Standartox: Standardizing Toxicity Data
Abstract
:1. Summary
2. Data Description
2.1. Filters
2.2. Aggregation
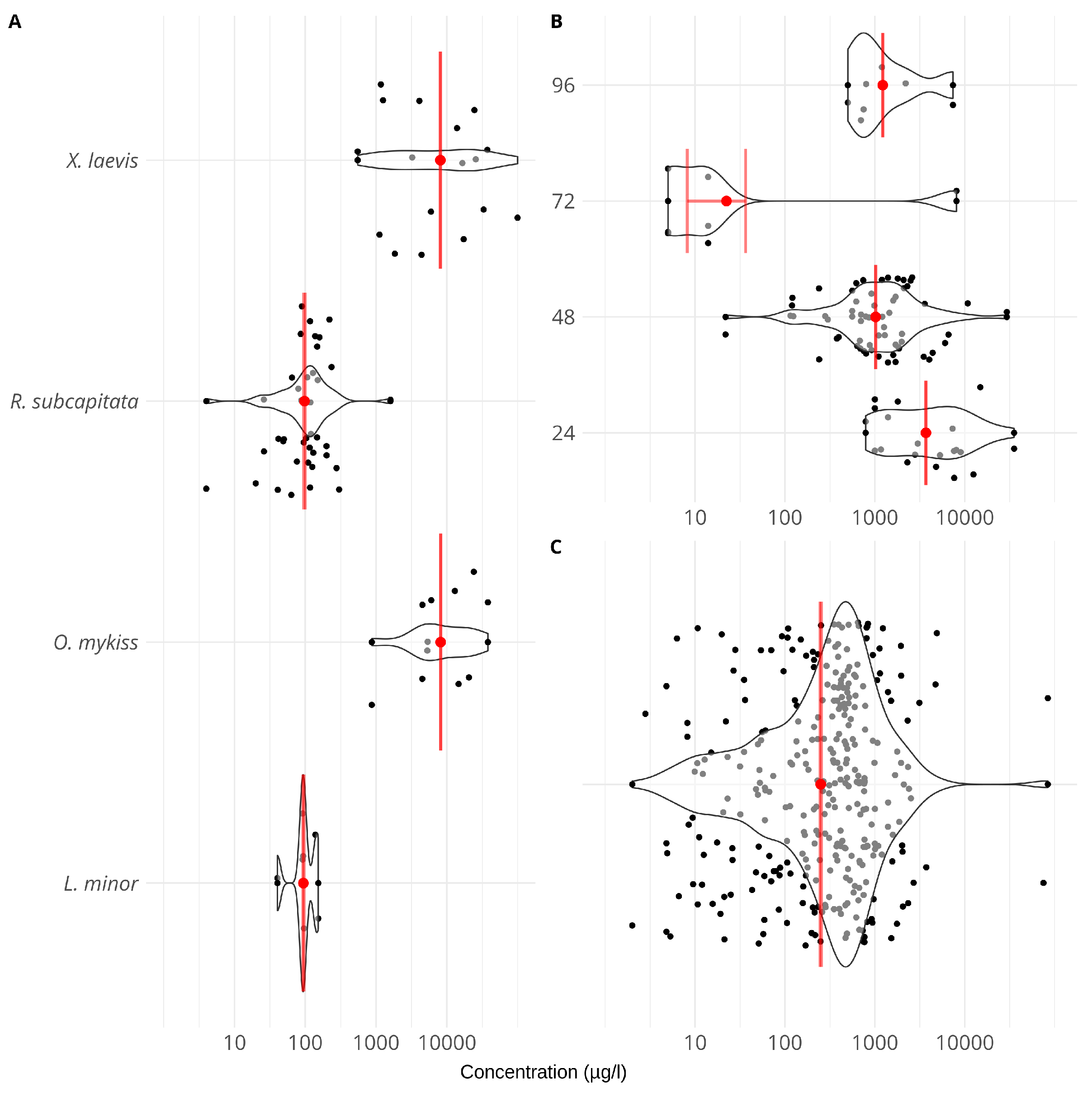
2.3. Accuracy Assessment
2.4. Perspectives
3. Methods
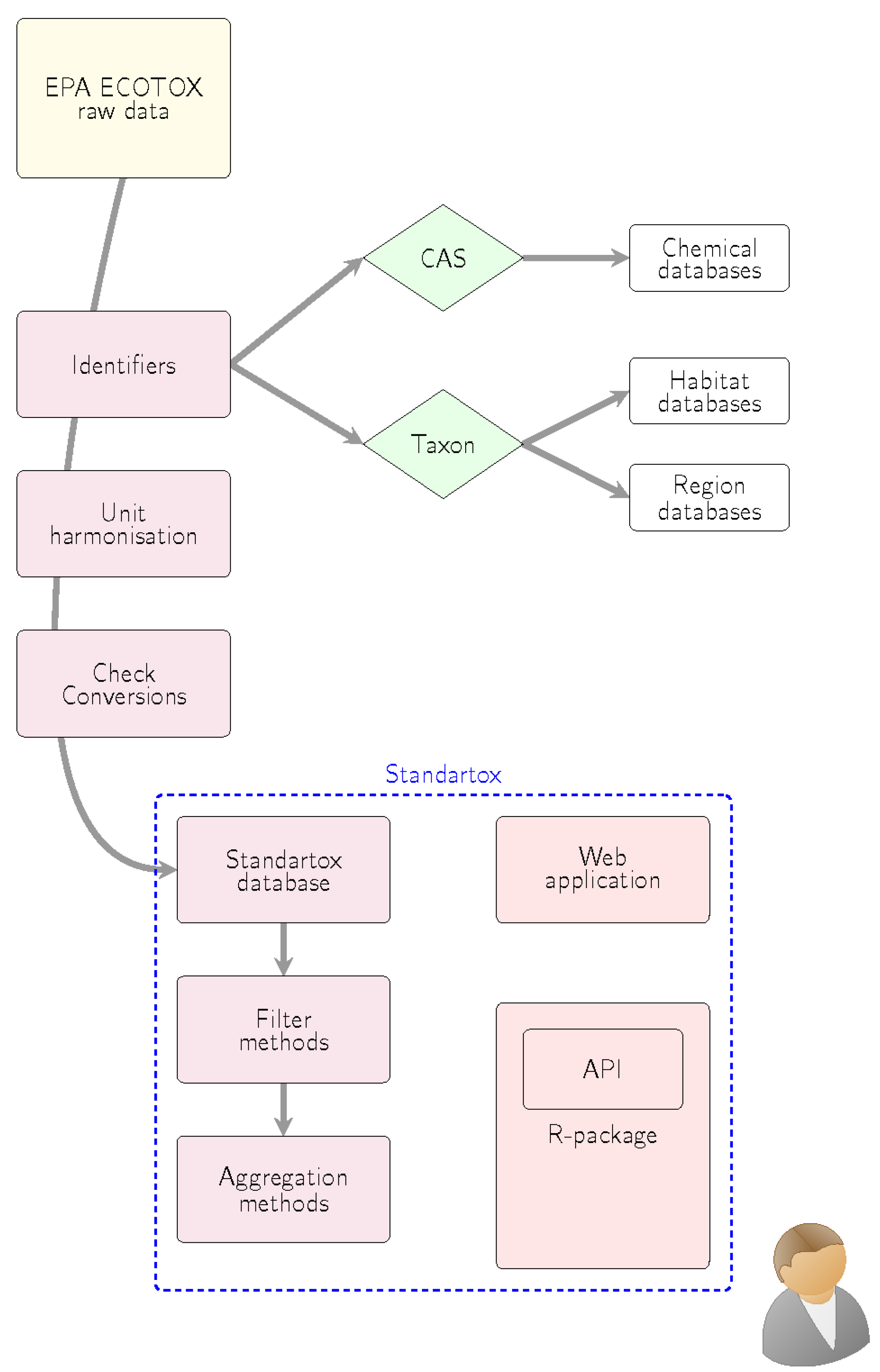
3.1. Processing
3.2. Application Methods
4. User Notes
| Listing 1: Sample code to access the Standartox database through the API and the the R package standartox. stx_catalog() returns a catalog of possible filter and aggregation parameters. stx_query() returns the Standartox object, a list of the filtered and the aggregated data as well as a meta data entry. Example for XX50 tests on the chemical glyphosate (CAS number: 1071-83-6) and the taxon Oncorhynchus lasting 24 h to 120 h. |
 |
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| API | Application programming interface |
| CAS | Chemical abstracts service registry number |
| ChEBI | Chemical Entities of Biological Interest database |
| CIR | Chemical Identifier Resolver service |
| E/LC/D50 | Half maximal effective/lethal concentration/dose |
| XX50 | Summarizes E/LC/D50 (Table A2) |
| ECOTOX | US EPA ECOTOXicology Knowledgebase |
| GBIF | Global Biodiversity Information Facility |
| JSON | JavaScript Object Notation file format |
| LOEC/L | Lowest observed effect concentrations/levels |
| LOEX | Summarizes LOEC/L (Table A2) |
| NOEC/L | No observed effect concentrations/levels |
| NOEX | Summarizes NOEC/L (Table A2) |
| PPDB | Pesticides Properties DataBase |
| QSAR | Quantitative structure-activity relationship |
| REST | Representational state transfer software architectural style |
| SSD | Species sensitivity distribution |
| TU | Toxic unit |
| WoRMS | World Register of Marine Species |
Appendix A
| Database | URL |
|---|---|
| Chemical Entities of Biological Interest (ChEBI) | https://www.ebi.ac.uk/chebi |
| Chemical Identifier Resolver | https://cactus.nci.nih.gov/chemical/structure |
| ChemSpider | http://www.chemspider.com |
| Eurostat | https://ec.europa.eu/eurostat/home |
| PubChem | https://pubchem.ncbi.nlm.nih.gov |
| Wikidata | https://www.wikidata.org/wiki/Wikidata:Main_Page |
| Global Biodiversity Information Facility (GBIF) | https://www.gbif.org |
| World Register of Marine Species (WoRMS) | http://marinespecies.org |
| freshwaterecology.info | https://www.freshwaterecology.info |
| Standartox Endpoint | Ecotox Endpoint | Ecotox Endpoint Description |
|---|---|---|
| XX50 | LC50 | Lethal concentration to 50% of test organisms |
