Myeloma Cells Deplete Bone Marrow Glutamine and Inhibit Osteoblast Differentiation Limiting Asparagine Availability
Simple Summary
Abstract
1. Introduction
2. Results
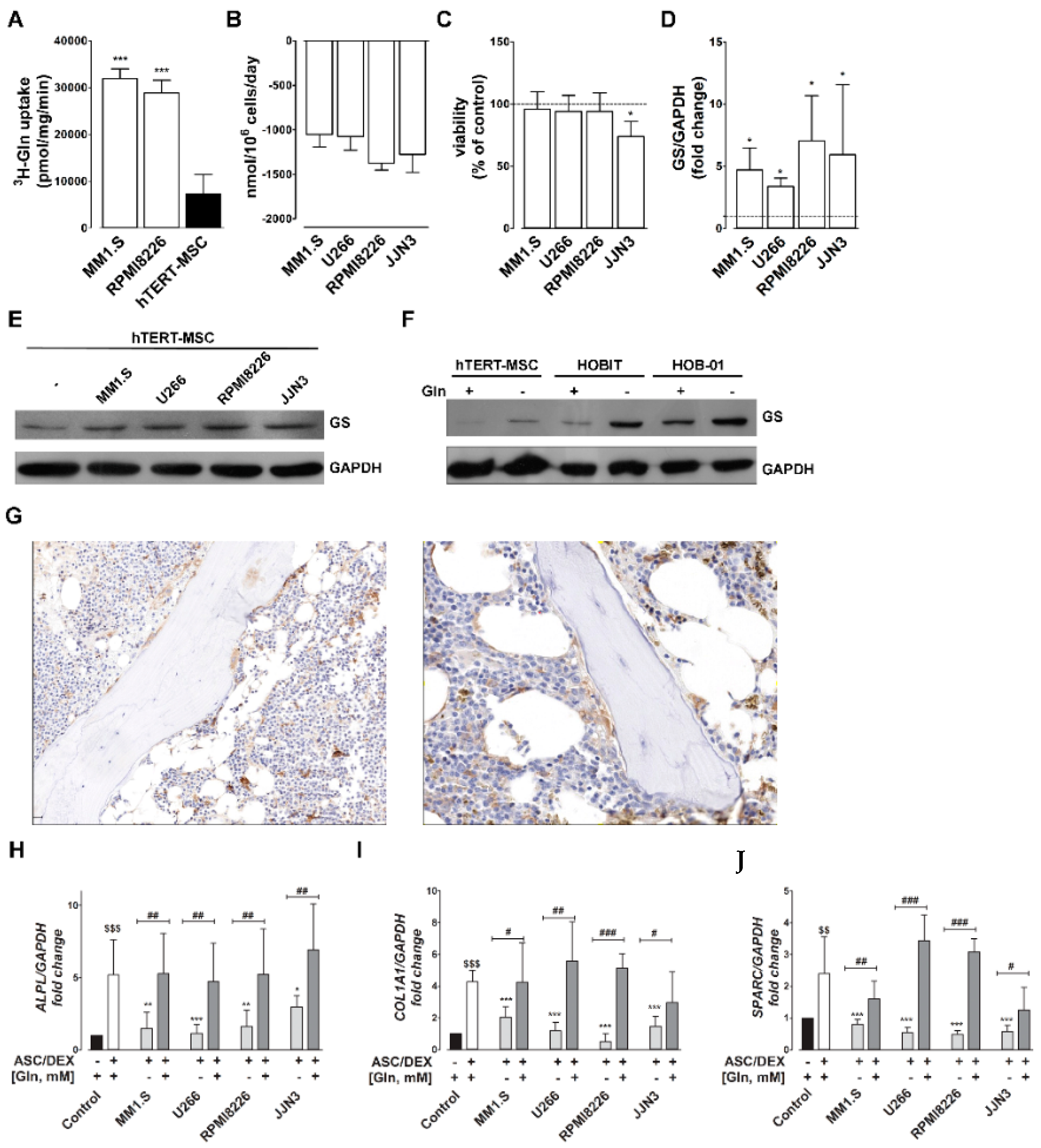
2.1. Gln Consumption by MM Cells Induces GS in MSCs and Inhibits Their OB Differentiation
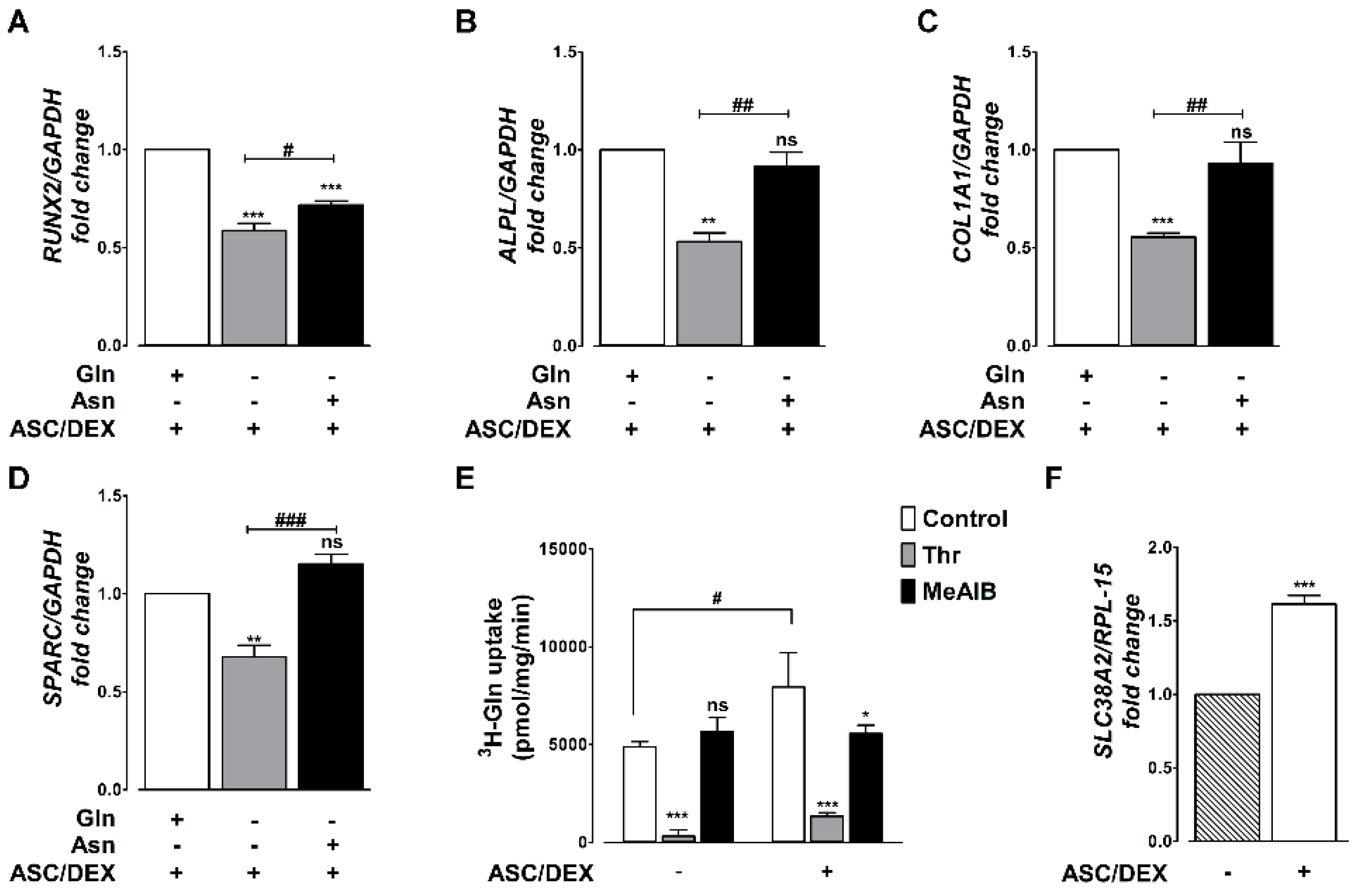
2.2. Gln Deprivation Hinders the Expression of OB Markers in MSCs
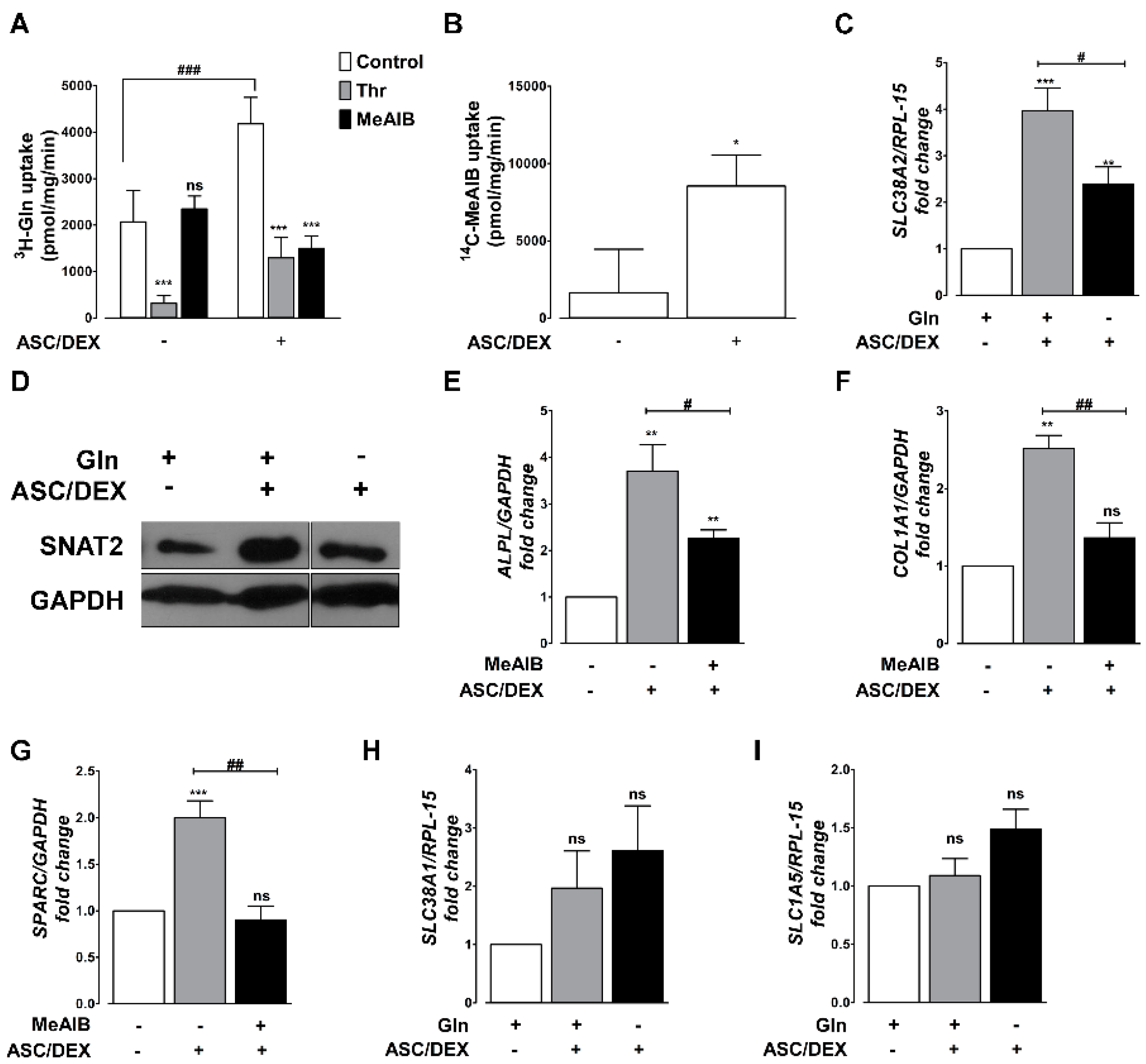
2.3. OB Differentiation Is Associated with the Induction of the SNAT2 Transporter
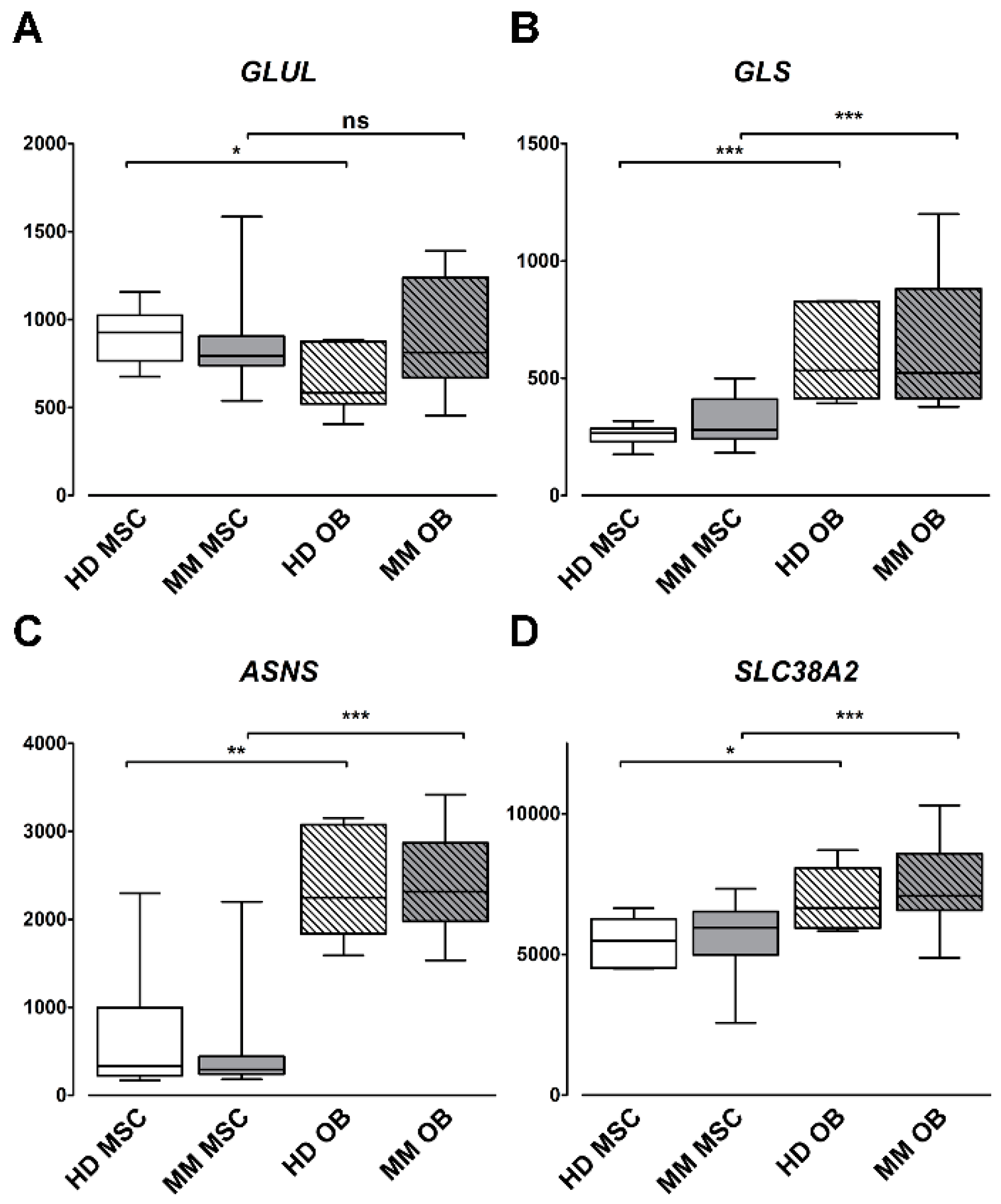
2.4. Gln-Related Gene Expression by Primary Human OBs and MSCs in MM Patients and HD
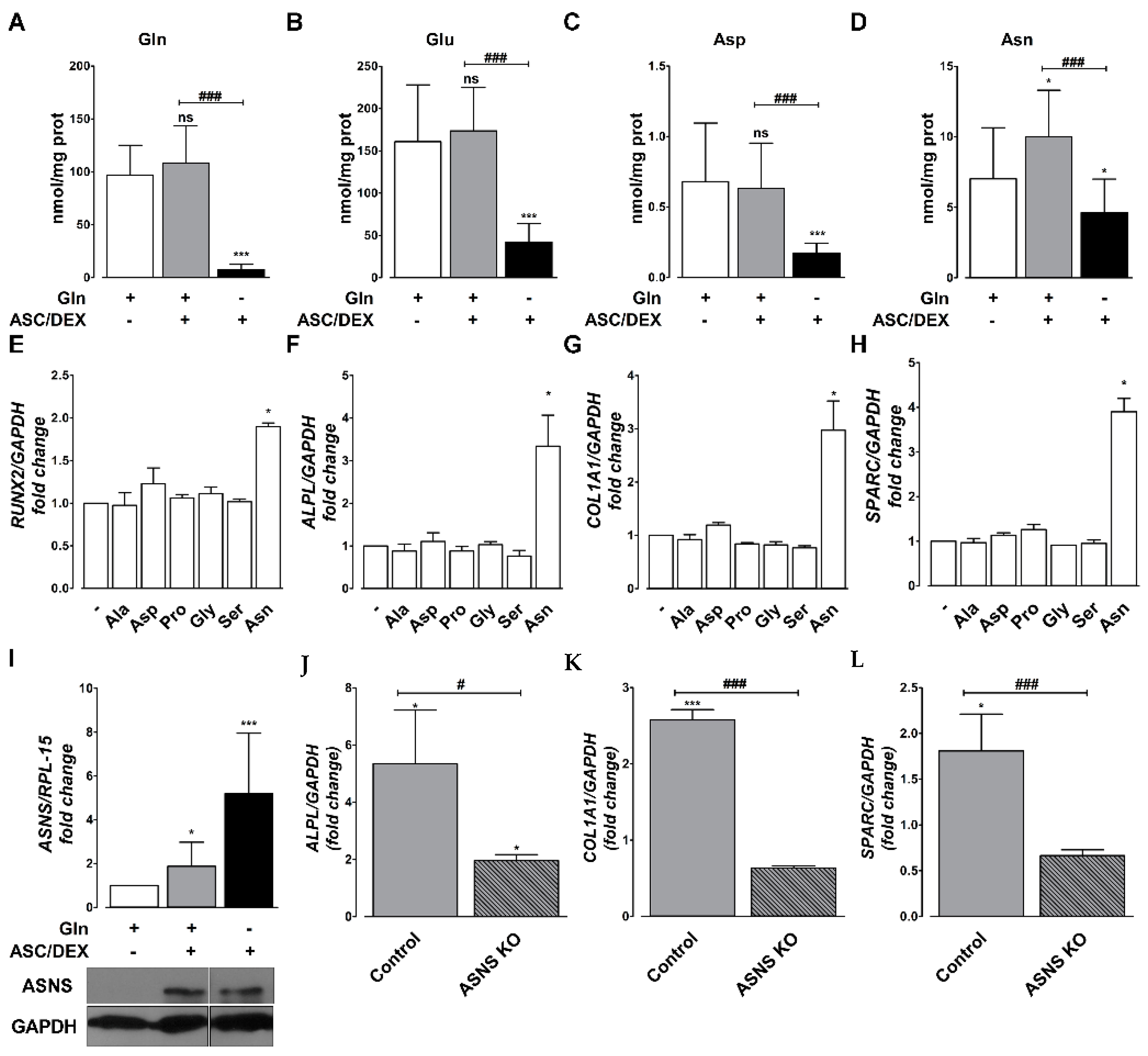
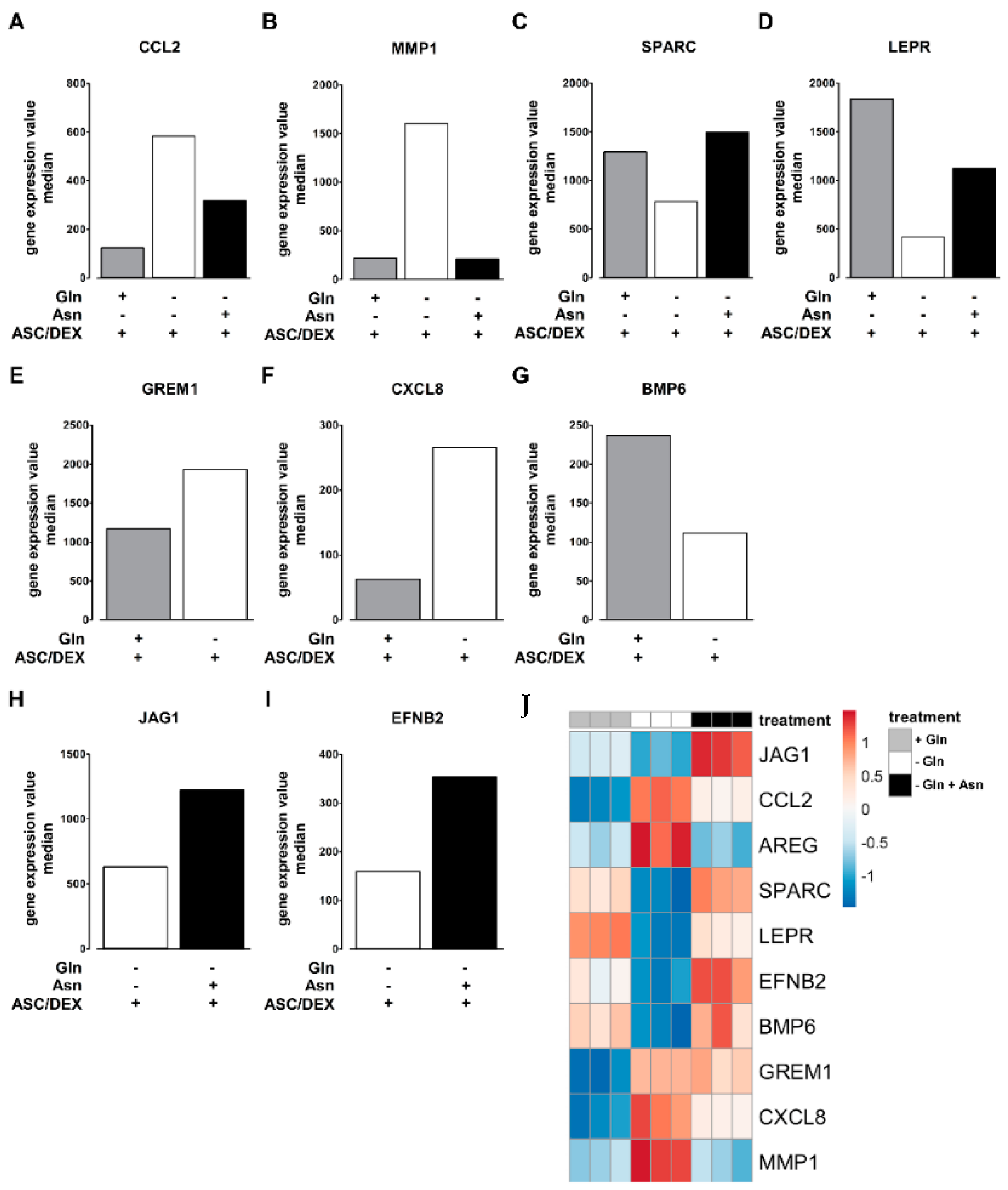
2.5. Asn Content Increases during OB Differentiation of MSCs
2.6. Asn Supplementation Corrects Changes in Transcriptional Profiles of hTERT-MSCs Induced by Gln Deprivation
2.7. Amino Acid Dependence of OB Differentiation of Primary BM MSCs
3. Discussions
4. Materials and Methods
4.1. Patients
4.2. Reagents, Cells, and Cell Culture Conditions
4.2.1. Primary MSCs
4.2.2. Co-Culture Experiments
4.2.3. OB Differentiation Experiments
4.3. GLUL (Glutamine Synthetase) Knockdown in hTERT-MSCs
4.4. ASNS Knockout in hTERT-MSCs
4.5. Immunohistochemistry
4.6. Western Blot
4.7. Real Time-PCR Analysis
4.8. AminoAacid Uptake
4.9. Alkaline Phosphatase (ALP) Staining and Activity
4.10. Liquid Chromatography Tandem Mass Spectrometry (LC–MS/MS)
