Extremophilic Microorganisms for the Treatment of Toxic Pollutants in the Environment
Abstract
:1. Introduction
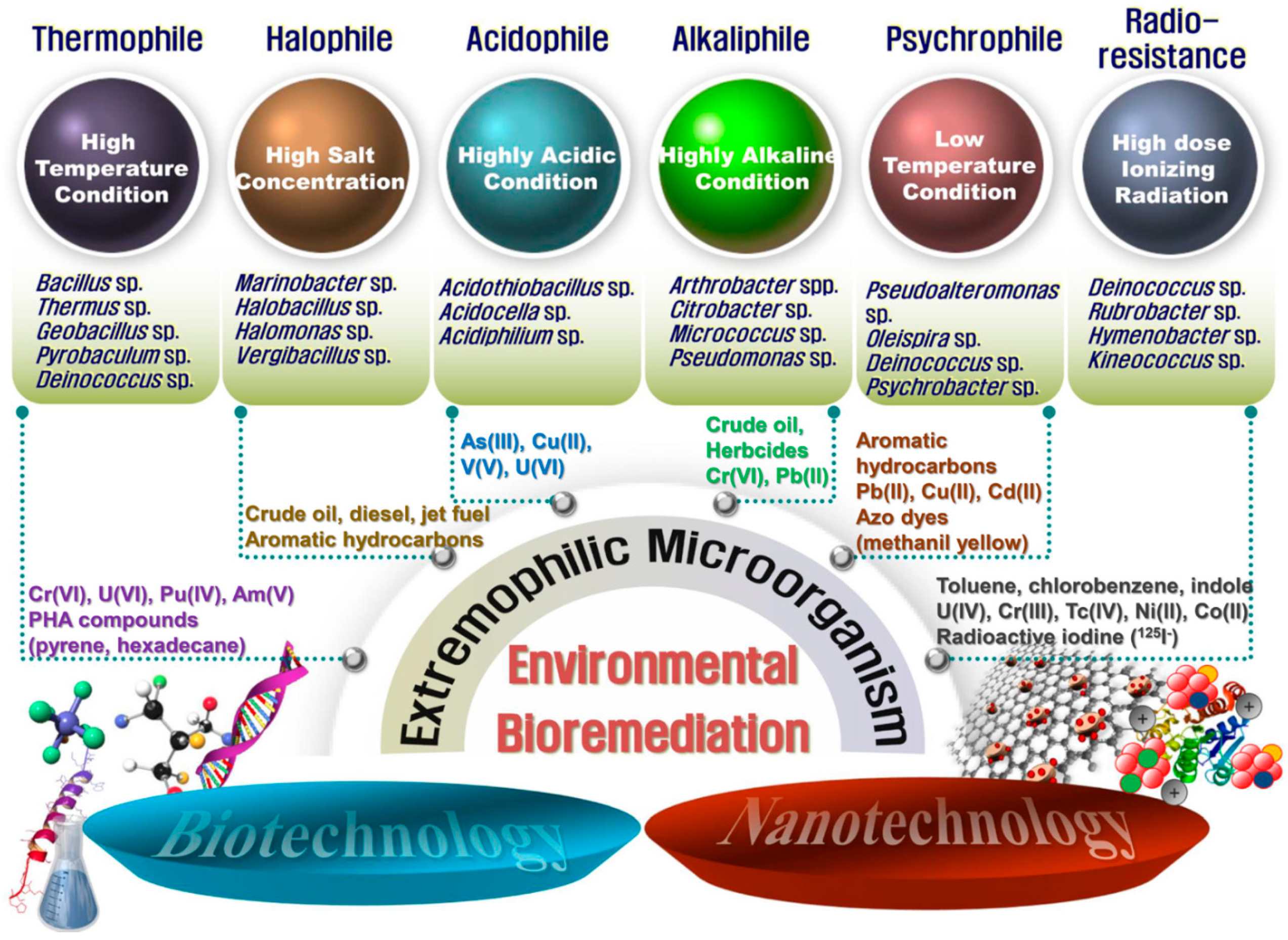
2. Survival Strategies of Extremophilic Microorganisms under Extreme Conditions
2.1. Acidophilic and Alkaliphilic Microorganisms
2.2. Halophilic Microorganisms
2.3. Psychrophilic and Thermophilic Microorganisms
2.4. Radiophilic Microorganisms
3. Bioremediation Using Extremophiles
3.1. Treatment of Heavy Metal Pollutants
3.2. Biodegradation of Organic Pollutants
3.3. Microbial Treatment of Radioactive Waste
4. The Future Direction
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Muddemann, T.; Haupt, D.; Sievers, M.; Kunz, U. Electrochemical reactors for wastewater treatment. ChemBioEng Rev. 2019, 6, 142–156. [Google Scholar] [CrossRef]
- Ouyang, W.; Chen, T.; Shi, Y.; Tong, L.; Chen, Y.; Wang, W.; Yang, J.; Xue, J. Physico-chemical process. Water Environ. Res. 2019, 91, 1350–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, Q.; Nomura, Y.; Fukahori, S.; Mizuno, T.; Tanaka, H.; Fujisawa, T. Innovative treatment of organic contaminants in reverse osmosis concentrate from water reuse: A mini review. Curr. Pollut. Rep. 2019, 5, 294–307. [Google Scholar] [CrossRef]
- Gebreeyessus, G.D. Status of hybrid membrane-ion-exchange systems for desalination: A comprehensive review. Appl. Water. Sci. 2019, 9, 135. [Google Scholar] [CrossRef] [Green Version]
- Abdelfattah, A.; Hossain, M.I.; Cheng, L. High-strength wastewater treatment using microbial biofilm reactor: A critical review. World J. Microb. Biot. 2020, 36, 75. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wu, D.; Ren, H. Bioremediation of oil contaminated soil using agricultural wastes via microbial consortium. Sci. Rep. 2020, 10, 9188. [Google Scholar] [CrossRef] [PubMed]
- Dixit, R.W.; Malaviya, D.; Pandiyan, K.; Singh, U.B.; Sahu, A.; Shukia, R.; Singh, B.P.; Rai, J.P.; Sharma, P.K.; Lade, H.; et al. Bioremediation of heavy metals from soil and aquatic environment: An overview of principles and criteria of fundamental processes. Sustainability 2015, 7, 2189–2212. [Google Scholar] [CrossRef] [Green Version]
- Mishra, G.K. Microbes in heavy metal bioremediation: A review on current trends and patents. Recent Pat. Biotechnol. 2017, 11, 188–196. [Google Scholar] [CrossRef]
- Diep, P.; Mahadevan, R.; Yakunin, A.F. Heavy metal removal by bioaccumulation using genetically engineered microorganisms. Front. Bioeng. Biotechnol. 2018, 6, 157. [Google Scholar] [CrossRef] [Green Version]
- Igiri, B.E.; Okoduwa, S.I.R.; Idoko, G.O.; Akabuogu, E.P.; Adeyi, A.O.; Ejiogu, I.K. Toxicity and bioremediation of heavy metals contaminated ecosystem from Tannery wastewater: A review. J. Toxicol. 2018, 2018, e2568038. [Google Scholar] [CrossRef]
- Singh, S.; Kang, S.H.; Mulchandani, A.; Chen, W. Bioremediation: Environmental clean-up through pathway engineering. Curr. Opin. Biotechnol. 2008, 19, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Kiadehi, M.S.H.; Amoozegar, M.A.; Asad, S.; Siroosi, M. Exploring the potential of halophilic archaea for the decolorization of azo dyes. Water Sci. Technol. 2018, 77, 1602–1611. [Google Scholar] [CrossRef] [PubMed]
- Azubuike, C.C.; Chikere, C.B.; Okpokwasili, G.C. Bioremediation techniques-classification based on site of application: Principles, advantages, limitations and prospects. World J. Microbiol. Biotechnol. 2016, 32, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waigi, M.G.; Sun, K.; Gao, Y.Z. Sphingomonads in microbe-assisted phytoremediation: Tackling soil pollution. Trends Biotechnol. 2017, 35, 883–899. [Google Scholar] [CrossRef] [PubMed]
- Tkacv, R.; Matrosova, V.Y.; Grichenko, O.E.; Gostinčar, C.; Volpe, R.P.; Klimenkova, P.; Gaidamakova, E.K.; Zhou, C.E.; Stewart, B.J.; Lyman, M.G.; et al. Prospects for fungal bioremediation of acidic radioactive waste sites: Characterization and genome sequence of Rhodotorula taiwanensis MD1149. Front. Microbiol. 2018, 8, 2528. [Google Scholar] [CrossRef]
- Ali, N.; Dashti, N.; Khanafer, M.; Al-Awadhi, H.; Radwan, S. Bioremediation of soils saturated with spilled crude oil. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Rambabu, K.; Banat, F.; Pham, Q.M.; Ho, S.-H.; Ren, N.-Q.; Show, P.L. Biological remediation of acid mine drainage: Review of past trends and current outlook. Environ. Sci. Technol. 2020, 2, 100024. [Google Scholar] [CrossRef]
- Cleary, A.; Lloyd, J.R.; Newsome, L.; Shaw, S.; Boothman, C.; Boshoff, G.; Atherton, N.; Morris, K. Bioremediation of strontium and technetium contaminated groundwater using glycerol phosphate. Chem. Geol. 2019, 509, 213–222. [Google Scholar] [CrossRef]