| XX50 | LD50 | Lethal dose to 50% of test organisms |
| XX50 | EC50 | Effective concentration to 50% of test organisms |
| XX50 | ED50 | Effective dose to 50% of test organisms |
| XX50 | IC50 | Inhibition concentration to 50% of test organisms |
| XX50 | ID50 | Inhibition dose to 50% of test organisms |
| XX50 | ET50 | Effective response time to 50% of test organisms |
| XX50 | LT50 | Time to 50% mortality of test organisms |
| NOEX | NOEC | No-observable-effect-concentration |
| NOEX | NOEL | No-observable-effect-level |
| LOEX | LOEC | Lowest observable effect concentration |
| LOEX | LOEL | Lowest-observable-effect-level |
| Endpoint | HTTP Method | Request | Response |
|---|---|---|---|
| /catalog | POST | Standartox version string | Catalog object (JSON) |
| /filter | POST | Standartox filter parameters | Filtered Standartox data (serialized) |
| /meta | POST | Standartox version string | Meta data on request (JSON) |
Appendix B
| Package | Description | Citation |
|---|---|---|
| bib2df | Parse a BibTeX File to a Data Frame | [60] |
| countrycode | Convert Country Names and Country Codes | [61] |
| cowplot | Streamlined Plot Theme and Plot Annotations for ’ggplot2’ | [62] |
| data.table | Extension of ‘data.frame‘ | [63] |
| DBI | R Database Interface | [64] |
| dbreport | Automated reports from tables | [65] |
| devtools | Tools to Make Developing R Packages Easier | [66] |
| doParallel | Foreach Parallel Adaptor for the ’parallel’ Package | [67] |
| DT | A Wrapper of the JavaScript Library ’DataTables’ | [68] |
| foreach | Provides Foreach Looping Construct for R | [69] |
| fst | Lightning Fast Serialization of Data Frames for R | [59] |
| ggplot2 | Create Elegant Data Visualisations Using the Grammar of Graphics | [70] |
| httr | Tools for Working with URLs and HTTP | [71] |
| jsonlite | A Robust, High Performance JSON Parser and Generator for R | [72] |
| knitr | A General-Purpose Package for Dynamic Report Generation in R | [73] |
| openxlsx | Read, Write and Edit xlsx Files | [74] |
| plotly | Create Interactive Web Graphics via ’plotly.js’ | [75] |
| plumber | An API Generator for R | [58] |
| R.utils | Various Programming Utilities | [76] |
| RColorBrewer | ColorBrewer Palettes | [77] |
| reactlog | Reactivity Visualizer for ’shiny’ | [78] |
| readxl | Read Excel Files | [79] |
| rgbif | Interface to the Global ’Biodiversity’ Information Facility API | [80] |
| RPostgreSQL | R Interface to the ’PostgreSQL’ Database System | [81] |
