. Statistical Analysis
3. Results
3.1. Polyphenol Phytochemical Composition
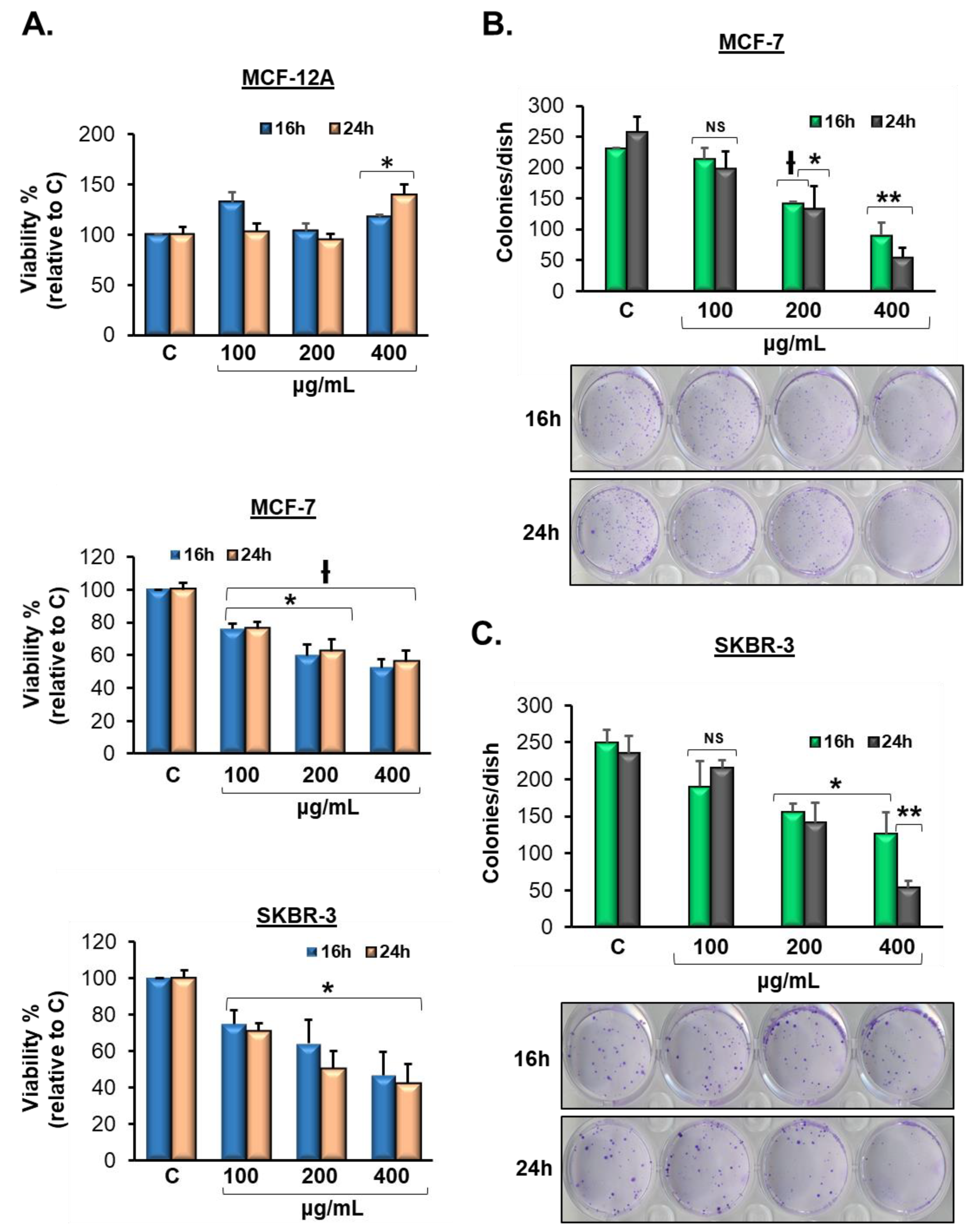
3.2. Vermentino Extract Lowers Cell Viability in Breast Cancer Cell Lines
3.3. Vermentino Extract inhibits Clonogenicity in Breast Cancer Cell Lines
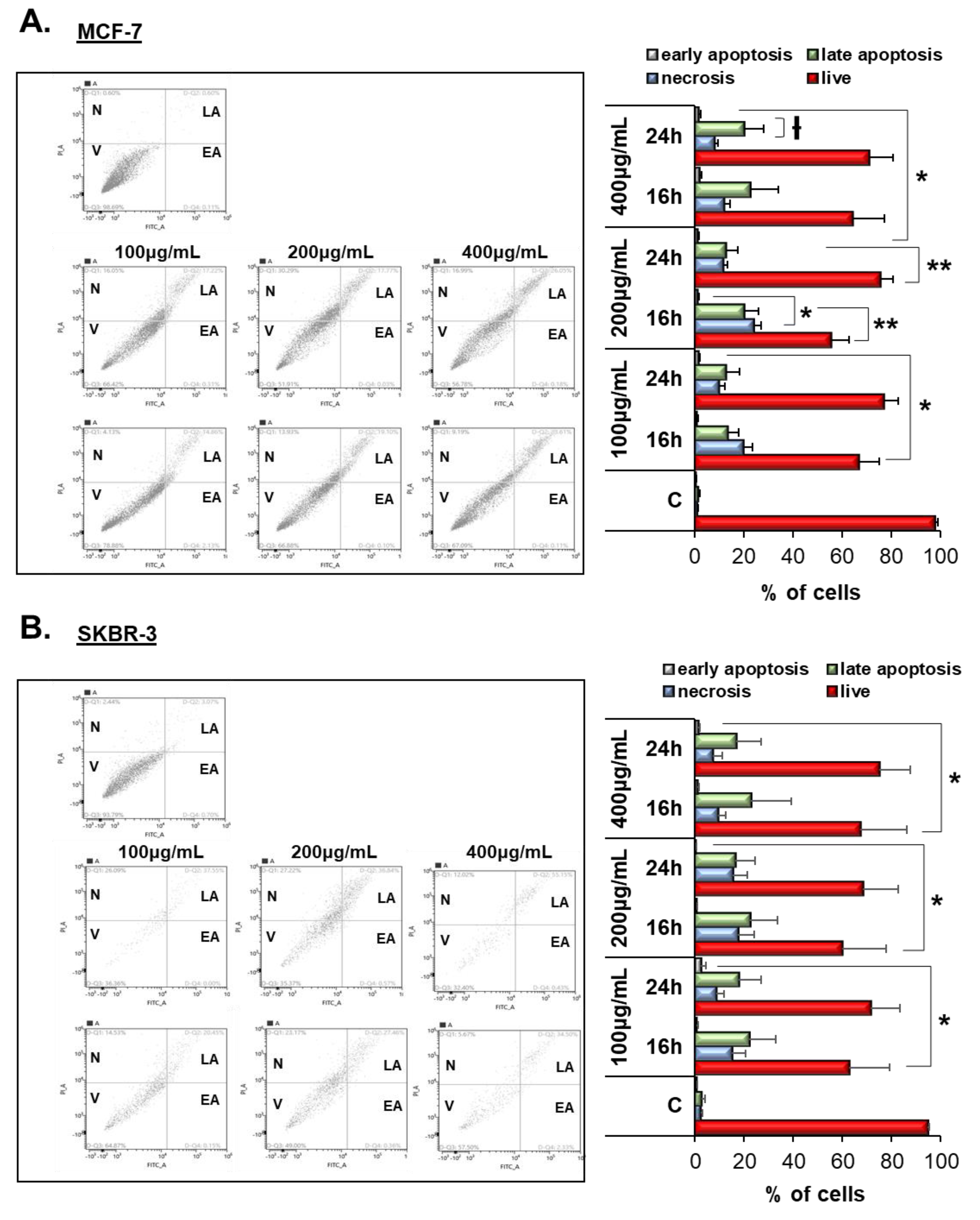
3.4. Vermentino Extract Induced Late Apoptosis/Necrosis in Breast Cancer Cell Lines
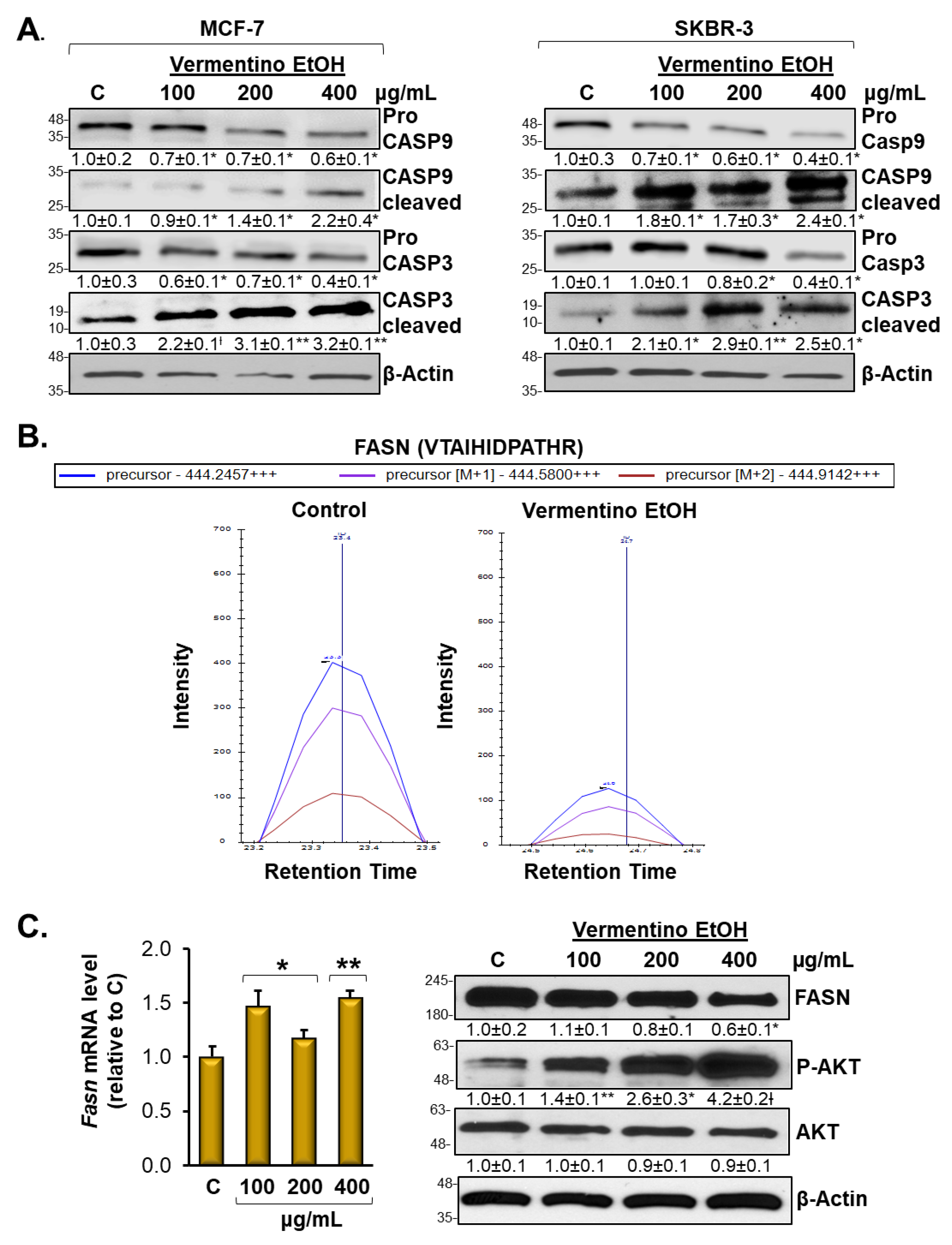
3.5. Vermentino Extract Induced CASP-9 and CASP-3 in Breast Cancer Cell Lines
3.6. Vermentino Extract Lowers FASN Protein Level and Promoted AKT1 Signaling in Breast Cancer Cell Lines
3.7. Vermentino Extract Lowered FASN Protein Expression by Activating Proteasomal Degradation
3.8. Effects of Vermentino Extract on Proteasomal Activity
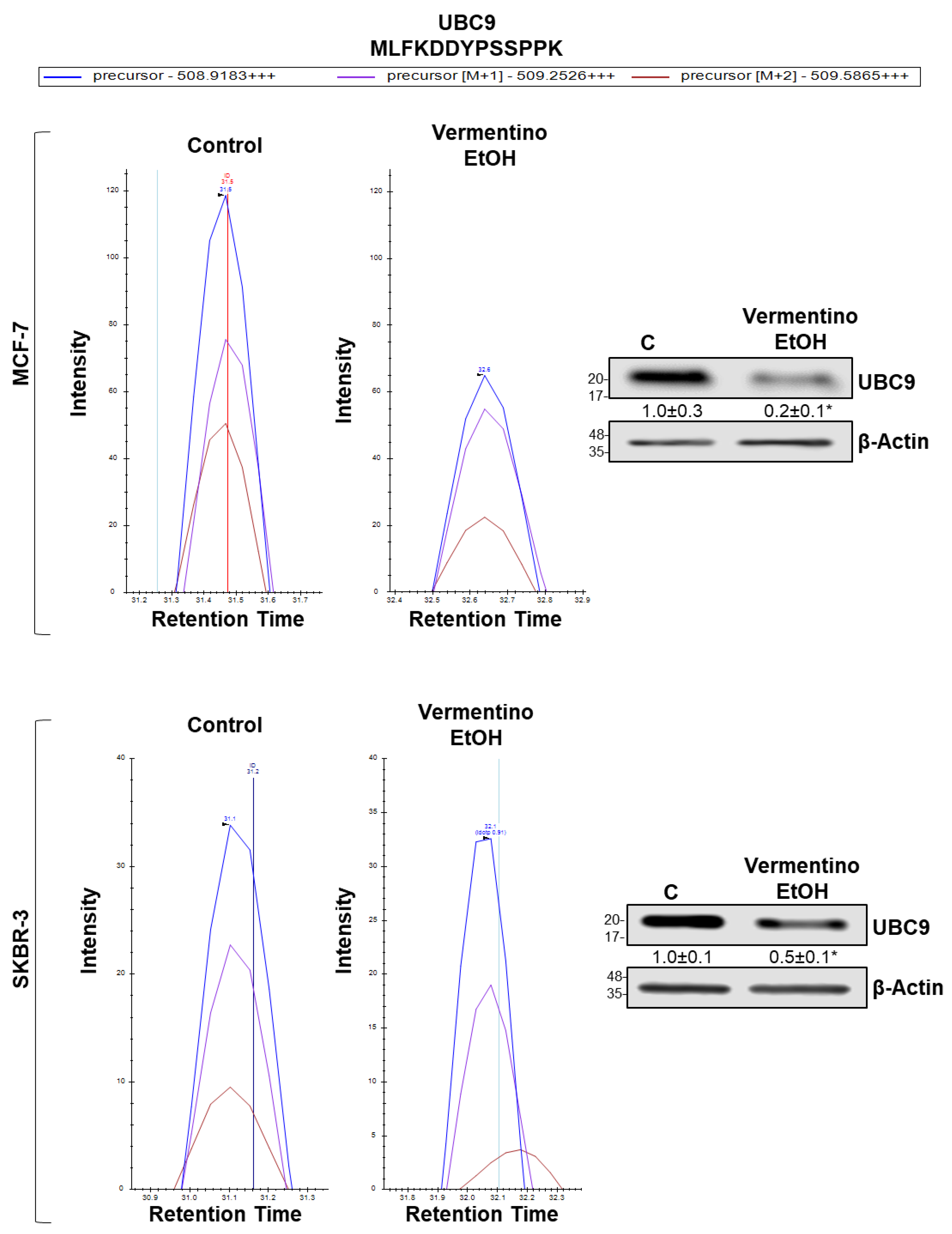
3.9. Vermentino Hydroalcoholic Extract Lowered UBC9 Protein Level in Human Breast Cancer Cell Lines
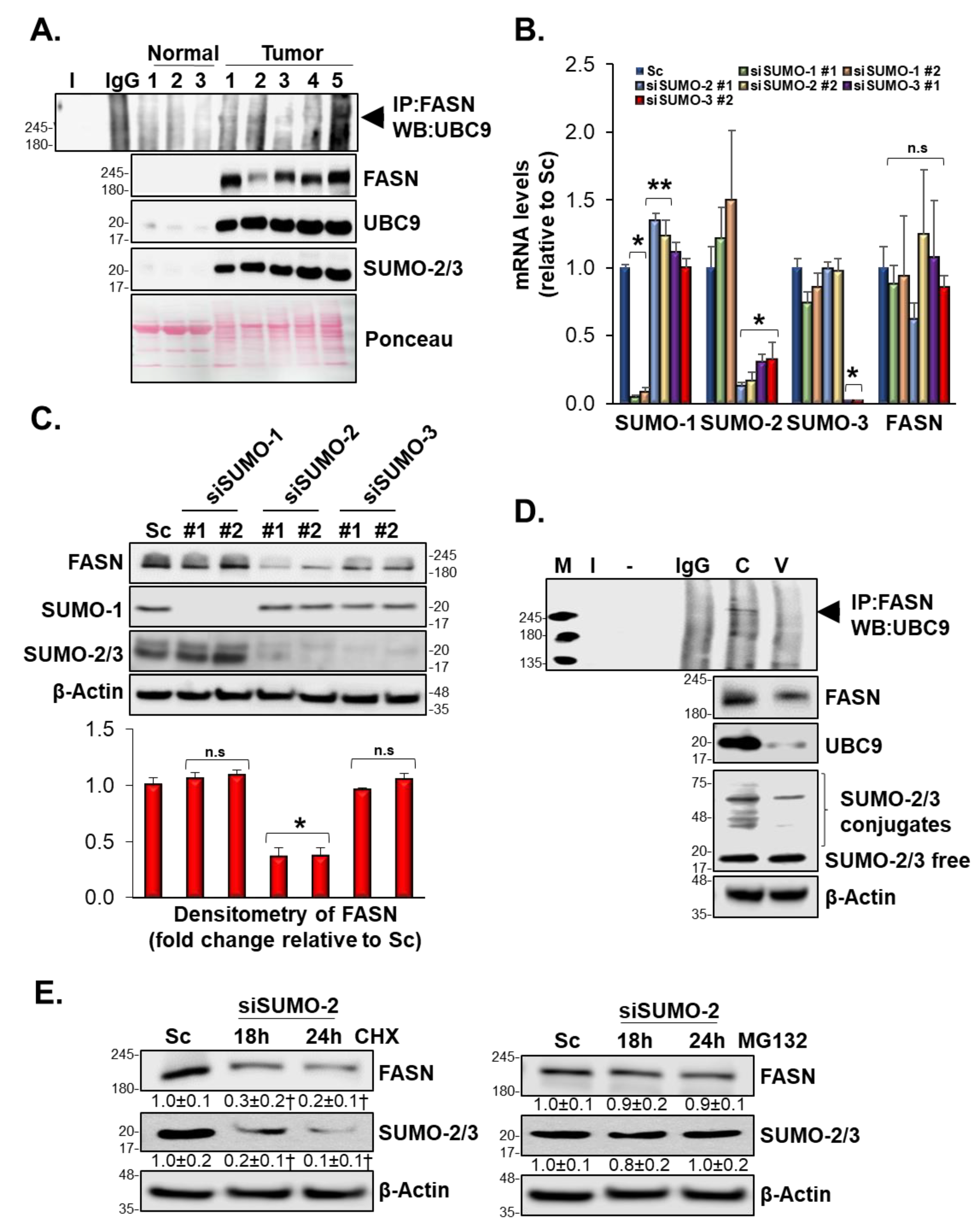
3.10. SUMOylation Mediated FASN Protein Stability
4. Discussion
5. Conclusions
Availability of Data and Materials
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| FASN | fatty-acid synthase |
| Acetyl-CoA | acetyl coenzyme A |
| Malonyl-CoA | malonic acid |
| SREBF1 | sterol regulatory element binding transcription factor 1 |
| SREBP1c | sterol regulatory element binding protein 1c |
| AKT | protein kinase B |
| CHX | cycloheximide |
| HPLC-UV | high performance liquid chromatography ultraviolet |
| LC–MS | liquid chromatography–mass spectrometry |
| LC-HRMS | liquid chromatography high resolution mass spectrometry |
| MS | mass spectrometry |
| FWHM | full width at half maximum |
| IT | injection time |
| PBS | phosphate-buffered saline |
| RIPA | radioimmunoprecipitation assay buffer |
| SUMO | small ubiquitin-like modifier |
| UBC9 | Ubiquitin-conjugating enzyme 9 |
| CASP-3 | caspase-3 |
| CASP-9 | caspase-9 |
| PCR | polymerase chain reaction |
| HPRT1 | hypoxanthine phosphoribosyl-transferase |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| V-FITC | annexin V fluorescein isothiocyanate |
| PI | propidium iodide |
| FASTA | text-based format for representing protein sequences |