- Siliakus, M.F.; Van der Oost, J.; Kengen, S.W.M. Adaptations of archaeal and bacterial membranes to variations in temperature, pH and pressure. Extremophiles 2017, 21, 651–670. [Google Scholar] [CrossRef]
- Rastädter, K.; Wurm, D.J.; Spadiut, O.; Quehenberger, J. The cell membrane of Sulfolobus spp.–homeoviscous adaptation and biotechnological applications. Int. J. Mol. Sci. 2020, 21, 3935. [Google Scholar] [CrossRef]
- Vergara, E.; Neira, G.; González, C.; Cortez, D.; Dopson, M.; Holmes, D.S. Evolution of predicted acid resistance mechanisms in the extremely acidophilic Leptospirillum genus. Genes 2019, 11, 389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, N.; Liu, L. Microbial response to acid stress: Mechanisms and applications. Appl. Microbiol. Biotechnol. 2020, 104, 51–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvador-Castell, M.; Tourte, M.; Ogar, P.M. In search for the membrane regulators of archaea. Int. J. Mol. Sci. 2019, 20, 4434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golyshina, O.V.; Tran, H.; Reva, O.N.; Lemak, S.; Yakunin, A.F.; Goesmann, A.; Nechitaylo, T.Y.; LaCono, V.; Smedile, F.; Slesarev, A.; et al. Metabolic and evolutionary patterns in the extreme acidophilic archaeon Ferroplasma acidiphilum YT. Sci. Rep. 2017, 7, 3682. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Dopson, M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 2007, 15, 165–171. [Google Scholar] [CrossRef]
- Aono, R.; Ito, M.; Machida, T. Contribution of the cell wall component teichuronopeptide to pH homeostasis and alkaliphily in the alkaliphile Bacillus lentus C-125. J. Bacteriol. 1999, 181, 6600–6606. [Google Scholar] [CrossRef] [Green Version]
- Aono, R. Assignment of facultatively alkaliphilic Bacillus sp. strain C-125 to Bacillus lentus group 3. Int. J. Syst. Bacteriol. 1995, 45, 582–585. [Google Scholar] [CrossRef]
- Calamita, H.G.; Ehringer, W.D.; Koch, A.L.; Doyle, R.J. Evidence that the cell wall of Bacillus subtilis is protonated during respiration. Proc. Natl. Acad. Sci. USA 2001, 98, 15260–15263. [Google Scholar] [CrossRef] [Green Version]
- Padan, E.; Bibi, E.; Ito, M.; Krulwich, T.A. Alkaline pH homeostasis in bacteria: New insights. Biochim. Biophys. Acta Biomembr. 2005, 2, 67–88. [Google Scholar] [CrossRef] [Green Version]
- Kitada, M.; Kosono, S.; Kudo, T. The Na+/H+ antiporter of alkaliphilic Bacillus sp. Extremophiles 2000, 4, 253–258. [Google Scholar] [CrossRef]
- Fang, H.; Qin, X.-Y.; Zhang, K.-D.; Nie, Y.; Wu, X.-L. Role of the Group 2 Mrp sodium/proton antiporter in rapid response to high alkaline shock in the alkaline- and salt-tolerant Dietzia sp. DQ12-45-1b. Appl. Microbiol. Biotechnol. 2018, 102, 3765–3777. [Google Scholar] [CrossRef] [PubMed]
- Matsuno, T.; Goto, T.; Ogami, S.; Morimoto, H.; Yamazaki, K.; Inoue, N.; Matsuyama, H.; Yoshimune, K.; Yumoto, I. Formation of proton motive force under low-aeration alkaline conditions in alkaliphilic bacteria. Front. Microbiol. 2018, 9, 2331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stancik, L.M.; Stancik, D.M.; Schmidt, B.; Barnhart, D.M.; Yoncheva, Y.N.; Slonczewski, J.L. pH-Dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 2002, 184, 4246–4258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slonczewski, J.L.; Fujisawa, M.; Dopson, M.; Krulwich, T.A. Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv. Microb. Physiol. 2009, 55, 1–79. [Google Scholar] [CrossRef] [PubMed]
- Wernick, D.G.; Pontrelli, S.P.; Pollock, A.W.; Liao, J.C. Sustainable biorefining in wastewater by engineered extreme alkaliphile Bacillus marmarensis. Sci. Rep. 2016, 6, 20224. [Google Scholar] [CrossRef]
- Setati, M.E. Diversity and industrial potential of hydrolase-producing halophilic/halotolerant eubacteria. Afr. J. Biotechnol. 2010, 9, 1555–1560. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Plemenitaš, A.; Oren, A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 2018, 42, 353–375. [Google Scholar] [CrossRef]
- Starhl, H.; Greie, J.-C. The extremely halophilic archaeon Halobacterium salinarum R1 responds to potassium limitation by expression of the K+-transporting KdpFABC P-type ATPase and by a decrease in intracellular K+. Extremophiles 2008, 12, 741–752. [Google Scholar] [CrossRef]
- Engel, M.B.; Catchpole, H.R. A microprobe analysis of inorganic elements in Halobacterium salinarum. Cell Biol. Int. 2005, 29, 616–622. [Google Scholar] [CrossRef]
- Coker, J.A.; DasSarma, P.; Kumar, J.; Müller, J.A.; DasSarma, S. Transcriptional profiling of model Archeon Halobacterium sp. NRC-1: Responses to changes in salinity and temperature. Saline Syst. 2007, 3, 6. [Google Scholar] [CrossRef] [Green Version]