| rvest | Easily Harvest (Scrape) Web Pages | [82] |
| scales | Scale Functions for Visualization | [83] |
| shiny | Web Application Framework for R | [57] |
| shinydashboard | Create Dashboards with ’Shiny’ | [84] |
| shinydashboardPlus | Add More ’AdminLTE2’ Components to ’shinydashboard’ | [85] |
| shinyjs | Easily Improve the User Experience of Your Shiny Apps in Seconds | [86] |
| shinyWidgets | Custom Inputs Widgets for Shiny | [87] |
| stringi | Character String Processing Facilities | [88] |
| stringr | Simple, Consistent Wrappers for Common String Operations | [89] |
| taxize | Taxonomic Information from Around the Web | [90] |
| treemap | Treemap Visualization | [91] |
| treemapify | Draw Treemaps in ’ggplot2’ | [92] |
| udunits2 | Udunits-2 Bindings for R | [93] |
| webchem | Chemical Information from the Web | [94] |
References
- Breithaupt, H. The Costs of REACH. REACH Is Largely Welcomed, but the Requirement to Test Existing Chemicals for Adverse Effects Is Not Good News for All. EMBO Rep. 2006, 7, 968–971. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, R.P. The Challenge of Micropollutants in Aquatic Systems. Science 2006, 313, 1072–1077. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, R.B.; von der Ohe, P.C.; Rasmussen, J.; Kefford, B.J.; Beketov, M.A.; Schulz, R.; Liess, M. Thresholds for the Effects of Pesticides on Invertebrate Communities and Leaf Breakdown in Stream Ecosystems. Environ. Sci. Technol. 2012, 46, 5134–5142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malaj, E.; von der Ohe, P.C.; Grote, M.; Kühne, R.; Mondy, C.P.; Usseglio-Polatera, P.; Brack, W.; Schäfer, R.B. Organic Chemicals Jeopardize the Health of Freshwater Ecosystems on the Continental Scale. Proc. Natl. Acad. Sci. USA 2014, 111, 9549–9554. [Google Scholar] [CrossRef] [Green Version]
- Hallmann, C.A.; Foppen, R.P.B.; van Turnhout, C.A.M.; de Kroon, H.; Jongejans, E. Declines in Insectivorous Birds Are Associated with High Neonicotinoid Concentrations. Nature 2014, 511, 341–343. [Google Scholar] [CrossRef]
- Barra Caracciolo, A.; Topp, E.; Grenni, P. Pharmaceuticals in the Environment: Biodegradation and Effects on Natural Microbial Communities. A Review. J. Pharm. Biomed. Anal. 2015, 106, 25–36. [Google Scholar] [CrossRef]
- Johnston, E.L.; Mayer-Pinto, M.; Crowe, T.P. REVIEW: Chemical Contaminant Effects on Marine Ecosystem Functioning. J. Appl. Ecol. 2015, 52, 140–149. [Google Scholar] [CrossRef] [Green Version]
