In Silico Selection and In Vitro Evaluation of New Molecules That Inhibit the Adhesion of Streptococcus mutans through Antigen I/II
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structure-Based Virtual Screening
2.1.1. Target Proteins Selection
2.1.2. Molecule Selection
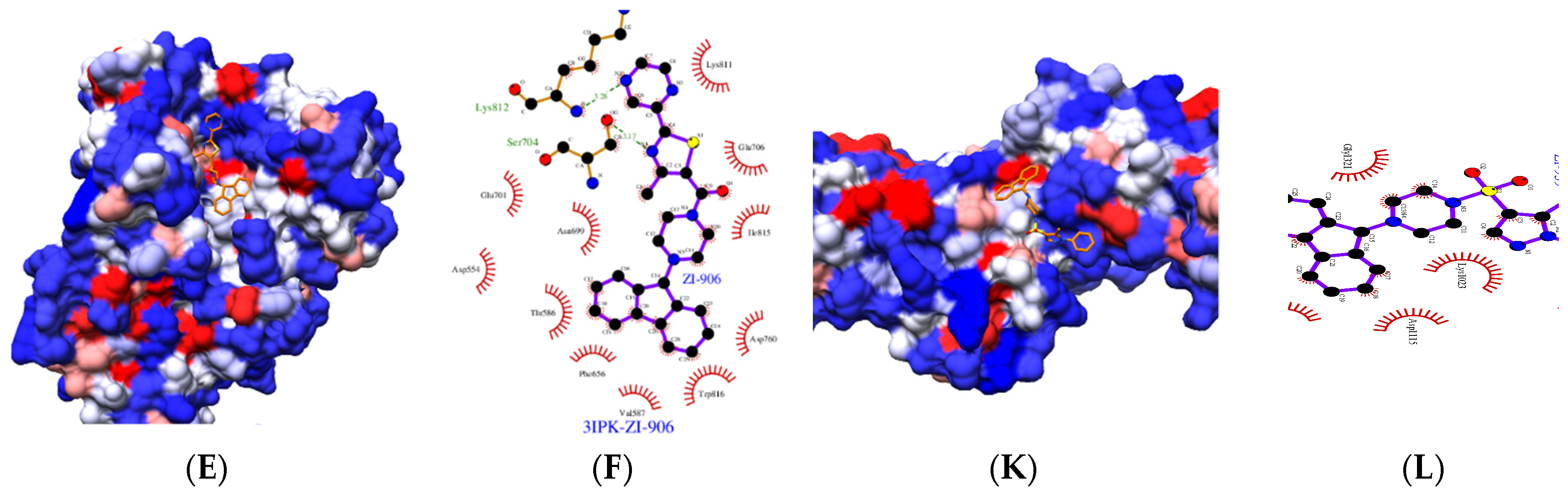
2.1.3. Molecule–Protein Interaction and Solubility Analysis
2.2. In Vitro Assays
2.2.1. Cytotoxicity and Antimicrobial Assays
2.2.2. Adhesion Assays
2.2.3. Molecular Dynamics Simulations (MD)
3. Materials and Methods
3.1. Structure-Based Virtual Screening
3.1.1. Target Proteins Selection
3.1.2. Molecule Selection
3.1.3. Molecule–Protein Interaction and Solubility Analysis
3.2. In Vitro Assays
3.2.1. Cytotoxicity and Antimicrobial Assays
3.2.2. Adhesion Assay
3.2.3. Data Analysis
3.2.4. Scanning Electron Microscopy (SEM)
3.2.5. Molecular Dynamics Simulations (MD)
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kyu, H.H.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, regional, and national disability-adjusted life-years (DALYs) for 359 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018. [Google Scholar] [CrossRef] [Green Version]
- Vos, T.; Allen, C.; Arora, M.; Barber, R.M.; Brown, A.; Carter, A.; Casey, D.C.; Charlson, F.J.; Chen, A.Z.; Coggeshall, M.; et al. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016. [Google Scholar] [CrossRef] [Green Version]
- Lu, M.; Xuan, S.; Wang, Z. Oral microbiota: A new view of body health. Food Sci. Hum. Wellness 2019, 8, 8–15. [Google Scholar] [CrossRef]
- Tanzer, J.M. Dental Caries is a Transmissible Infectious Disease: The Keyes and Fitzgerald Revolution. J. Dent. Res. 1995, 74, 1536–1542. [Google Scholar] [CrossRef]
- Selwitz, R.H.; Ismail, A.I.; Pitts, N.B. Dental caries. Lancet 2007, 369, 51–59. [Google Scholar] [CrossRef]
- Kolenbrander, P.E.; Palmer, R.J.; Periasamy, S.; Jakubovics, N.S. Oral multispecies biofilm development and the key role of cell-cell distance. Nat. Rev. Microbiol. 2010, 8, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Moschioni, M.; Pansegrau, W.; Barocchi, M.A. Adhesion determinants of the Streptococcus species. Microb. Biotechnol. 2010, 3, 370–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto-Nakano, M. Role of Streptococcus mutans surface proteins for biofilm formation. Jpn. Dent. Sci. Rev. 2018, 54, 22–29. [Google Scholar] [CrossRef]
- Krzyściak, W.; Jurczak, A.; Kościelniak, D.; Bystrowska, B.; Skalniak, A. The virulence of Streptococcus mutans and the ability to form biofilms. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 499–515. [Google Scholar] [CrossRef] [Green Version]
- Scharnow, A.M.; Solinski, A.E.; Wuest, W.M. Targeting: S. mutans biofilms: A perspective on preventing dental caries. Medchemcomm 2019, 10, 1057–1067. [Google Scholar] [CrossRef]