- Corral, P.; Amoozegar, M.A.; Ventosa, A. Halophiles and their biomolecules: Recent advances and future applications in biomedicine. Mar. Drugs 2020, 18, 33. [Google Scholar] [CrossRef] [Green Version]
- Reed, C.J.; Bushnell, S.; Evilia, C. Circular dichroism and fluorescence spectroscopy of cysteine-tRNA synthetase from Halobacterium salinarum ssp. NRC-1 demonstrates that group I cations are particularly effective in providing structure and stability to this halophilic protein. PLoS ONE 2014, 9, e89452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brininger, C.; Spradlin, S.; Cobani, L.; Evilia, C. The more adaptive to change, the more likely you are to survive: Protein adaptation in extremophiles. Semin. Cell Dev. Biol. 2018, 84, 158–169. [Google Scholar] [CrossRef]
- Elcock, A.H.; McCammon, J.A. Electrostatic contributions to the stability of halophilic proteins. J. Mol. Biol. 1998, 280, 731–748. [Google Scholar] [CrossRef]
- León, M.J.; Hoffmann, T.; Sánchez-Porro, C.; Heider, J.; Ventosa, A.; Bremer, E. Compatible solute synthesis and imported by the moderate halophile Spiribacter salinus: Physiology and genomics. Front. Microbiol. 2018, 9, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Borgne, S.; Paniagua, D.; Vazques-Duhalt, R. Biodegradation of organic pollutants by halophilic bacteria and archaea. J. Mol. Microbiol. Biotechnol. 2008, 15, 74–92. [Google Scholar] [CrossRef] [PubMed]
- Vandrich, J.; Pfeiffer, F.; Alfaro-Espinoza, G.; Kunte, H.J. Contribution of mechanosensitive channels to osmoadaptation and ectoine excretion in Halomonas elongata. Extremophile 2020, 24, 421–432. [Google Scholar] [CrossRef] [Green Version]
- Collins, T.; Margesin, R. Psychrophilic lifestyles: Mechanisms of adaptation and biotechnological tools. Appl. Microbiol. Biotechnol. 2019, 103, 2857–2871. [Google Scholar] [CrossRef]
- De Maayer, P.; Anderson, D.; Cary, C.; Cowan, D.A. Some like it cold: Understanding the survival strategies of psychrophiles. EMBO Rep. 2014, 15, 508–517. [Google Scholar] [CrossRef]
- Yoshimune, K.; Galkin, A.; Kulakova, L.; Yoshimura, T.; Esaki, N. Cold-active DnaK of an Antarctic psychrotroph Shewanella sp. Ac10 supporting the growth of dnaK-null mutant of Escherichia coli at cold temperatures. Extremophiles 2005, 9, 145–150. [Google Scholar] [CrossRef]
- Białkowska, A.; Majewska, E.; Olczak, A.; Twarda-Clapa, A. Ice binding proteins: Diverse biological roles and applications in different types of industry. Biomolecules 2020, 10, 274. [Google Scholar] [CrossRef] [Green Version]
- Ranawat, P.; Rawat, S. Stress response physiology of thermophiles. Arch. Microbiol. 2017, 199, 391–414. [Google Scholar] [CrossRef]
- Sprott, G.D.; Meloche, M.; Richards, J.C. Proportions of diether, macrocyclic diether, and tetraether lipids in Methanococcus jannaschii grown at different temperatures. J. Bacteriol. 1991, 173, 3907–3910. [Google Scholar] [CrossRef] [Green Version]
- Mansilla, M.C.; Cybulski, L.E.; Albanesi, D.; de Mendoza, D. Control of membrane lipid fluidity by molecular thermosensors. J. Bacteriol. 2004, 186, 6681–6688. [Google Scholar] [CrossRef] [Green Version]
- Valenti, A.; Perugino, G.; Rossi, M.; Ciaramella, M. Positive supercoiling in thermophiles and mesophiles: Of the good and evil. Biochem. Soc. Trans. 2011, 39, 58–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sieck, G. Life at the extreme: Physiological adaptation. Physiology 2015, 30, 84–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Cen, Z.; Zhao, J. The survival mechanisms of thermophiles at high temperatures: An angle of omics. Physiology 2015, 30, 97–106. [Google Scholar] [CrossRef]
- Ferreira, A.C.; Nobre, M.F.; Rainey, F.A.; Silva, M.T.; Wait, R.; Burghardt, J.; Chung, A.P.; Da Costa, M.S. Deinococcus geothermalis sp. nov. and Deinococcus murrayi sp. nov., two extremely radiation-resistant and slightly thermophilic species from hot springs. Int. J. Syst. Evol. 1997, 47, 939–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Kim, M.C.; Wang, L.; Zhu, G.; Zhang, Y.; Huang, Y.; Wei, Z.; Danzeng, W.; Peng, F. Deinococcus taklimakanensis sp. nov. isolated from desert soil. Int. J. Syst. Evol. 2017, 67, 4311–4316. [Google Scholar] [CrossRef]
- Srinivasan, S.; Lim, S.Y.; Lim, J.-H.; Jung, H.-Y.; Kim, M.K. Deinococcus rubrus sp. nov., a bacterium isolated from Antarctic coastal sea water. J. Microbiol. Biotechnol. 2017, 27, 535–541. [Google Scholar] [CrossRef]
- Park, M.R.; Song, J.H.; Nam, G.G.; Joung, Y.C.; Zhao, L.; Kim, M.-K.; Cho, J.C. Deinococcus lacus sp. nov., a gamma radiation-resistant bacterium isolated from an artificial freshwater pond. Int. J. Syst. Evol. 2018, 68, 1372–1377. [Google Scholar] [CrossRef] [PubMed]