- Peters, K.; Bundschuh, M.; Schäfer, R. Review on the Effects of Toxicants on Freshwater Ecosystem Functions. Environ. Pollut. 2013, 180, 324–329. [Google Scholar] [CrossRef]
- Van der Sluijs, J.P.; Simon-Delso, N.; Goulson, D.; Maxim, L.; Bonmatin, J.M.; Belzunces, L.P. Neonicotinoids, Bee Disorders and the Sustainability of Pollinator Services. Curr. Opin. Environ. Sustain. 2013, 5, 293–305. [Google Scholar] [CrossRef]
- Yamamuro, M.; Komuro, T.; Kamiya, H.; Kato, T.; Hasegawa, H.; Kameda, Y. Neonicotinoids Disrupt Aquatic Food Webs and Decrease Fishery Yields. Science 2019, 366, 620–623. [Google Scholar] [CrossRef]
- Steffen, W.; Crutzen, P.J.; McNeill, J.R. The Anthropocene: Are Humans Now Overwhelming the Great Forces of Nature. AMBIO J. Hum. Environ. 2007, 36, 614–621. [Google Scholar] [CrossRef]
- Steffen, W.; Richardson, K.; Rockstrom, J.; Cornell, S.E.; Fetzer, I.; Bennett, E.M.; Biggs, R.; Carpenter, S.R.; de Vries, W.; de Wit, C.A.; et al. Planetary Boundaries: Guiding Human Development on a Changing Planet. Science 2015, 347, 1259855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernhardt, E.S.; Rosi, E.J.; Gessner, M.O. Synthetic Chemicals as Agents of Global Change. Front. Ecol. Environ. 2017, 15, 84–90. [Google Scholar] [CrossRef]
- Rosa, W. (Ed.) Transforming Our World: The 2030 Agenda for Sustainable Development. In A New Era in Global Health; Springer Publishing Company: New York, NY, USA, 2017. [Google Scholar] [CrossRef]
- Beketov, M.A.; Kefford, B.J.; Schäfer, R.B.; Liess, M. Pesticides Reduce Regional Biodiversity of Stream Invertebrates. Proc. Natl. Acad. Sci. USA 2013, 110, 11039–11043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schäfer, R.B.; Liess, M.; Altenburger, R.; Filser, J.; Hollert, H.; Roß-Nickoll, M.; Schäffer, A.; Scheringer, M. Future Pesticide Risk Assessment: Narrowing the Gap between Intention and Reality. Environ. Sci. Eur. 2019, 31, 21. [Google Scholar] [CrossRef] [Green Version]
- Morrissey, C.A.; Mineau, P.; Devries, J.H.; Sanchez-Bayo, F.; Liess, M.; Cavallaro, M.C.; Liber, K. Neonicotinoid Contamination of Global Surface Waters and Associated Risk to Aquatic Invertebrates: A Review. Environ. Int. 2015, 74, 291–303. [Google Scholar] [CrossRef]
- ECOTOX User Guide: ECOTOXicology Knowledgebase System. Version 5.0. Available online: https://www.epa.gov/ecotox (accessed on 1 February 2020).
- Umweltbundesamt. ETOX: Information System Ecotoxicology and Environmental Quality Targets. 2019. Available online: https://webetox.uba.de/webETOX (accessed on 18 December 2019).