| Chym-L | chymotrypsin-like |
| Tr-L | trypsin-like |
| Casp-L | caspase-like |
| siRNA | small interfering RNA |
| M | precursor isotopic |
| PI3K | phosphoinositide 3-kinase |
| HSP90 | heat shock protein 90 |
| TRAP-1 | tumor necrosis factor receptor associated protein 1 |
References
- Pilleron, S.; Sarfati, D.; Janssen-Heijnen, M.; Vignat, J.; Ferlay, J.; Bray, F.; Soerjomataram, I. Global cancer incidence in older adults, 2012 and 2035: A population-based study. Int. J. Cancer 2019, 144, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Guarneri, V.; Conte, P. Metastatic breast cancer: Therapeutic options according to molecular subtypes and prior adjuvant therapy. Oncologist 2009, 14, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Giró-Perafita, A.; Sarrats, A.; Pérez-Bueno, F.; Oliveras, G.; Buxó, M.; Brunet, J.; Viñas, G.; Miquel, T.P. Fatty acid synthase expression and its association with clinico-histopathological features in triple-negative breast cancer. Oncotarget 2017, 8, 74391–74405. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Tu, Y.; Simpson, P.J.; Kuhajda, F.P. Malonyl-CoA decarboxylase inhibition is selectively cytotoxic to human breast cancer cells. Oncogene 2009, 28, 2979–2987. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Huang, W.; Zheng, M.; Xue, Y.; Yang, J.; Liu, W.; Han, S. The mTOR inhibitor rapamycin synergizes with a fatty acid synthase inhibitor to induce cytotoxicity in ER/HER2-positive breast cancer cells. PLoS ONE 2014, 9, e97697. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Feng, D.; Wang, Q. Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP1. J. Clin. Investig. 2012, 122, 2417–2427. [Google Scholar] [CrossRef]
- Wang, H.; Luo, Q.F.; Peng, A.F.; Long, X.H.; Wang, T.F.; Liu, Z.L.; Zhang, G.M.; Zhou, R.P.; Gao, S.; Zhou, Y.; et al. Positive feedback regulation between Akt phosphorylation and fatty acid synthase expression in osteosarcoma. Int. J. Mol. Med. 2014, 33, 633–639. [Google Scholar] [CrossRef] [Green Version]
- Filaire, E.; Dupuis, C.; Galvaing, G.; Aubreton, S.; Laurent, H.; Richard, R.; Filaire, M. Lung cancer: What are the links with oxidative stress, physical activity and nutrition. Lung Cancer 2013, 82, 383–389. [Google Scholar] [CrossRef]