- Tanner, K.; Molina-Menor, E.; Latorre-Pérez, A.; Vidal-Verdú, À.; Vilanova, C.; Peretó, J.; Porcar, M. Extremophilic microbial communities on photovoltaic panel surfaces: A two-year study. Microb. Biotechnol. 2020, 13, 1819–1830. [Google Scholar] [CrossRef]
- Slade, D.; Radman, M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. 2011, 75, 133–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radjpurohit, Y.S.; Bihani, S.C.; Waldor, M.K.; Misra, H.S. Phosphorylation of Deinococcus radiodurans RecA regulates its activity and may contribute to radioresistance. J. Biol. Chem. 2016, 291, 16672–16685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, M.; Earl, A.M.; Howell, H.A.; Park, M.J.; Eisen, J.A.; Peterson, S.N.; Battista, J.R. Analysis of Deinococcus radiodurans’s transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics 2004, 168, 21–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, S.Y.; Jung, J.H.; Blanchard, L.; de Groot, A. Conservative and diversity of radiation and oxidative stress resistance mechanisms in Deinococcus radiodurans. FEMS Microbiol. Rev. 2019, 43, 19–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, M.; Xiao, A.; Zhang, Z.; Huang, H.; Jiang, L. The diversity and commonalities of the radiation-resistance mechanisms of Deinococcus and its up-to-date applications. AMB Express. 2019, 9, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Floc’h, K.; Lacroix, F.; Servant, P.; Wong, Y.-S.; Kelman, J.-P.; Bourgeois, D.; Timmins, J. Cell morphology and nucleoid dynamic in dividing Deinococcus radiodurans. Nat. Commun. 2019, 10, 3815. [Google Scholar] [CrossRef] [Green Version]
- Jeong, S.-W.; Jung, J.H.; Kim, M.K.; Seo, H.S.; Lim, H.-M.; Lim, S.Y. The three catalases in Deinococcus radiodurans: Only two show catalase activity. Biochem. Biophys. Res. Commun. 2016, 469, 443–448. [Google Scholar] [CrossRef]
- Maqbool, I.; Sudharsan, M.; Kanimozhi, G.; Alrashood, S.T.; Khan, H.A.; Prasad, N.R. Crude cell-free extract from Deinococcus radiodurans exhibit anticancer activity by inducing apoptosis in triple-negative breast cancer cells. Front. Cell Dev. Biol. 2020, 8, 707. [Google Scholar] [CrossRef]
- Choi, J.Y.; Lee, K.J.; Lee, P.C. Characterization of carotenoid biosynthesis in newly isolated Deinococcus sp. AJ005 and investigation of the effects of environmental conditions on cell growth and carotenoid biosynthesis. Mar. Drugs 2019, 17, 705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daly, M.J.; Gaidamakova, E.K.; Matrosova, V.Y.; Kiang, J.G.; Fukumoto, R.; Lee, D.-Y.; Wehr, N.B.; Viteri, G.A.; Berlett, B.S.; Levine, R.L. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS ONE 2010, 5, e12570. [Google Scholar] [CrossRef]
- Santos, S.P.; Yang, Y.; Rosa, M.T.G.; Rodrigues, M.A.A.; De La Tour, C.B.; Sommer, S.; Teixeira, M.; Carrondo, M.A.; Cloetens, P.; Abreu, I.A.; et al. The interplay between Mn and Fe in Deinococcus radiodurans triggers cellular protection during paraquat-induced oxidative stress. Sci. Rep. 2019, 9, 17217. [Google Scholar] [CrossRef] [PubMed]
- Robinson, C.K.; Webb, K.; Kaur, A.; Jaruga, P.; Dizdaroglu, M.; Baliga, N.S.; Place, A.; DiRuggiero, J. A major role for nonenzymatic antioxidant processes in the radioresistance of Halobacterium salinarum. J. Bacteriol. 2011, 193, 1653–1662. [Google Scholar] [CrossRef] [Green Version]
- Gumulya, Y.; Boxall, N.J.; Khaleque, H.N.; Santala, V.; Carlson, R.P.; Kaksonen, A.H. In a quest for engineering acidophiles for biomining applications: Challenges and opportunities. Genes 2018, 9, 116. [Google Scholar] [CrossRef] [Green Version]
- Navarro, C.A.; Von Bernath, D.; Jerez, C.A. Heavy metal resistance strategies of acidophilic bacteria and their acquisition: Importance for biomining and bioremediation. Biol. Res. 2013, 46, 363–371. [Google Scholar] [CrossRef] [Green Version]
- Saavedra, A.; Aguirre, P.; Gentina, J.C. Biooxidation of iron by Acidithiobacillus ferroxidans in the presence of D-galactose: Understanding its influence on the production of EPS and cell tolerance to high concentration of iron. Front. Microbiol. 2020, 11, 759. [Google Scholar] [CrossRef] [Green Version]
- Jafari, M.; Abdollahi, H.; Shafaei, S.Z.; Gharabaghi, M.; Jafari, H.; Akcil, A. Acidophilic bioleaching: A review on the process and effect of organic-inorganic reagents and materials on its efficiency. Min. Proc. Ext. Met. Rev. 2019, 2, 87–107. [Google Scholar] [CrossRef]
- Brierley, J.A. A perspective on developments in biohydrometallurgy. Hydrometallurgy 2008, 94, 2–7. [Google Scholar] [CrossRef]