- Lewis, K.A.; Tzilivakis, J.; Warner, D.J.; Green, A. An International Database for Pesticide Risk Assessments and Management. Hum. Ecol. Risk Assess. Int. J. 2016, 22, 1050–1064. [Google Scholar] [CrossRef] [Green Version]
- Health and Environmental Sciences Institute (HESI). EnviroTox Database & Tools; Version 1.1.0; HESI: Washington, DC, USA, 2019. [Google Scholar]
- Connors, K.A.; Beasley, A.; Barron, M.G.; Belanger, S.E.; Bonnell, M.; Brill, J.L.; de Zwart, D.; Kienzler, A.; Krailler, J.; Otter, R.; et al. Creation of a Curated Aquatic Toxicology Database: EnviroTox. Environ. Toxicol. Chem. 2019, 38, 1062–1073. [Google Scholar] [CrossRef] [Green Version]
- Mark, U.; Solbé, J. Analysis of the Ecetoc Aquatic Toxicity (EAT) Database V— The Relevance of Daphnia Magna as a Representative Test Species. Chemosphere 1998, 36, 155–166. [Google Scholar] [CrossRef]
- Malaj, E.; Grote, M.; Schäfer, R.B.; Brack, W.; von der Ohe, P.C. Physiological Sensitivity of Freshwater Macroinvertebrates to Heavy Metals. Environ. Toxicol. Chem. 2012, 31, 1754–1764. [Google Scholar] [CrossRef]
- US EPA. ECOTOX Knowledgebase; US EPA: Washington, DC, USA, 2019.
- Posthuma, L.; Suter, G.W.; Traas, T.P. (Eds.) Species Sensitivity Distributions in Ecotoxicology; Environmental and Ecological Risk Assessment; Lewis Publishers: Boca Raton, FL, USA, 2002. [Google Scholar]
- Kefford, B.J.; Marchant, R.; Schäfer, R.B.; Metzeling, L.; Dunlop, J.E.; Choy, S.C.; Goonan, P. The Definition of Species Richness Used by Species Sensitivity Distributions Approximates Observed Effects of Salinity on Stream Macroinvertebrates. Environ. Pollut. 2011, 159, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, R.B.; Pettigrove, V.; Rose, G.; Allinson, G.; Wightwick, A.; von der Ohe, P.C.; Shimeta, J.; Kühne, R.; Kefford, B.J. Effects of Pesticides Monitored with Three Sampling Methods in 24 Sites on Macroinvertebrates and Microorganisms. Environ. Sci. Technol. 2011, 45, 1665–1672. [Google Scholar] [CrossRef] [PubMed]
- OECD. OECD Guidelines for the Testing of Chemicals; OECD: Paris, France, 2020. [Google Scholar]
- Hartung, T.; Rovida, C. Chemical Regulators Have Overreached. Nature 2009, 460, 1080–1081. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Leith, K.F.; Bowerman, W.W.; Wierda, M.R.; Best, D.A.; Grubb, T.G.; Sikarske, J.G. A Comparison of Techniques for Assessing Central Tendency in Left-Censored Data Using PCB and p,P′DDE Contaminant Concentrations from Michigan’s Bald Eagle Biosentinel Program. Chemosphere 2010, 80, 7–12. [Google Scholar] [CrossRef]
- Posthuma, L.; van Gils, J.; Zijp, M.C.; van de Meent, D.; de Zwart, D. Species Sensitivity Distributions for Use in Environmental Protection, Assessment, and Management of Aquatic Ecosystems for 12 386 Chemicals. Environ. Toxicol. Chem. 2019, 38, 905–917. [Google Scholar] [CrossRef] [Green Version]
- UFZ Department of Ecological Chemistry. ChemProp 6.5. 2016. Available online: http://www.ufz.de/ecochem/chemprop (accessed on 1 February 2016).
- Schüürmann, G.; Ebert, R.U.; Kühne, R. Quantitative Read-Across for Predicting the Acute Fish Toxicity of Organic Compounds. Environ. Sci. Technol. 2011, 45, 4616–4622. [Google Scholar] [CrossRef]
- Malaj, E.; Guénard, G.; Schäfer, R.B.; von der Ohe, P.C. Evolutionary Patterns and Physicochemical Properties Explain Macroinvertebrate Sensitivity to Heavy Metals. Ecol. Appl. 2016, 26, 1249–1259. [Google Scholar] [CrossRef]
- Van den Berg, S.J.P.; Baveco, H.; Butler, E.; De Laender, F.; Focks, A.; Franco, A.; Rendal, C.; Van den Brink, P.J. Modeling the Sensitivity of Aquatic Macroinvertebrates to Chemicals Using Traits. Environ. Sci. Technol. 2019, 53, 6025–6034. [Google Scholar] [CrossRef] [Green Version]
- Martin, T.M.; Young, D.M. Prediction of the Acute Toxicity (96-h LC 50) of Organic Compounds to the Fathead Minnow (PimephalesPromelas) Using A Group Contribution Method. Chem. Res. Toxicol. 2001, 14, 1378–1385. [Google Scholar] [CrossRef]
- Williams, A.J.; Grulke, C.M.; Edwards, J.; McEachran, A.D.; Mansouri, K.; Baker, N.C.; Patlewicz, G.; Shah, I.; Wambaugh, J.F.; Judson, R.S.; et al. The CompTox Chemistry Dashboard: A Community Data Resource for Environmental Chemistry. J. Cheminform. 2017, 9, 61. [Google Scholar] [CrossRef]