- Tan, B.L.; Norhaizan, M.E.; Chan, L.C. ROS-Mediated Mitochondrial Pathway is Required for Manilkara zapota (L.) P. Royen Leaf Methanol Extract Inducing Apoptosis in the Modulation of Caspase Activation and EGFR/NF-κB Activities of HeLa Human Cervical Cancer Cells. Evid. Based Complement. Altern. Med. 2018, 2018, 6578648. [Google Scholar] [CrossRef]
- Sathishkumar, P.; Gu, F.L.; Zhan, Q.; Palvannan, T.; Mohd Yusoff, A.R. Flavonoids mediated “Green nanomaterials: A novel nanomedicine system to treat various disease-curent trends and future perspective. Matter. Lett. 2018, 210, 26–30. [Google Scholar] [CrossRef]
- Govindaraju, K.; Ingels, A.; Hasan, M.N.; Sun, D.; Mathieu, V.; Masi, M.; Evidente, A.; Kornienko, A. Synthetic analogues of the montanine-type alkaloids with activity against apoptosis-resistant cancer cells. Bioorg. Med. Chem. Lett. 2018, 28, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.R.; Zhou, W.; Zhang, H.M.; Guo, Q.S.; Yang, W.; Li, B.J.; Sun, Z.H.; Gao, S.H.; Cui, R.J. Modulation of Multiple Signaling Pathways of the Plant-Derived Natural Products in Cancer. Front. Oncol. 2019, 9, 1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Xiao, B.; Hou, L.; Zhou, G.; Mo, B.; Yao, D. Bioactive molecule Icariin inhibited proliferation while enhanced apoptosis and autophagy of rat airway smooth muscle cells in vitro. Cytotechnology 2019, 71, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Abd El Maksoud, A.I.; Taher, R.F.; Gaara, A.H.; Abdelrazik, E.; Keshk, O.S.; Elawdan, K.A.; Morsy, S.E.; Salah, A.; Khalil, H. Selective Regulation of B-Raf Dependent K-Ras/Mitogen-Activated Protein by Natural Occurring Multi-kinase Inhibitors in Cancer Cells. Front. Oncol. 2019, 9, 1220. [Google Scholar] [CrossRef] [PubMed]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Xu, J.; Gong, X.; Yang, M.; Zhang, C.; Li, M. Biosynthesis, Chemistry, and Pharmacology of Polyphenols from Chinese Salvia Species: A Review. Molecules 2019, 24, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferhi, S.; Santaniello, S.; Zerizer, S.; Cruciani, S.; Fadda, A.; Sanna, D.; Dore, A.; Maioli, M.; D’hallewin, G. Total Phenols from Grape Leaves Counteract Cell Proliferation and Modulate Apoptosis-Related Gene Expression in MCF-7 and HepG2 Human Cancer Cell Lines. Molecules 2019, 24, 612. [Google Scholar] [CrossRef] [Green Version]