4.11. Gene Expression Profiles Analysis
4.12. Statistical Analysis
4.13. Study Approval
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maiso, P.; Huynh, D.; Moschetta, M.; Sacco, A.; Aljawai, Y.; Mishima, Y.; Asara, J.M.; Roccaro, A.M.; Kimmelman, A.C.; Ghobrial, I.M. Metabolic signature identifies novel targets for drug resistance in multiple myeloma. Cancer Res. 2015, 75, 2071–2082. [Google Scholar] [CrossRef]
- Rizzieri, D.; Paul, B.; Kang, Y. Metabolic alterations and the potential for targeting metabolic pathways in the treatment of multiple myeloma. J. Cancer Metastasis Treat. 2019, 5. [Google Scholar] [CrossRef]
- Bolzoni, M.; Chiu, M.; Accardi, F.; Vescovini, R.; Airoldi, I.; Storti, P.; Todoerti, K.; Agnelli, L.; Missale, G.; Andreoli, R.; et al. Dependence on glutamine uptake and glutamine addiction characterize myeloma cells: A new attractive target. Blood 2016, 128, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Puchades-Carrasco, L.; Lecumberri, R.; Martinez-Lopez, J.; Lahuerta, J.J.; Mateos, M.V.; Prosper, F.; San-Miguel, J.F.; Pineda-Lucena, A. Multiple myeloma patients have a specific serum metabolomic profile that changes after achieving complete remission. Clin. Cancer Res. 2013, 19, 4770–4779. [Google Scholar] [CrossRef]
- Barille, S.; Collette, M.; Bataille, R.; Amiot, M. Myeloma cells upregulate interleukin-6 secretion in osteoblastic cells through cell-to-cell contact but downregulate osteocalcin. Blood 1995, 86, 3151–3159. [Google Scholar] [CrossRef]