- Judson, R.; Richard, A.; Dix, D.; Houck, K.; Elloumi, F.; Martin, M.; Cathey, T.; Transue, T.R.; Spencer, R.; Wolf, M. ACToR—Aggregated Computational Toxicology Resource. Toxicol. Appl. Pharmacol. 2008, 233, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Judson, R.S.; Martin, M.T.; Egeghy, P.; Gangwal, S.; Reif, D.M.; Kothiya, P.; Wolf, M.; Cathey, T.; Transue, T.; Smith, D.; et al. Aggregating Data for Computational Toxicology Applications: The U.S. Environmental Protection Agency (EPA) Aggregated Computational Toxicology Resource (ACToR) System. Int. J. Mol. Sci. 2012, 13, 1805–1831. [Google Scholar] [CrossRef] [PubMed]
- Kienzler, A.; Barron, M.G.; Belanger, S.E.; Beasley, A.; Embry, M.R. Mode of Action (MOA) Assignment Classifications for Ecotoxicology: An Evaluation of Approaches. Environ. Sci. Technol. 2017, 51, 10203–10211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenkrantz, R.T.; Cedergreen, N.; Baun, A.; Kusk, K.O. Influence of pH, Light Cycle, and Temperature on Ecotoxicity of Four Sulfonylurea Herbicides towards Lemna Gibba. Ecotoxicology 2013, 22, 33–41. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Zhou, D.; Wang, P.; Li, L. Temperature Affects Cadmium-Induced Phytotoxicity Involved in Subcellular Cadmium Distribution and Oxidative Stress in Wheat Roots. Ecotoxicol. Environ. Saf. 2011, 74, 2029–2035. [Google Scholar] [CrossRef]
- Compson, Z.G.; Monk, W.A.; Curry, C.J.; Gravel, D.; Bush, A.; Baker, C.J.; Al Manir, M.S.; Riazanov, A.; Hajibabaei, M.; Shokralla, S.; et al. Linking DNA Metabarcoding and Text Mining to Create Network-Based Biomonitoring Tools: A Case Study on Boreal Wetland Macroinvertebrate Communities. Adv. Ecol. Res. 2018, 59, 33–74. [Google Scholar] [CrossRef]
- Wood, A. Compendium of Pesticide Common Names. 2019. Available online: http://www.alanwood.net/pesticides (accessed on 1 April 2020).
- Hastings, J.; Owen, G.; Dekker, A.; Ennis, M.; Kale, N.; Muthukrishnan, V.; Turner, S.; Swainston, N.; Mendes, P.; Steinbeck, C. ChEBI in 2016: Improved Services and an Expanding Collection of Metabolites. Nucleic Acids Res. 2016, 44, D1214–D1219. [Google Scholar] [CrossRef]
- National Institutes of Health (NIH). Chemical Identifier Resolver; NIH: Bethesda, MD, USA, 2019. [Google Scholar]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound Databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef]
- European Commission. Eurostat; European Commission: Brussels, Belgium, 2019. [Google Scholar]
- Vrandečić, D.; Krötzsch, M. Wikidata: A Free Collaborative Knowledgebase. Commun. ACM 2014, 57, 78–85. [Google Scholar] [CrossRef]
- WoRMS Editorial Board. World Register of Marine Species (WoRMS); WoRMS Editorial Board: Worms, Germany, 2020; Available online: http://www.marinespecies.org (accessed on 1 April 2020).
- GBIF: The Global Biodiversity Information Facility. What Is GBIF? 2020. Available online: https://www.gbif.org/what-is-gbif (accessed on 1 April 2020).
- Schmidt-Kloiber, A.; Hering, D. www.Freshwaterecology.Info—An Online Tool That Unifies, Standardises and Codifies More than 20,000 European Freshwater Organisms and Their Ecological Preferences. Ecol. Indic. 2015, 53, 271–282. [Google Scholar] [CrossRef]
- Rainbow, P.S. Trace Metal Concentrations in Aquatic Invertebrates: Why and so What? Environ. Pollut. 2002, 120, 497–507. [Google Scholar] [CrossRef]
- Buchwalter, D.B.; Luoma, S.N. Differences in Dissolved Cadmium and Zinc Uptake among Stream Insects: Mechanistic Explanations. Environ. Sci. Technol. 2005, 39, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Cheng, J.; Allaire, J.; Xie, Y.; McPherson, J. Shiny: Web Application Framework for R, R package version 1.4.0.2; 2020. Available online: https://CRAN.R-project.org/package=shiny (accessed on 1 April 2020).
- Trestle Technology, LLC. plumber: An API Generator for R, R package version 0.4.6; Trestle Technology, LLC: Dallas, TX, USA, 2018; Available online: https://CRAN.R-project.org/package=plumber (accessed on 1 April 2020).