- Barberis, A.; Spissu, Y.; Fadda, A.; Azara, E.; Serra, P.A. Simultaneous amperometric detection of ascorbic acid and antioxidant capacity in orange, blueberry and kiwi juice, by a telemetric system coupled with a fullerene- or nanotubesmodified ascorbate subtractive biosensor. Biosens. Bioelectron. 2015, 6715, 214–223. [Google Scholar] [CrossRef]
- Pascale, R.M.; Simile, M.M.; De Miglio, M.R.; Muroni, M.R.; Calvisi, D.F.; Asara, G.; Casabona, D.; Frau, M.; Seddaiu, M.A.; Feo, F. Cell cycle deregulation in liver lesions of rats with and without genetic predisposition to hepatocarcinogenesis. Hepatology 2002, 35, 1341–1350. [Google Scholar] [CrossRef]
- Robertson, C.; Ronald, C.B. TANDEM: Matching proteins with mass spectra. Bioinformatics 2004, 20, 1466–1467. [Google Scholar]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Vitek, O.; Nesvizhskii, A.I. A statistical model-building perspective to identification of MS/MS spectra with PeptideProphet. BMC Bioinformatics. 2012, 13, S1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar]
- Schilling, B.; Rardin, M.J.; MacLean, B.X.; Zawadzka, A.M.; Frewen, B.E.; Cusack, M.P.; Sorensen, D.J.; Bereman, M.S.; Jing, E.; Wu, C.C.; et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: Application to protein acetylation and phosphorylation. Mol. Cell. Proteom. 2012, 11, 202–214. [Google Scholar] [CrossRef] [Green Version]
- Choi, M.; Chang, C.Y.; Clough, T.; Broudy, D.; Killeen, T.; MacLean, B.; Vitek, O. MSstats: An R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 2014, 30, 2524–2526. [Google Scholar] [CrossRef] [Green Version]
- Lou, Y.; Yao, J.; Zereshki, A.; Dou, Z.; Ahmed, K.; Wang, H.; Hu, J.; Wang, Y.; Yao, X. NEK2A interacts with MAD1 and possibly functions as a novel integrator of the spindle checkpoint signaling. J. Biol. Chem. 2004, 279, 20049–20057. [Google Scholar] [CrossRef] [Green Version]