- Chen, P.; Yan, L.; Leng, F.; Nan, W.; Yue, X.; Zheng, Y.; Feng, N.; Li, H. Bioleaching of realgar by Acidithiobacillus ferrooxidans using ferrous iron and elemental sulfur as the sole and mixed energy sources. Bioresour. Technol. 2011, 102, 3260–3267. [Google Scholar] [CrossRef]
- Zhang, S.; Yan, L.; Xing, W.; Chen, P.; Zhang, Y.; Wang, W. Acidothiobacillus ferrooxidans and its potential application. Extremophiles 2018, 22, 563–579. [Google Scholar] [CrossRef] [PubMed]
- Romero-González, M.; Nwaobi, B.C.; Hufton, J.M.; Gilmour, D.J. Ex-situ bioremediation of U(VI) from contaminated mine water using Acidothiobacillus ferrooxidans strains. Front. Environ. Sci. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Jameson, E.; Rowe, O.F.; Hallberg, K.B.; Johnson, D.B. Sulfidogenesis and selective precipitation of metals at low pH mediated by Acidithiobacillus spp. and acidophilic sulfate-reducing bacteria. Hydrometallurgy 2010, 104, 488–493. [Google Scholar] [CrossRef]
- Okibe, N.; Maki, M.; Nakayama, D.; Sasaki, K. Microbial recovery of vanadium by the acidophilic bacterium, Acidocella aromatica. Biotechnol. Lett. 2016, 38, 1475–1481. [Google Scholar] [CrossRef]
- Chakravarty, R.; Banerjee, P.C. Mechanism of cadmium binding on the cell wall of an acidophilic bacterium. Bioresour. Technol. 2012, 108, 176–183. [Google Scholar] [CrossRef]
- Beolchini, F.; Dell’Anno, A.; De Propris, L.; Ubaldini, S.; Cerrone, F.; Danovaro, R. Auto- and heterotrophic acidophilic bacteria enhance the bioremediation efficiency of sediments contaminated by heavy metals. Chemosphere 2009, 74, 1321–1326. [Google Scholar] [CrossRef]
- Gupta, A.; Sar, P. Characterization and application of an anaerobic, iron and sulfate reducing bacterial culture in enhanced bioremediation of acid mine drainage impacted soil. J. Environ. Sci. Health C 2020, 4, 464–482. [Google Scholar] [CrossRef]
- Abd-Elnaby, H.; Abou-Elela, G.M.; El-Sersy, N.A. Cadmium resisting bacteria in Alexandria Eastern Harbor (Egypt) and optimization of cadmium bioaccumulation by Vibrio harveyi. Afr. J. Biotechnol. 2011, 10, 3412–3423. [Google Scholar] [CrossRef]
- Iyer, A.; Mody, K.; Jha, B. Biosorption of heavy metals by a marine bacterium. Mar. Pollut. Bull. 2005, 50, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Ӧzdemir, S.; Kilinc, E.; Poli, A.; Nicolaus, B. Biosorption of heavy metals (Cd2+, Cu2+, Co2+, and Mn2+) by thermophilic bacteria, Geobacillus thermantarcticus and Anoxybacillus amylolyticus: Equilibrium and kinetic studies. Bioremediat. J. 2013, 17, 86–96. [Google Scholar] [CrossRef]
- Eslami, N.; Kermanshahi, R.K.; Erfan, M. Studying the stability of S-layer protein of Lactobacillus acdiophilius ATCC 4356 in simulated gastrointestinal fluids using SDS-PAGE and circular dichroism. Iran J. Pharm. Res. 2013, 12, 47–56. [Google Scholar] [PubMed]
- Liu, S.; Zheng, Y.; Ma, Y.; Sarwar, A.; Zhao, X.; Luo, T.; Yang, Z. Evaluation and proteomic analysis of lead adsorption by lactic acid bacteria. Int. J. Mol. Sci. 2019, 20, 5540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerbino, E.; Mobili, P.; Tymczyszyn, E.; Fausto, R.; Gómez-Zavaglia, A. FTIR spectroscopy structural analysis of the interaction between Lactobacillus kefir S-layers and metal ions. J. Mol. Struct. 2011, 1–3, 186–192. [Google Scholar] [CrossRef] [Green Version]
- Kashefi, K.; Lovely, D.R. Reduction of Fe(III), Mn(IV), and toxic metals at 100 °C by Pyrobacculum islandicum. Appl. Environ. Microbiol. 2000, 66, 1050–1056. [Google Scholar] [CrossRef] [Green Version]
- Feitkenhauer, H.; Muller, R.; Markl, H. f. Degradation of polycyclic aromatic hydrocarbons and long chain alkanes at 60-70 degrees C by Thermus and Bacillus spp. Biodegradation 2003, 14, 367–372. [Google Scholar] [CrossRef]
- Nazina, T.N.; Tourova, T.P.; Poltaraus, A.B.; Novikova, E.V.; Grigoryan, A.A.; Ivanova, A.E.; Lysenko, A.M.; Petrunyaka, V.V.; Osipov, G.A.; Belyaev, S.S.; et al. Taxonomic study of aerobic thermophilic bacilli: Descriptions of Geobacillus subterraneus gen. nov., sp. nov., and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. th. Int. J. Syst. Evol. Microbiol. 2001, 51, 433–446. [Google Scholar] [CrossRef] [Green Version]
- Feng, L.; Wang, W.; Cheng, J.; Ren, Y.; Zhao, G.; Gao, C.; Tang, Y.; Liu, X.; Han, W.; Peng, X.; et al. Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc. Natl. Acad. Sci. USA 2007, 104, 5602–5607. [Google Scholar] [CrossRef] [Green Version]
- Sood, N.; Lal, B. (2008). Isolation and characterization of a potential paraffin-wax degrading thermophilic bacterial strain Geobacillus kaustophilus TERI NSM for application in oil wells with paraffin deposition problems. Chemosphere 2008, 70, 1445–1451. [Google Scholar] [CrossRef]