- Klik, M. fst: Lightning Fast Serialization of Data Frames for R, R package version 0.9.2; 2020. Available online: https://CRAN.R-project.org/package=fst (accessed on 1 April 2020).
- Ottolinger, P. bib2df: Parse a BibTeX File to a Data Frame, R package version 1.1.1; 2019. Available online: https://CRAN.R-project.org/package=bib2df (accessed on 1 April 2020).
- Arel-Bundock, V. Countrycode: Convert Country Names and Country Codes, R package version 1.1.1; 2020. Available online: https://CRAN.R-project.org/package=countrycode (accessed on 1 April 2020).
- Wilke, C.O. cowplot: Streamlined Plot Theme and Plot Annotations for ‘ggplot2’, R package version 0.9.4; 2019. Available online: https://CRAN.R-project.org/package=cowplot (accessed on 1 April 2020).
- Dowle, M.; Srinivasan, A. data.table: Extension of ‘data.frame’, R package version 1.12.8; 2019. Available online: https://CRAN.R-project.org/package=data.table (accessed on 1 April 2020).
- R Special Interest Group on Databases (R-SIG-DB); Wickham, H.; Müller, K. DBI: R Database Interface, R package version 1.1.0; 2019. Available online: https://CRAN.R-project.org/package=DBI (accessed on 1 April 2020).
- Scharmüller, A. dbreport: Automated Reports from Tables, R package version 0.0.0.9007; 2020. Available online: https://github.com/andschar/dbreport (accessed on 1 April 2020).
- Wickham, H.; Hester, J.; Chang, W. devtools: Tools to Make Developing R Packages Easier, R package version 2.2.1; 2019. Available online: https://CRAN.R-project.org/package=devtools (accessed on 1 April 2020).
- Corporation, M.; Weston, S. doParallel: Foreach Parallel Adaptor for the ‘Parallel’ Package, R package version 1.0.14; 2018. Available online: https://CRAN.R-project.org/package=doParallel (accessed on 1 April 2020).
- Xie, Y.; Cheng, J.; Tan, X. DT: A Wrapper of the JavaScript Library ‘DataTables’, R package version 0.7; 2019. Available online: https://CRAN.R-project.org/package=DT (accessed on 1 April 2020).
- Microsoft; Weston, S. foreach: Provides Foreach Looping Construct for R, R package version 1.4.4; 2017. Available online: https://CRAN.R-project.org/package=foreach (accessed on 1 April 2020).
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D. ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics, R package version 3.3.0; 2020. Available online: https://CRAN.R-project.org/package=ggplot2 (accessed on 1 April 2020).
- Wickham, H. httr: Tools for Working with URLs and HTTP, R package version 1.4.1; 2019. Available online: https://CRAN.R-project.org/package=httr (accessed on 1 April 2020).
- Ooms, J. jsonlite: A Robust, High Performance JSON Parser and Generator for R, R package version 1.6.1; 2020. Available online: https://CRAN.R-project.org/package=jsonlite (accessed on 1 April 2020).
- Xie, Y. knitr: A General-Purpose Package for Dynamic Report Generation in R, R package version 1.28; 2020. Available online: https://CRAN.R-project.org/package=knitr (accessed on 1 April 2020).
- Schauberger, P.; Walker, A. openxlsx: Read, Write and Edit xlsx Files, R package version 4.1.4; 2019. Available online: https://CRAN.R-project.org/package=openxlsx (accessed on 1 April 2020).
- Sievert, C.; Parmer, C.; Hocking, T.; Chamberlain, S.; Ram, K.; Corvellec, M.; Despouy, P. plotly: Create Interactive Web Graphics via ‘plotly.js’, R package version 4.9.0; 2019. Available online: https://CRAN.R-project.org/package=plotly (accessed on 1 April 2020).
- Bengtsson, H. utils: Various Programming Utilities, R package version 2.9.0; 2019. Available online: https://CRAN.R-project.org/package=R.utils (accessed on 1 April 2020).
- Neuwirth, E. RColorBrewer: ColorBrewer Palettes, R package version 1.1-2; 2014. Available online: https://CRAN.R-project.org/package=RColorBrewer (accessed on 1 April 2020).
- Schloerke, B. reactlog: Reactivity Visualizer for ‘Shiny’, R package version 1.0.0; 2019. Available online: https://CRAN.R-project.org/package=reactlog (accessed on 1 April 2020).
- Wickham, H.; Bryan, J. readxl: Read Excel Files, R package version 1.3.1; 2019. Available online: https://CRAN.R-project.org/package=readxl (accessed on 1 April 2020).
- Chamberlain, S. rgbif: Interface to the Global ‘Biodiversity’ Information Facility API, R package version 1.3; 2019. Available online: https://CRAN.R-project.org/package=rgbif (accessed on 1 April 2020).
- Conway, J.; Eddelbuettel, D.; Nishiyama, T.; Prayaga, S.K.; Tiffin, N. RPostgreSQL: R Interface to the ‘PostgreSQL’ Database System, R package version 0.6-2; 2017. Available online: https://CRAN.R-project.org/package=RPostgreSQL (accessed on 1 April 2020).