- Vizcaíno, J.A.; Deutsch, E.W.; Wang, R.; Csordas, A.; Reisinger, F.; Ríos, D.; Dianes, J.A.; Sun, Z.; Farrah, T.; Bandeira, N.; et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014, 32, 223–226. [Google Scholar] [CrossRef]
- Kang, B.H.; Plescia, J.; Dohi, T.; Rosa, J.; Doxsey, S.J.; Altieri, D.C. Regulation of tumor cell mitochondrial homeostasis by an organelle-specific Hsp90 chaperone network. Cell 2007, 131, 257–270. [Google Scholar] [CrossRef]
- Matassa, D.S.; Agliarulo, I.; Avolio, R.; Landriscina, M.; Esposito, F. TRAP1 Regulation of Cancer Metabolism: Dual Role as Oncogene or Tumor Suppressor. Genes (Basel) 2018, 9, E195. [Google Scholar] [CrossRef] [Green Version]
- Johnson, E.S.; Blogel, G. Ubc9 is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 1997, 272, 26799–26802. [Google Scholar] [CrossRef] [Green Version]
- Manza, L.L.; Codreanu, S.G.; Stamer, S.L.; Smith, D.L.; Wells, K.S.; Roberts, R.L.; Liebler, D.C. Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem. Res. Toxicol. 2004, 17, 1706–1715. [Google Scholar] [CrossRef] [PubMed]
- Rabellino, A.; Khanna, K.K. The implication of the SUMOylation pathway in breast cancer pathogenesis and treatment. Crit. Rev. Biochem. Mol. Biol. 2020, 18, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhang, Y.; Jiang, X.; Zhang, H.; Gao, Z.; Li, Y.; Fu, R.; Li, L.; Li, J.; Cui, H.; et al. ROS-mediated activation and mitochondrial translocation of CaMKII contributes to Drp1-dependent mitochondrial fission and apoptosis in triple-negative breast cancer cells by isorhamnetin and chloroquine. J. Exp. Clin. Cancer Res. 2019, 38, 225. [Google Scholar] [CrossRef] [PubMed]
- Tor, Y.S.; Yazan, L.S.; Foo, J.B.; Wibowo, A.; Ismail, N.; Cheah, Y.K.; Abdullah, R.; Ismail, M.; Ismail, I.S.; Yeap, S.K. Induction of apoptosis in MCF-7 via oxidative stress generation, mitochondria-dependent and caspase-indipendent pathway by ethyl acetate extract of Dillenia sufruticosa and its chemical profile. PLoS ONE 2015, 10, e0127441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomig, T.H.B.; Cavalli, I.J.; Souza, R.L.R.; Lucena, A.C.R.; Batista, M.; Machado, K.C.; Marchini, F.K.; Marchi, F.A.; Lima, R.S.; Urban, C.A.; et al. High-throughput mass spectrometry and bioinformatics analysis of breast cancer proteomic data. Data Brief. 