- Sun, Y.; Ning, Z.; Yang, F.; Li, X. Characteristics of newly isolated Geobacillus sp. ZY-10 degrading hydrocarbons in crude oil. Pol. J. Microbiol. 2015, 64, 253–263. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Liu, J.; Li, R.; Shen, B. Isolation of a thermophilic bacterium, Geobacillus sp. SH-1, capable of degrading aliphatic hydrocarbons and naphthalene simultaneously, and identification of its naphthalene degrading pathway. Bioresour. Technol. 2012, 124, 83–89. [Google Scholar] [CrossRef]
- Elumalai, P.; Parthipan, P.; Karthikeyan, O.P.; Rajasekar, A. Enzyme-mediated biodegradation of long-chain n-alkanes (C32 and C40) by thermophilic bacteria. 3 Biotech. 2017, 7, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, X.; Yang, B.; Shen, J.; Du, N. Biodegradation of crude oil by an Arctic psychrotrophic bacterium Pseudoalteromomas sp. P29. Curr. Microbiol. 2009, 59, 341–345. [Google Scholar] [CrossRef]
- Gentile, G.; Bonsignore, M.; Santisi, S.; Catalfamo, M.; Giuliano, L.; Genovese, L.; Yakimov, M.M.; Denaro, R.; Genovese, M.; Cappello, S. Biodegradation potentiality of psychrophilic bacterial strain Oleispira antarctica RB-8(T). Mar. Pollut. Bull. 2016, 105, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Cui, Z.; Li, Q.; Xu, G.; Jia, X.; Zheng, L. Marinobacter nanhaiticus sp. nov., polycyclic aromatic hydrocarbon-degrading bacterium isolated from the sediment of the South China Sea. Antonie Van Leeuwenhoek 2013, 103, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Al-Mailem, D.M.; Eliyas, M.; Radwan, S.S. Oil-bioremediation potential of two hydrocarbonoclastic, diazotrophic Marinobacter strains from hypersaline areas along the Arabian Gulf coasts. Extremophiles 2013, 17, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, I.M.; Konova, S.V.; Sigida, E.N.; Lyubun, E.V.; Muratova, A.Y.; Fedonenko, Y.P.; Elbanna, K. Bioremediation potential of a halophilic Halobacillus sp. strain EG1HP4QL: Exopolysaccharide production, crude oil degradation, and heavy metal tolerance. Extremophile 2020, 24, 157–166. [Google Scholar] [CrossRef]
- Gutierrez, T.; Berry, D.; Yang, T.; Mishamandani, S.; McKay, L.; Teske, A.; Aitken, M.D. Role of exopolysaccharide (EPS) in the fate of the oil released during the Deepwater Horizon oil spill. PLoS ONE 2013, 8, e67717. [Google Scholar] [CrossRef]
- Sadegh, H.; Ali, G.A.M.; Gupta, V.K.; Makhlouf, A.S.H.M.; Shahryari-ghoshekandi, R.; Nadagouda, M.N.; Sillanpää, M.; Megiel, E. The role of nanomaterials as effective adsorbents and their applications in wastewater treatment. J. Nanostruct. Chem. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.-H.; Jeon, J.H.; Hong, S.H.; Rhim, W.-K.; Lee, Y.-S.; Youn, H.W.; Chung, J.-K.; Lee, M.C.; Lee, D.S.; Kang, K.W.; et al. Tumor targeting and imaging using cyclic RGD-PEGlated gold nanoparticle probes with directly conjugated iodine-125. Small 2011, 7, 2052–2060. [Google Scholar] [CrossRef]
- Choi, M.H.; Shim, H.E.; Yun, S.J.; Park, S.H.; Choi, D.S.; Jang, B.-S.; Choi, Y.J.; Jeon, J.H. Gold-nanoparticle-immobilized desalting column for highly efficient and specific removal of radioactive iodine in aqueous media. Acs Appl. Mater. Interspace 2016, 8, 29227–29231. [Google Scholar] [CrossRef]
- Mushtaq, S.; Yun, S.-J.; Yang, J.E.; Jeong, S.-W.; Shim, H.E.; Choi, M.H.; Park, S.H.; Choi, Y.J.; Jeon, J.H. Efficient and selective removal of radioactive iodine anions using engineered nanocomposite membranes. Environ. Sci. Nano 2017, 4, 2157–2163. [Google Scholar] [CrossRef]
- Lloyd, J.R. Microbial reduction of metals and radionuclides. FEMS Microbiol. Rev. 2003, 27, 411–425. [Google Scholar] [CrossRef]
- Kumar, R.; Singh, S.; Singh, O.V. Bioremediation of radionuclides: Emerging technologies. OMICS 2007, 11, 295–304. [Google Scholar] [CrossRef]
- Prakash, D.; Gabani, P.; Chandel, A.K.; Ronen, Z.; Singh, O.V. Bioremediation: A genuine technology to remediate radionuclides from the environment. Microb. Biotechnol. 2013, 6, 349–360. [Google Scholar] [CrossRef]
- Shukla, A.; Parmar, P.; Sarar, M. Radiation, radionuclides and bacteria: An in-perspective review. J. Environ. Radioact. 2017, 180, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Wildung, R.E.; Gorby, Y.A.; Krupka, K.M.; Hess, N.J.; Li, S.W.; Plymale, A.E.; McKinley, J.P.; Fredrickson, J.K. Effect of electron donor and solution chemistry on products of dissimilatory reduction of technetium by Shewanella putrefaciens. Appl. Environ. Microbiol. 2000, 66, 2451–2460. [Google Scholar] [CrossRef] [Green Version]
- Istok, J.D.; Senko, J.M.; Krumholz, L.R.; Watson, D.; Bogle, M.A.; Peacock, A.; Chang, Y.-J.; White, D.C. In situ bioreduction of technetium and uranium in a nitrate-contaminated aquifer. Environ. Sci. Technol. 2004, 38, 468–475. [Google Scholar] [CrossRef]