- Mitchell, T.J. The pathogenesis of streptococcal infections: From Tooth decay to meningitis. Nat. Rev. Microbiol. 2003, 1, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Lamont, R.J.; Demuth, D.R.; Davis, C.A.; Malamud, D.; Rosan, B. Salivary-agglutinin-mediated adherence of Streptococcus mutans to early plaque bacteria. Infect. Immun. 1991, 59, 3446–3450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakubovics, N.S.; Strömberg, N.; Van Dolleweerd, C.J.; Kelly, C.G.; Jenkinson, H.F. Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol. Microbiol. 2005, 55, 1591–1605. [Google Scholar] [CrossRef] [PubMed]
- Pecharki, D.; Petersen, F.C.; Assev, S.; Scheie, A.A. Involvement of antigen I/II surface proteins in Streptococcus mutans and Streptococcus intermedius biofilm formation. Oral Microbiol. Immunol. 2005, 20, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Crowley, P.J.; Brady, L.J.; Michalek, S.M.; Bleiweis, A.S. Virulence of a spaP mutant of Streptococcus mutans in a gnotobiotic rat model. Infect. Immun. 1999, 67, 1201–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsushita, K.; Nisizawa, T.; Nagaoka, S.; Kawagoe, M.; Koga, T. Identification of antigenic epitopes in a surface protein antigen of Streptococcus mutans in humans. Infect. Immun. 1994, 62, 4034–4042. [Google Scholar] [CrossRef] [Green Version]
- Robinette, R.A.; Heim, K.P.; Oli, M.W.; Crowley, P.J.; McArthur, W.P.; Brady, L.J. Alterations in immunodominance of Streptococcus mutans AgI/II: Lessons learned from immunomodulatory antibodies. Vaccine 2014, 32, 375–382. [Google Scholar] [CrossRef] [Green Version]
- Batista, M.T.; Souza, R.D.; Ferreira, E.L.; Robinette, R.; Crowley, P.J.; Rodrigues, J.F.; Jeannine Brady, L.; Ferreira, L.C.S.; Ferreira, R.C.C. Immunogenicity and in vitro and in vivo protective effects of antibodies targeting a recombinant form of the Streptococcus mutans P1 surface protein. Infect. Immun. 2014, 82, 4978–4988. [Google Scholar] [CrossRef] [Green Version]
- Jenkinson, H.F.; Demuth, D.R. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol. Microbiol. 1997, 23, 183–190. [Google Scholar] [CrossRef]
- Larson, M.R.; Rajashankar, K.R.; Crowley, P.J.; Kelly, C.; Mitchell, T.J.; Brady, L.J.; Deivanayagam, C. Crystal structure of the C-terminal region of Streptococcus mutans antigen I/II and characterization of salivary agglutinin adherence domains. J. Biol. Chem. 2011, 286, 21657–21666. [Google Scholar] [CrossRef] [Green Version]
- Larson, M.R.; Rajashankar, K.R.; Patel, M.H.; Robinette, R.A.; Crowley, P.J.; Michalek, S.; Brady, L.J.; Deivanayagam, C. Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of alpha- and PPII-helices. Proc. Natl. Acad. Sci. USA 2010, 107, 5983–5988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brady, L.J.; Maddocks, S.E.; Larson, M.R.; Forsgren, N.; Persson, K.; Deivanayagam, C.C.; Jenkinson, H.F. The changing faces of Streptococcus antigen I/II polypeptide family adhesins: MicroReview. Mol. Microbiol. 2010, 77, 276–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Scoffield, J.; Wu, R.; Deivanayagam, C.; Zou, J.; Wu, H. Antigen I/II mediates interactions between Streptococcus mutans and Candida albicans. Mol. Oral Microbiol. 2018, 176, 139–148. [Google Scholar] [CrossRef]