2019, 25, 104125. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Deng, R.; Zhu, H.H.; Zhang, S.S.; Zhu, C.; Montminy, M.; Davis, R.; Feng, G.S. Modulation of Fatty Acid Synthase degradation by concerted action of p38 MAP kinase, E3 ligase COP1, and SH2Tyrosine Phosphatase Shp2. J. Biol. Chem. 2013, 288, 3823–3830. [Google Scholar] [CrossRef] [Green Version]
- Bueno, M.J.; Jimenez-Renard, V.; Samino, S.; Capellades, J.; Junza, A.; López-Rodríguez, M.L.; Garcia-Carceles, J.; Lopez-Fabuel, I.; Bolaños, J.P.; Chandel, N.S.; et al. Essentiality of fatty acid synthase in the 2D to anchorage-independent growth transition in transforming cells. Nat. Commun. 2019, 10, 5011. [Google Scholar] [CrossRef]
- Schley, P.D.; Jijon, H.B.; Robinson, L.E.; Field, C.J. Mechanisms of omega-3 fatty acid-induced growth inhibition in MDA-MB-231 human breast cancer cells. Breast Cancer Res. Treat. 2005, 92, 187–195. [Google Scholar] [CrossRef]
- Lee, J.S.; Sul, J.Y.; Park, J.B. Fatty acid synthase inhibition by amentoflavone suppresses HER2/neu (erbB2) oncogene in SKBR3 human breast cancer cells. Phytother. Res. 2013, 27, 713–720. [Google Scholar] [CrossRef]
- Adedara, I.A.; Olabiyi, B.F.; Idris, U.F.; Onibiyo, E.M.; Farombi, E.O. Taurine reverse sodium fluoride-mediated increase in inflammation, caspase-3 activity, and oxidative damage along the brain-pituitary-gonad axis in male rats. Can. J. Physiol. Pharm. 2017, 95, 1019–1029. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, H.; Liu, Y. Anti-Inflammatory and Apoptotic Signaling Effect of Fucoxanthin on Benzo(A)PyreneInduced Lung Cancer in Mice. J. Environ. Pathol. Toxicol. Oncol. 2019, 38, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Baumann, J.; Wong, J.; Sun, Y.; Conklin, D.S. Palmitate-induced ER stress increases trastuzumab sensitivity in HER2/neupositive breast cancer cells. BMC Cancer 2016, 16, 551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menendez, J.A.; Lupu, R. Fatty acid synthase regulates estrogen receptor-α signaling in breast cancer cells. Oncogenesis 2017, 6, e299. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Fang, S.; Chen, Y.; Yang, Z.; Yuan, Y.; Zhang, J.; Ye, L.; Gu, W. Inhibition of FASN suppresses the malignant biological behavior of non-small cell lung cancer cells via deregulating glucose metabolism and AKT/ERK pathway. Lipids Health Dis. 2019, 18, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Hu, J.; Sheng, L.; Yuan, M.; Wu, Y.; Chen, L.; Wang, G.; Qiu, Z. Ellagic acid ameliorates AKT-driven hepatic steatosis in mice by suppressing de novo lipogenesis via the AKT/SREBP-1/FASN pathway. Food Funct. 2019, 10, 34103420. [Google Scholar] [CrossRef] [PubMed]
- Gardner, R.C.; Assinder, S.J.; Christie, G. Characterization of peptidyl boronic acid inhibitors of mammalian 20 S and 26 S proteasomes and their inhibition of proteasomes in cultured cells. Biochem. J. 2000, 346, 447–454. [Google Scholar] [CrossRef]
- Little, J.L.; Wheeler, F.B.; Koumenis, C.; Kridel, S.J. Disruption of Crosstalk between the Fatty Acid Synthesis and Proteasome Pathways Enhances Unfolded Protein Response Signaling and Cell Death. Mol. Cancer Ther. 2008, 7, 3816–3824. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, S.; Tsutsumi, S.; Muhlebach, G.; Sourbier, C.; Lee, M.J.; Lee, S.; Vartholomaiou, E.; Tatokoro, M.; Beebe, K.; Miyajima, N. Molecular chaperone TRAP1 regulates a metabolic switch between mitochondrial respiration and aerobic glycolysis. Proc. Natl. Acad. Sci. USA 2013, 110, 1604–1612. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Tan, M.; Cai, Q. The Warburg effect in tumor progression: Mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. 2015, 356, 156–164. [Google Scholar] [CrossRef] [Green Version]
- Romanenko, A.M.; Kinoshita, A.; Wanibuchi, H.; Wei, M.; Zaparin, W.K.; Vinnichenko, W.I.; Vozianov, A.F.; Fukushima, S. Involvement of ubiquitination and sumoylation in bladder lesions induced by persistent long-term low dose ionizing radiation in humans. J. Urol. 2006, 175, 739–743. [Google Scholar] [CrossRef]

| Compound | Vermentino (EtOH/H20) |
|---|---|
| Myricetin 3-O glucoside | 0.11 ± 0.01 |
| Eridictyol 7-glucoside | 0.25 ± 0.03 |