- Capp, J.P.; Bataille, R. Multiple Myeloma Exemplifies a Model of Cancer Based on Tissue Disruption as the Initiator Event. Front. Oncol. 2018, 8, 355. [Google Scholar] [CrossRef] [PubMed]
- Roodman, G.D. Pathogenesis of myeloma bone disease. Leukemia 2009, 23, 435–441. [Google Scholar] [CrossRef]
- Toscani, D.; Bolzoni, M.; Accardi, F.; Aversa, F.; Giuliani, N. The osteoblastic niche in the context of multiple myeloma. Ann. N. Y. Acad. Sci. 2015, 1335, 45–62. [Google Scholar] [CrossRef]
- Terpos, E.; Ntanasis-Stathopoulos, I.; Gavriatopoulou, M.; Dimopoulos, M.A. Pathogenesis of bone disease in multiple myeloma: From bench to bedside. Blood Cancer J. 2018, 8, 7. [Google Scholar] [CrossRef]
- Giuliani, N.; Colla, S.; Morandi, F.; Lazzaretti, M.; Sala, R.; Bonomini, S.; Grano, M.; Colucci, S.; Svaldi, M.; Rizzoli, V. Myeloma cells block RUNX2/CBFA1 activity in human bone marrow osteoblast progenitors and inhibit osteoblast formation and differentiation. Blood 2005, 106, 2472–2483. [Google Scholar] [CrossRef]
- Giuliani, N.; Rizzoli, V. Myeloma cells and bone marrow osteoblast interactions: Role in the development of osteolytic lesions in multiple myeloma. Leuk. Lymphoma 2007, 48, 2323–2329. [Google Scholar] [CrossRef]
- Raje, N.; Roodman, G.D. Advances in the biology and treatment of bone disease in multiple myeloma. Clin. Cancer Res. 2011, 17, 1278–1286. [Google Scholar] [CrossRef]