- Yang, J.; Deng, D.; Brandt, B.W.; Nazmi, K.; Wu, Y.; Crielaard, W.; Ligtenberg, A.J.M. Diversity of SpaP in genetic and salivary agglutinin mediated adherence among Streptococcus mutans strains. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Esberg, A.; Löfgren-Burström, A.; Öhman, U.; Strömberg, N. Host and bacterial phenotype variation in adhesion of Streptococcus mutans to matched human hosts. Infect. Immun. 2012, 80, 3869–3879. [Google Scholar] [CrossRef] [Green Version]
- Janakiram, C.; Deepan Kumar, C.V.; Joseph, J. Xylitol in preventing dental caries: A systematic review and meta-analyses. J. Nat. Sci. Biol. Med. 2017, 8, 16–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riley, P.; Moore, D.; Ahmed, F.; Sharif, M.O.; Worthington, H.V. Xylitol-containing products for preventing dental caries in children and adults. Cochrane Database Syst. Rev. 2015, 3, CD010743. [Google Scholar] [CrossRef]
- Autio-Gold, J. The role of chlorhexidine in caries prevention. Oper. Dent. 2008, 33, 710–716. [Google Scholar] [CrossRef]
- Walsh, T.; Oliveira-Neto, J.M.; Moore, D. Chlorhexidine treatment for the prevention of dental caries in children and adolescents. Cochrane Database Syst. Rev. 2015, 2015. [Google Scholar] [CrossRef]
- Koga, T.; Oho, T.; Shimazaki, Y.; Nakano, Y. Immunization against dental caries. Vaccine 2002, 20, 2027–2044. [Google Scholar] [CrossRef]
- Oh, D.H.; Chen, X.; Daliri, E.B.M.; Kim, N.; Kim, J.R.; Yoo, D. Microbial etiology and prevention of dental caries: Exploiting natural products to inhibit cariogenic biofilms. Pathogens 2020, 9, 569. [Google Scholar] [CrossRef]
- Ren, Z.; Chen, L.; Li, J.; Li, Y. Inhibition of Streptococcus mutans polysaccharide synthesis by molecules targeting glycosyltransferase activity. J. Oral Microbiol. 2016, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tada, A.; Nakayama-Imaohji, H.; Yamasaki, H.; Hasibul, K.; Yoneda, S.; Uchida, K.; Nariya, H.; Suzuki, M.; Miyake, M.; Kuwahara, T. Cleansing effect of acidic L-arginine on human oral biofilm. BMC Oral Health 2016, 16, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Madrid Troconis, C.C.; Perez Puello, S.D.C. Nanocomplejo De Fosfopéptido De Caseína-Fosfato De Calcio Amorfo (Cpp-Acp) En Odontología: Estado Del Arte. Rev. Fac. Odontol. 2019, 30, 248–263. [Google Scholar] [CrossRef]
- Bijle, M.N.A.; Ekambaram, M.; Lo, E.C.M.; Yiu, C.K.Y. The combined antimicrobial effect of arginine and fluoride toothpaste. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; He, J.; Wang, L.; Zhou, S.; Peng, X.; Huang, S.; Zheng, L.; Cheng, L.; Hao, Y.; Li, J.; et al. Ecological Effect of Arginine on Oral Microbiota. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrer, M.D.; López-López, A.; Nicolescu, T.; Perez-Vilaplana, S.; Boix-Amorós, A.; Dzidic, M.; Garcia, S.; Artacho, A.; Llena, C.; Mira, A. Topic Application of the Probiotic Streptococcus dentisani Improves Clinical and Microbiological Parameters Associated with Oral Health. Front. Cell. Infect. Microbiol. 2020, 10, 465. [Google Scholar] [CrossRef]