- Panak, P.J.; Nitsche, H. Interaction of aerobic soil bacteria with plutonium(VI). Radiochim. Acta. 2001, 89, 499–504. [Google Scholar] [CrossRef]
- Kim, S.-J.; Koh, D.-C.; Park, S.-J.; Cha, I.-T.; Park, J.-W.; Na, J.-H.; Roh, Y.; Ko, K.-S.; Kim, K.J.; Rhee, S.-K. Molecular analysis of spatial variation of iron-reducing bacteria in riverine alluvial aquifers of the Mankyeong River. J. Microbiol. 2012, 50, 207–217. [Google Scholar] [CrossRef]
- Anderson, R.T.; Vrionis, H.A.; Ortiz-Bernard, I.; Resch, C.T.; Long, P.E.; Dayvault, R.; Karp, K.; Metzler, D.R.; Peacock, A.; White, D.C.; et al. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 2003, 69, 5884–5891. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, A.C.; Nobre, M.F.; Moore, E.; Rainey, F.A.; Battista, J.R.; Da Costa, M.S. Characterization and radiation resistance of new isolates of Rubrobacter radiotolerans and Rubrobacter xylanophilus. Extremophiles 1999, 3, 235–238. [Google Scholar] [CrossRef]
- Billi, D.; Friedmann, E.I.; Hofer, K.G.; Caiola, M.G.; Ocampo-Friedmann, R. Ionizing-radiation resistance in the desiccation-tolerant cyanobacterium Chroococcidiopsis. Appl. Environ. Microbiol. 2000, 66, 1489–1492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brim, H.; Mcfarlan, S.C.; Fredrickson, J.K.; Minton, K.W.; Zhai, M.; Wackett, L.P.; Daly, M.J. Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nat. Biotechnol. 2000, 18, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Daly, M.J. Engineering radiation-resistant bacteria for environmental biotechnology. Curr. Opin. Biotechnol. 2000, 11, 280–285. [Google Scholar] [CrossRef]
- Jeong, S.-W.; Choi, Y.J. Research perspective of an extremophilic bacterium, Deinococcus radiodurans on bioremediation of radioactive wastes. Appl. Chem. Eng. 2017, 28, 133–140. [Google Scholar] [CrossRef]
- Fredrickson, J.K.; Kostandarithes, H.M.; Li, S.W.; Plymale, A.E.; Daly, M.J. Reduction of Fe(III), Cr(VI), U(VI), and Tc(VII) by Deinococcus radiodurans R1. Appl. Environ. Microbiol. 2000, 66, 2006–2011. [Google Scholar] [CrossRef] [Green Version]
- Appukuttan, D.; Rao, A.S.; Apte, S.K. Engineering of Deinococcus radiodurans R1 for bioprecipitation of uranium from dilute nuclear waste. Appl. Environ. Microbiol. 2006, 72, 7873–7878. [Google Scholar] [CrossRef] [Green Version]
- Misra, C.S.; Appukuttan, D.; Kantamreddi, V.S.S.; Rao, A.S.; Apte, S.K. Recombinant, D. radiodurans cells for bioremediation of heavy metals from acidic/neutral aqueous wastes. Bioeng. Bugs 2012, 3, 44–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appukuttan, D.; Seetharam, C.; Padma, N.; Rao, A.S.; Apte, S.K. PhoN-expressing, lyophilized, recombinant Deinococcus radiodurans cells for uranium bioprecipitation. J. Biotechnol. 2011, 154, 285–290. [Google Scholar] [CrossRef]
- Gogada, R.; Singh, S.S.; Lunavat, S.K.; Pamarthi, M.M.; Rodrigue, A.; Vadivelu, B.; Phanithi, P.-B.; Gopala, V.; Apte, S.K. Engineered Deinococcus radiodurans R1 with NiCoT genes for bioremoval of trace cobalt from spent decontamination solutions of nuclear power reactors. Appl. Microbiol. Biotechnol. 2015, 99, 9203–9213. [Google Scholar] [CrossRef]
- Choi, M.H.; Jeong, S.-W.; Shim, H.E.; Yun, S.J.; Mushtaq, S.; Choi, D.S.; Jang, B.-S.; Jung, E.Y.; Choi, Y.J.; Jeon, J.H. Efficient bioremediation of radioactive iodine using biogenic gold nanomaterial-containing radiation-resistant bacterium, Deinococcus radiodurans R1. Chem. Comm. 2017, 53, 3937–3940. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, R.R.; Shaiwale, N.S.; Deobagkar, D.N.; Deobagkar, D.D. Synthesis and extracellular accumulation of silver nanoparticles by employing radiation-resistant Deinococcus radiodurans, their characterization, and determination of bioactivity. Int. J. Nanomed. 2015, 10, 963–974. [Google Scholar] [CrossRef] [Green Version]
- Beeler, E.; Singh, O.V. Extremophiles as sources of inorganic bio-nanoparticles. World, J. Microbiol. Biotechnol. 2016, 32, 156. [Google Scholar] [CrossRef]
- Li, J.; Li, Q.; Ma, X.; Tian, B.; Yu, J.; Dai, S.; Weng, Y.; Hua, Y. Biosynthesis of gold nanoparticles by the extreme bacterium Deinococcus radiodurans and an evaluation of their antibacterial properties. Int. J. Nanomed. 2016, 11, 5931–5944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, A.; Contreras, L.M.; Keitz, B.K. Imposed environmental stresses facilitate cell-free nanoparticle formation by Deinococcus radiodurans. Appl. Environ. Microbiol. 2017, 83, e00798-17. [Google Scholar] [CrossRef] [Green Version]