- Brown, P.M.; Hutchison, J.D.; Crockett, J.C. Absence of glutamine supplementation prevents differentiation of murine calvarial osteoblasts to a mineralizing phenotype. Calcif. Tissue Int. 2011, 89, 472–482. [Google Scholar] [CrossRef]
- Huang, T.; Liu, R.; Fu, X.; Yao, D.; Yang, M.; Liu, Q.; Lu, W.W.; Wu, C.; Guan, M. Aging Reduces an ERRalpha-Directed Mitochondrial Glutaminase Expression Suppressing Glutamine Anaplerosis and Osteogenic Differentiation of Mesenchymal Stem Cells. Stem Cells 2017, 35, 411–424. [Google Scholar] [CrossRef]
- Karner, C.M.; Esen, E.; Okunade, A.L.; Patterson, B.W.; Long, F. Increased glutamine catabolism mediates bone anabolism in response to WNT signaling. J. Clin. Investig. 2015, 125, 551–562. [Google Scholar] [CrossRef]
- Yu, Y.; Newman, H.; Shen, L.; Sharma, D.; Hu, G.; Mirando, A.J.; Zhang, H.; Knudsen, E.; Zhang, G.F.; Hilton, M.J.; et al. Glutamine Metabolism Regulates Proliferation and Lineage Allocation in Skeletal Stem Cells. Cell Metab. 2019, 29, 966–978 e964. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, Y.R.; Fan, X.L.; Lin, P.; Yang, H.; Chen, X.Z.; Xu, X.D. miR-206 inhibits osteogenic differentiation of bone marrow mesenchymal stem cells by targetting glutaminase. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Chiu, M.; Taurino, G.; Bianchi, M.G.; Ottaviani, L.; Andreoli, R.; Ciociola, T.; Lagrasta, C.A.M.; Tardito, S.; Bussolati, O. Oligodendroglioma Cells Lack Glutamine Synthetase and Are Auxotrophic for Glutamine, but Do not Depend on Glutamine Anaplerosis for Growth. Int. J. Mol. Sci. 2018, 19, 1099. [Google Scholar] [CrossRef]
- Todoerti, K.; Lisignoli, G.; Storti, P.; Agnelli, L.; Novara, F.; Manferdini, C.; Codeluppi, K.; Colla, S.; Crugnola, M.; Abeltino, M.; et al. Distinct transcriptional profiles characterize bone microenvironment mesenchymal cells rather than osteoblasts in relationship with multiple myeloma bone disease. Exp. Hematol. 2010, 38, 141–153. [Google Scholar] [CrossRef]
- Giuliani, N.; Rizzoli, V.; Roodman, G.D. Multiple myeloma bone disease: Pathophysiology of osteoblast inhibition. Blood 2006, 108, 3992–3996. [Google Scholar] [CrossRef]
- Patton, A.J.; Genever, P.G.; Birch, M.A.; Suva, L.J.; Skerry, T.M. Expression of an N-methyl-D-aspartate-type receptor by human and rat osteoblasts and osteoclasts suggests a novel glutamate signaling pathway in bone. Bone 1998, 22, 645–649. [Google Scholar] [CrossRef]
- Chiu, M.; Taurino, G.; Bianchi, M.G.; Dander, E.; Fallati, A.; Giuliani, N.; D’Amico, G.; Bussolati, O. Functional Consequences of Low Activity of Transport System A for Neutral Amino Acids in Human Bone Marrow Mesenchymal Stem Cells. Int. J. Mol. Sci. 2020, 21, 1899. [Google Scholar] [CrossRef]