- Mayr, L.M.; Fuerst, P. The Future of High-Throughput Screening. J. Biomol. Screen. 2008, 13, 443–448. [Google Scholar] [CrossRef]
- Mohs, R.C.; Greig, N.H. Drug discovery and development: Role of basic biological research. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2017, 3, 651–657. [Google Scholar] [CrossRef]
- Sinha, S.; Vohora, D. Drug Discovery and Development: An Overview; Elsevier Inc.: New Delhi, India, 2017; pp. 19–32. [Google Scholar]
- Barbosa, A.; Romário, D.; Avelar, S.; Gomes, G.; Albuquerque, A.R.; Gaudencio, T. In Silico Approach for the Identification of Potential Targets and Specific Antimicrobials for Streptococcus mutans. Adv. Biosci. Biotechnol. 2014, 5, 373–385. [Google Scholar]
- Ren, Z.; Cui, T.; Zeng, J.; Chen, L.; Zhang, W.; Xu, X.; Cheng, L.; Li, M.; Li, J.; Zhou, X.; et al. Molecule targeting glucosyltransferase inhibits Streptococcus mutans biofilm formation and virulence. Antimicrob. Agents Chemother. 2016, 60, 126–135. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Nijampatnam, B.; Hua, Z.; Nguyen, T.; Zou, J.; Cai, X.; Michalek, S.M.; Velu, S.E.; Wu, H. Structure-Based Discovery of Small Molecule Inhibitors of Cariogenic Virulence. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Troffer-Charlier, N.; Ogier, J.; Moras, D.; Cavarelli, J. Crystal structure of the V-region of streptococcus mutans antigen I/II at 2.4 Å resolution suggests a sugar preformed binding site. J. Mol. Biol. 2002, 318, 179–188. [Google Scholar] [CrossRef]
- Nylander, Å.; Forsgren, N.; Persson, K. Structure of the C-terminal domain of the surface antigen SpaP from the caries pathogen Streptococcus mutans. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Heim, K.P.; Crowley, P.J.; Long, J.R.; Kailasan, S.; McKenna, R.; Brady, L.J. An intramolecular lock facilitates folding and stabilizes the tertiary structure of streptococcus mutans adhesin p1. Proc. Natl. Acad. Sci. USA 2014, 111, 15711–15716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lionta, E.; Spyrou, G.; Vassilatis, D.K.; Cournia, Z. Send Orders for Reprints to [email protected] Structure-Based Virtual Screening for Drug Discovery: Principles, Applications and Recent Advances. Curr. Top. Med. Chem. 2014, 14, 1923–1938. [Google Scholar] [CrossRef]
- Gazgalis, D.; Zaka, M.; Zaka, M.; Abbasi, B.H.; Logothetis, D.E.; Mezei, M.; Cui, M. Protein Binding Pocket Optimization for Virtual High-Throughput Screening (vHTS) Drug Discovery. ACS Omega 2020, 5, 14297–14307. [Google Scholar] [CrossRef]
- Huang, B. MetaPocket: A Meta Approach to Improve Protein Ligand Binding Site Prediction. Omi. J. Integr. Biol. 2009, 13, 325–330. [Google Scholar] [CrossRef]
- Yang, J.; Roy, A.; Zhang, Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 2013, 29, 2588–2595. [Google Scholar] [CrossRef]