- Kitjanukit, S.; Sasaki, K.; Okibe, N. Production of highly catalytic, archaeal Pd(0) bionanoparticles using Sulfolobus tokodaii. Extremophiles 2019, 20, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Okibe, N.; Nakayama, D.; Matsumoto, T. Palladium bionanoparticles production from acidic Pd(II) solutions and spent catalyst leachate using acidophilic Fe(II)-reducing bacteria. Extremophiles 2017, 21, 1091–1100. [Google Scholar] [CrossRef]
- Crini, G.; Lichtfouse, E. Advantages and disadvantages of techniques used for wastewater treatment. Environ. Chem. Lett. 2019, 17, 145–155. [Google Scholar] [CrossRef]
- Ren, J.; Lee, J.G.; Na, D.K. Recent advances in genetic engineering tools based on synthetic biology. J. Microbiol. 2020, 58, 1–10. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, D.; Wang, X.; Tao, H.; Feng, E.; Zhu, L.; Pan, C.; Wang, B.; Liu, C.; Liu, X.; et al. Highly efficient genome engineering in Bacillus anthracis and Bacillus cereus using CRISPR/Cas9 system. Front. Microbiol. 2019, 10, 1932. [Google Scholar] [CrossRef] [Green Version]
- Marques, C.R. Extremophilic microfactories: Application in metal and radionuclide bioremediation. Front. Microbiol. 2018, 9, 1191. [Google Scholar] [CrossRef] [PubMed]
- Giovanella, P.; Vieira, G.A.L.; Ramos Otero, I.V.; Pellizzer, E.P.; de Jesus Fontes, B.; Sette, L.D. Metal and organic pollutants bioremediation by extremophile microorganisms. J. Hazard. Mater. 2020, 382, 121024. [Google Scholar] [CrossRef] [PubMed]
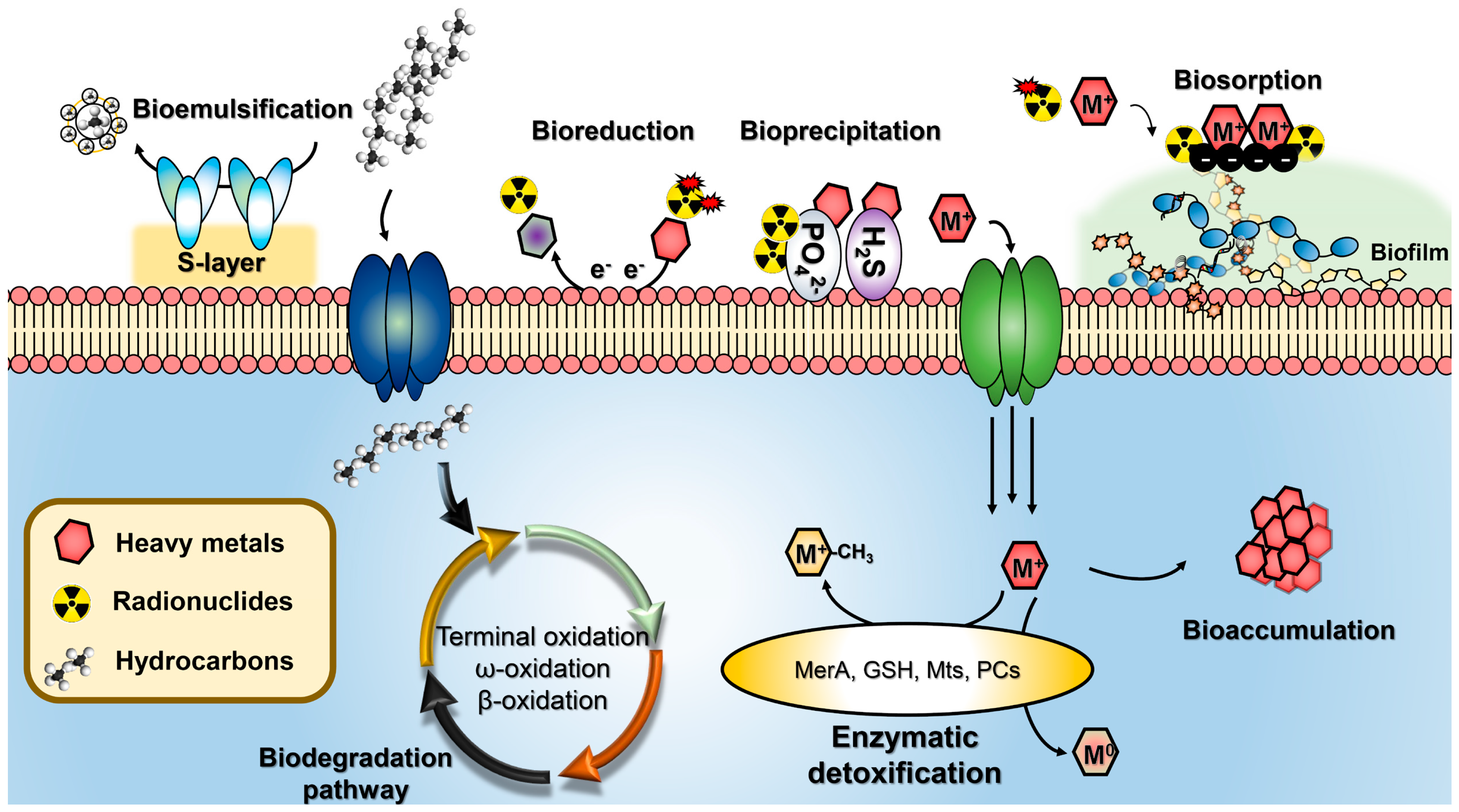
| Heavy Metal | Method/Mechanism | Extremophile | Resistance 1 | Removal Efficiency (Initial Concentration) 2 | Reference |
|---|---|---|---|---|---|
| As(III) | Bioleaching 3 | Acidothiobacillus ferrooxidans BY-3 | Low pH (pH < 1.8) | 35.9% (ND) | [80] |
| U(VI) | Bioleaching | At. ferrooxidans | Low pH (pH 1.5–4.5) | 50% (100 mg/L) | [82] |
| Cu(II) | Bioprecipitation | Acidothiobacillus ferrivorans | Low pH (pH 2.5) | >99% (50 mM) | [83] |
| V(V) | Bioreduction | Acidocella aromatica | Low pH (pH 2.5) | 70% (1 mM) | [84] |
| Cd(II) | Biosorption | Acidiphilium symbioticum H8 | ND | 248.62 mg Cd(II)/g biomass (250 mg/L) | [85] |
| Bioaccumulation | Vibrio harveyi | 60 mg/L MIC | 84% (30–60 mg/L) | [88] | |
| Biosorption | Enterobacter cloaceae | ND | 65% (100 mg/L) | [89] | |
| Biosorption | Geobacillus thermantarcticus, Anoxybacillus amylolyticus | High temperature (80 °C) | 85.4%, 74.1% (50 mg/L) | [90] | |
| Cr(VI) | Bioreduction | Pyrobaculum islandicum | High temperature (100 °C) | 100% (600 μM) | [94] |
| Hydrocarbons | Extremophile | Resistance | Removal Efficiency (Initial Concentration) | Reference |
|---|---|---|---|---|
| acenaphthene, fluoranthene, pyrene, benzo[e]pyrene | Bacillus spp., Thermus sp. | High temperature (60–70 °C) | 35–77% (30–60 mg/L) | [95] |
| Pentadecane, octadecane, octacosane | Geobacillus sp. SH-1 | High temperature (70 °C) | >70% (100 mg/L) | [100] |
| Rotricontane, tetracotane | Geobacillus thermoparaffinivorans, Geobacillus stearothermophillus, Bacillus licheniformis | High temperature (50 °C) | >87% (1 g/L) | [101] |
| Mixed oil | Pseudoalteromonas sp. P29 | Low temperature (5 °C) | 90% (2 g/L) | [102] |
| Diesel, jet fuel, crude oil | Oleispira antarctica RB-8T | Low temperature (4–15 °C) | 53.7–79.4% (1 g/L) | [103] |
| Biphenyl, phenanthrene, anthracene, naphthalene | Marinobacter sedimentalis, Marinobacter falvimaris, Marinobacter nanhaiticus | High salinity (5M NaCl) | 70–90% (0.2–3 g/L) | [104,105] |
| Radionuclide | Extremophile | Resistance | Removal Efficiency | Reference |
|---|---|---|---|---|
| U(VI), Cr(VI), Tc(VII) | Deinococcus geothermalis | Radiation (12 kGy), high temperature (55 °C) | >90% | [123] |
| D. radiodurans | ND | 95–100% | [126] | |
| U(VI) | D. radiodurans expressing PhoN | Radiation (6 kGy) | >90% | [127] |
| Co-60 | D. radiodurans expressing NiCoT | Radiation (6.4 kGy) | >60% | [130] |
| I-125 | D. radiodurans | Radiation (8 kGy) | >99% | [131] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, S.-W.; Choi, Y.J. Extremophilic Microorganisms for the Treatment of Toxic Pollutants in the Environment. Molecules 2020, 25, 4916. https://doi.org/10.3390/molecules25214916
Jeong S-W, Choi YJ. Extremophilic Microorganisms for the Treatment of Toxic Pollutants in the Environment. Molecules. 2020; 25(21):4916. https://doi.org/10.3390/molecules25214916
Chicago/Turabian StyleJeong, Sun-Wook, and Yong Jun Choi. 2020. "Extremophilic Microorganisms for the Treatment of Toxic Pollutants in the Environment" Molecules 25, no. 21: 4916. https://doi.org/10.3390/molecules25214916
APA StyleJeong, S.-W., & Choi, Y. J. (2020). Extremophilic Microorganisms for the Treatment of Toxic Pollutants in the Environment. Molecules, 25(21), 4916. https://doi.org/10.3390/molecules25214916