- Menchini, R.J.; Chaudhry, F.A. Multifaceted regulation of the system A transporter Slc38a2 suggests nanoscale regulation of amino acid metabolism and cellular signaling. Neuropharmacology 2019, 161, 107789. [Google Scholar] [CrossRef]
- Chiu, M.; Taurino, G.; Bianchi, M.G.; Kilberg, M.S.; Bussolati, O. Asparagine Synthetase in Cancer: Beyond Acute Lymphoblastic Leukemia. Front. Oncol. 2019, 9, 1480. [Google Scholar] [CrossRef]
- Huang, H.; Vandekeere, S.; Kalucka, J.; Bierhansl, L.; Zecchin, A.; Bruning, U.; Visnagri, A.; Yuldasheva, N.; Goveia, J.; Cruys, B.; et al. Role of glutamine and interlinked asparagine metabolism in vessel formation. EMBO J. 2017, 36, 2334–2352. [Google Scholar] [CrossRef]
- Li, X.; Kumar, A.; Carmeliet, P. Metabolic Pathways Fueling the Endothelial Cell Drive. Annu. Rev. Physiol. 2019, 81, 483–503. [Google Scholar] [CrossRef]
- Jiang, J.; Pavlova, N.N.; Zhang, J. Asparagine, a critical limiting metabolite during glutamine starvation. Mol. Cell Oncol. 2018, 5, e1441633. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Hui, S.; Ghergurovich, J.M.; Fan, J.; Intlekofer, A.M.; White, R.M.; Rabinowitz, J.D.; Thompson, C.B.; Zhang, J. As Extracellular Glutamine Levels Decline, Asparagine Becomes an Essential Amino Acid. Cell Metab. 2018, 27, 428–438 e425. [Google Scholar] [CrossRef] [PubMed]
- Pant, A.; Cao, S.; Yang, Z. Asparagine Is a Critical Limiting Metabolite for Vaccinia Virus Protein Synthesis during Glutamine Deprivation. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Krall, A.S.; Xu, S.; Graeber, T.G.; Braas, D.; Christofk, H.R. Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat. Commun. 2016, 7, 11457. [Google Scholar] [CrossRef]