- El-Sayed, R.; Althagafi, I.I.; Ahmed, S.A. Fluorene Derivatives with Multi-addressable Properties: Synthesis, Characterization, and Reactivity. J. Surfactants Deterg. 2017, 20, 933–945. [Google Scholar] [CrossRef]
- Rathi, A.K.; Syed, R.; Shin, H.S.; Patel, R.V. Piperazine derivatives for therapeutic use: A patent review (2010-present). Expert Opin. Ther. Pat. 2016, 26, 777–797. [Google Scholar] [CrossRef] [PubMed]
- Hadži, D.; Kidrič, J.; Koller, J.; Mavri, J. The role of hydrogen bonding in drug-receptor interactions. J. Mol. Struct. 1990, 237, 139–150. [Google Scholar] [CrossRef]
- Kuhn, B.; Mohr, P.; Stahl, M. Intramolecular hydrogen bonding in medicinal chemistry. J. Med. Chem. 2010, 53, 2601–2611. [Google Scholar] [CrossRef] [PubMed]
- Caron, G.; Kihlberg, J.; Ermondi, G. Intramolecular hydrogen bonding: An opportunity for improved design in medicinal chemistry. Med. Res. Rev. 2019, 39, 1707–1729. [Google Scholar] [CrossRef] [PubMed]
- Yunta, M.J. It Is Important to Compute Intramolecular Hydrogen Bonding in Drug Design? Am. J. Model. Optim. 2017, 5, 24–57. [Google Scholar] [CrossRef] [Green Version]
- Ermondi, G.; Caron, G. Why we need to implement intramolecular hydrogen-bonding considerations in drug discovery. Future Med. Chem. 2016, 31, 48–49. [Google Scholar]
- Cottet-Rousselle, C.; Ronot, X.; Leverve, X.; Mayol, J.F. Cytometric assessment of mitochondria using fluorescent probes. Cytom. Part A 2011, 79, 405–425. [Google Scholar] [CrossRef]
- Rieger, A.M.; Nelson, K.L.; Konowalchuk, J.D.; Barreda, D.R. Modified annexin V/propidium iodide apoptosis assay for accurate assessment of cell death. J. Vis. Exp. 2011, 3–6. [Google Scholar] [CrossRef]
- Grivet, M.; Morrier, J.J.; Benay, G.; Barsotti, O. Effect of hydrophobicity on in vitro streptococcal adhesion to dental alloys. J. Mater. Sci. Mater. Med. 2000, 11, 637–642. [Google Scholar] [CrossRef]
- Wang, C.; van der Mei, H.C.; Busscher, H.J.; Ren, Y. Streptococcus mutans adhesion force sensing in multi-species oral biofilms. NPJ Biofilms Microbiomes 2020, 6, 1–9. [Google Scholar] [CrossRef]
- Wang, J.; Shi, Y.; Jing, S.; Dong, H.; Wang, D.; Wang, T. Astilbin Inhibits the Activity of Sortase A from Streptococcus mutans. Molecules 2019, 24, 465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, H.; Liang, D.F.; Bao, M.Y.; Sun, R.; Li, Y.Y.; Li, J.Z.; Wang, X.; Lu, K.M.; Bao, J.K. In silico identification of potential inhibitors targeting Streptococcus mutans sortase A. Int. J. Oral Sci. 2017, 9, 53–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, P.; Huang, P.; Chen, M.W. Curcumin reduces Streptococcus mutans biofilm formation by inhibiting sortase A activity. Arch. Oral Biol. 2013, 58, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Burgos-Morón, E.; Calderón-Montaño, J.M.; Salvador, J.; Robles, A.; López-Lázaro, M. The dark side of curcumin Estefanía. Int. J. Cancer 2010, 126, 1771–1775. [Google Scholar] [CrossRef] [PubMed]