- Wickham, H. rvest: Easily Harvest (Scrape) Web Pages, R package version 0.3.5; 2019. Available online: https://CRAN.R-project.org/package=rvest (accessed on 1 April 2020).
- Wickham, H.; Seidel, D. scales: Scale Functions for Visualization, R package version 1.1.0; 2019. Available online: https://CRAN.R-project.org/package=scales (accessed on 1 April 2020).
- Chang, W.; Borges Ribeiro, B. shinydashboard: Create Dashboards with ‘Shiny’, R package version 0.7.1; 2018. Available online: https://CRAN.R-project.org/package=shinydashboard (accessed on 1 April 2020).
- Granjon, D. shinydashboardPlus: Add More ‘AdminLTE2’ Components to ‘shinydashboard’, R package version 0.7.0; 2019. Available online: https://CRAN.R-project.org/package=shinydashboardPlus (accessed on 1 April 2020).
- Attali, D. shinyjs: Easily Improve the User Experience of Your Shiny Apps in Seconds, R package version 1.0; 2018. Available online: https://CRAN.R-project.org/package=shinyjs (accessed on 1 April 2020).
- Perrier, V.; Meyer, F.; Granjon, D. shinyWidgets: Custom Inputs Widgets for Shiny, R package version 0.4.8; 2019. Available online: https://CRAN.R-project.org/package=shinyWidgets (accessed on 1 April 2020).
- Gagolewski, M.; Tartanus, B. stringi: Character String Processing Facilities, R package version 1.4.6; 2020. Available online: https://CRAN.R-project.org/package=stringi (accessed on 1 April 2020).
- Wickham, H. stringr: Simple, Consistent Wrappers for Common String Operations, R package version 1.4.0; 2019. Available online: https://CRAN.R-project.org/package=stringr (accessed on 1 April 2020).
- Chamberlain, S.; Szoecs, E.; Foster, Z.; Arendsee, Z. taxize: Taxonomic Information from Around the Web, R package version 0.9.7; 2019. Available online: https://CRAN.R-project.org/package=taxize (accessed on 1 April 2020).
- Tennekes, M. treemap: Treemap Visualization, R package version 2.4-2; 2017. Available online: https://CRAN.R-project.org/package=treemap (accessed on 1 April 2020).
- Wilkins, D. treemapify: Draw Treemaps in ‘ggplot2’, R package version 2.5.3; 2019. Available online: https://CRAN.R-project.org/package=treemapify (accessed on 1 April 2020).
- Hiebert, J. udunits2: Udunits-2 Bindings for R, R package version 0.13; 2016. Available online: https://CRAN.R-project.org/package=udunits2 (accessed on 1 April 2020).
- Szöcs, E. webchem: Chemical Information from the Web. 2020. R package version 0.5.0; 2020. Available online: https://CRAN.R-project.org/package=webchem (accessed on 1 April 2020).


| Database | Filter | Aggregation, Selection | Access |
|---|---|---|---|
| Comptox [39] | Chemical | no | Web, file |
| Ecotox [25] | ALL | no | Web, file |
| EnviroTox [21] | ALL | chemical, organism | Web |
| Etox [19] | ALL | no | Web |
| Pesticides Properties DataBase (PPDB) [20] | fixed values | manual selection | Web, file |
| Standartox | ALL | chemical, organism | API, Web |
| Parameter | Example |
|---|---|
| cas | 7758987, 2921-88-2, 1912-24-9 |
| concentration_type | Active ingredient, Formulation |
| chemical_role | Antibiotic, Fungicide, Drug |
| chemical_class | Conazole, Neonicotinoid, Triazine |
| taxa | Oncorhynchus mykiss, Rattus norvegicus, Daphnia magna |
| habitat | Marine, Brackish, Freshwater |
| region | Europe, Africa, Asia |
| duration | 24, 96 |
| effect | Mortality, Population, Growth |
| endpoint | NOEX, XX50 |
| exposure | auquatic, diet |
| vers | 20191212 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scharmüller, A.; Schreiner, V.C.; Schäfer, R.B. Standartox: Standardizing Toxicity Data. Data 2020, 5, 46. https://doi.org/10.3390/data5020046
Scharmüller A, Schreiner VC, Schäfer RB. Standartox: Standardizing Toxicity Data. Data. 2020; 5(2):46. https://doi.org/10.3390/data5020046
Chicago/Turabian StyleScharmüller, Andreas, Verena C. Schreiner, and Ralf B. Schäfer. 2020. "Standartox: Standardizing Toxicity Data" Data 5, no. 2: 46. https://doi.org/10.3390/data5020046
APA StyleScharmüller, A., Schreiner, V. C., & Schäfer, R. B. (2020). Standartox: Standardizing Toxicity Data. Data, 5(2), 46. https://doi.org/10.3390/data5020046