| Quercetin 3-O rutinoside | 0.20 ± 0.02 |
| Quercetin 3-O galactoside | 1.29 ± 0.06 |
| Quercetin 3-O glucoside | 5.92 ± 0.10 |
| Kaempferol 3-O galactoside | 0.68 ± 0.03 |
| Kaempferol 3-O rutinoside | 0.08 ± 0.01 |
| Kaempferol 3-O glucoside | 1.64 ± 0.07 |
| Quercetin 3-O-(6 acetyl) glucoside | 0.19 ± 0.03 |
| Isorhamnetin glucoside | 8.00 ± 0.22 |
| Total | 18.36 |
| Position | Sequence | JASSA | SUMOsp | SUMOgo |
|---|---|---|---|---|
| K706 | QELKKVIREPKPRSARWLSTS | X | X | |
| K786 | KPSCTIIPLMKKDHRDNLEFF | X | X | X |
| K835 | RGTPLISPLIKWDHSLAWDVP | X | X | X |
| K1523 | AFRHFLLEEDKPEEPTAHAFV | X | X | X |
| K1752 | DLVLNSLAEEKLQASVRCLAT | X | X | |
| K2206 | EASELACPTPKEDGLAQQQTQ | X | X | X |
| K2471 | DYNLSQVCDGKVSVHVIEGDH | X | X |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Floris, A.; Mazarei, M.; Yang, X.; Robinson, A.E.; Zhou, J.; Barberis, A.; D’hallewin, G.; Azara, E.; Spissu, Y.; Iglesias-Ara, A.; et al. SUMOylation Protects FASN Against Proteasomal Degradation in Breast Cancer Cells Treated with Grape Leaf Extract. Biomolecules 2020, 10, 529. https://doi.org/10.3390/biom10040529
Floris A, Mazarei M, Yang X, Robinson AE, Zhou J, Barberis A, D’hallewin G, Azara E, Spissu Y, Iglesias-Ara A, et al. SUMOylation Protects FASN Against Proteasomal Degradation in Breast Cancer Cells Treated with Grape Leaf Extract. Biomolecules. 2020; 10(4):529. https://doi.org/10.3390/biom10040529
Chicago/Turabian StyleFloris, Andrea, Michael Mazarei, Xi Yang, Aaron Elias Robinson, Jennifer Zhou, Antonio Barberis, Guy D’hallewin, Emanuela Azara, Ylenia Spissu, Ainhoa Iglesias-Ara, and et al. 2020. "SUMOylation Protects FASN Against Proteasomal Degradation in Breast Cancer Cells Treated with Grape Leaf Extract" Biomolecules 10, no. 4: 529. https://doi.org/10.3390/biom10040529
APA StyleFloris, A., Mazarei, M., Yang, X., Robinson, A. E., Zhou, J., Barberis, A., D’hallewin, G., Azara, E., Spissu, Y., Iglesias-Ara, A., Orrù, S., & Tomasi, M. L. (2020). SUMOylation Protects FASN Against Proteasomal Degradation in Breast Cancer Cells Treated with Grape Leaf Extract. Biomolecules, 10(4), 529. https://doi.org/10.3390/biom10040529