- Chen, J.; Long, F. mTORC1 Signaling Promotes Osteoblast Differentiation from Preosteoblasts. PLoS ONE 2015, 10, e0130627. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. FEBS 2000, 267, 5421–5426. [Google Scholar] [CrossRef]
- Bolzoni, M.; Donofrio, G.; Storti, P.; Guasco, D.; Toscani, D.; Lazzaretti, M.; Bonomini, S.; Agnelli, L.; Capocefalo, A.; Dalla Palma, B.; et al. Myeloma cells inhibit non-canonical wnt co-receptor ror2 expression in human bone marrow osteoprogenitor cells: Effect of wnt5a/ror2 pathway activation on the osteogenic differentiation impairment induced by myeloma cells. Leukemia 2013, 27, 451–463. [Google Scholar] [CrossRef]
- Cong, L.; Zhang, F. Genome engineering using CRISPR-Cas9 system. Methods Mol. Biol. 2015, 1239, 197–217. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Storti, P.; Agnelli, L.; Palma, B.D.; Todoerti, K.; Marchica, V.; Accardi, F.; Sammarelli, G.; Deluca, F.; Toscani, D.; Costa, F.; et al. The transcriptomic profile of CD138(+) cells from patients with early progression from smoldering to active multiple myeloma remains substantially unchanged. Haematologica 2019, 104, e465–e469. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiu, M.; Toscani, D.; Marchica, V.; Taurino, G.; Costa, F.; Bianchi, M.G.; Andreoli, R.; Franceschi, V.; Storti, P.; Burroughs-Garcia, J.; et al. Myeloma Cells Deplete Bone Marrow Glutamine and Inhibit Osteoblast Differentiation Limiting Asparagine Availability. Cancers 2020, 12, 3267. https://doi.org/10.3390/cancers12113267
Chiu M, Toscani D, Marchica V, Taurino G, Costa F, Bianchi MG, Andreoli R, Franceschi V, Storti P, Burroughs-Garcia J, et al. Myeloma Cells Deplete Bone Marrow Glutamine and Inhibit Osteoblast Differentiation Limiting Asparagine Availability. Cancers. 2020; 12(11):3267. https://doi.org/10.3390/cancers12113267
Chicago/Turabian StyleChiu, Martina, Denise Toscani, Valentina Marchica, Giuseppe Taurino, Federica Costa, Massimiliano G. Bianchi, Roberta Andreoli, Valentina Franceschi, Paola Storti, Jessica Burroughs-Garcia, and et al. 2020. "Myeloma Cells Deplete Bone Marrow Glutamine and Inhibit Osteoblast Differentiation Limiting Asparagine Availability" Cancers 12, no. 11: 3267. https://doi.org/10.3390/cancers12113267
APA StyleChiu, M., Toscani, D., Marchica, V., Taurino, G., Costa, F., Bianchi, M. G., Andreoli, R., Franceschi, V., Storti, P., Burroughs-Garcia, J., Eufemiese, R. A., Dalla Palma, B., Campanini, N., Martella, E., Mancini, C., Shan, J., Kilberg, M. S., D’Amico, G., Dander, E., ... Giuliani, N. (2020). Myeloma Cells Deplete Bone Marrow Glutamine and Inhibit Osteoblast Differentiation Limiting Asparagine Availability. Cancers, 12(11), 3267. https://doi.org/10.3390/cancers12113267