- Cianfruglia, L.; Minnelli, C.; Laudadio, E.; Scirè, A.; Armeni, T. Side effects of curcumin: Epigenetic and antiproliferative implications for normal dermal fibroblast and breast cancer cells. Antioxidants 2019, 8, 382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, P.; Hu, P.; Zhou, S.Y.; Li, Q.; Chen, W.M. Morin inhibits sortase A and subsequent biofilm formation in streptococcus mutans. Curr. Microbiol. 2014, 68, 47–52. [Google Scholar] [CrossRef]
- Yang, J.Y.; Lee, H.S. Evaluation of antioxidant and antibacterial activities of morin isolated from mulberry fruits (Morus alba L.). J. Korean Soc. Appl. Biol. Chem. 2012, 55, 485–489. [Google Scholar] [CrossRef]
- Ye, Y.; Godzik, A. FATCAT: A web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 2004, 32, 582–585. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Sterling, T.; Irwin, J.J. ZINC 15—Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Rivera-Pérez, W.A.; Yépes-Pérez, A.F.; Martínez-Pabón, M.C. Molecular docking and in silico studies of the physicochemical properties of potential inhibitors for the phosphotransferase system of Streptococcus mutans. Arch. Oral Biol. 2019, 98, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfizer Inc. Material Safety Data Sheet Material Safety Data Sheet [Internet]. 2012. [cited 2019 Mar 20]. pp. 1–5. Available online: https://pfe-pfizercom-prod.s3.amazonaws.com/products/material_safety_data/PZ00719.pdf (accessed on 14 October 2020).
- Chen, D.; Oezguen, N.; Urvil, P.; Ferguson, C.; Dann, S.M.; Savidge, T.C. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biovia, D. Discovery Studio Modeling Environment, Release 2017; DassaultSystèmes: San Diego, CA, USA, 2016; Available online: http//accelrys.com/products/collaborative-science/biovia-discoverystudio/visualizationdownload.php (accessed on 14 November 2018).
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004. [Google Scholar] [CrossRef] [Green Version]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, L.; Tanzer, J.M.; MacAlister, T.J.; Tao, L.; Tanzer, J.M. Transformation Efficiency of EMS-induced Mutants of Streptococcus mutans of Altered Cell Shape. J. Dent. Res. 1993, 72, 1032–1039. [Google Scholar] [CrossRef]
- Chen, L.; Jia, L.; Zhang, Q.; Zhou, X.; Liu, Z.; Li, B.; Zhu, Z.; Wang, F.; Yu, C.; Zhang, Q.; et al. A novel antimicrobial peptide against dental-caries-associated bacteria. Anaerobe 2017, 47, 165–172. [Google Scholar] [CrossRef]
- Esberg, A.; Sheng, N.; Mårell, L.; Claesson, R.; Persson, K.; Borén, T.; Strömberg, N. EBioMedicine Streptococcus Mutans Adhesin Biotypes that Match and Predict Individual Caries Development. EBioMedicine 2017, 24, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]







| Number of Molecules Interaction Pockets | Molecules | Library | 3IPK | 3QE5 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | P2 | P3 | P1 | P2 | P3 | |||||||||
| COACH | MET | COACH | MET | COACH | MET | COACH | MET | COACH | MET | COACH | MET | |||
| Not applicable | ZINC68568370 | NAT | −12.8 | −12.8 | −7.5 | −9 | −9.4 | −10.9 | −8.5 | −8.8 | −8.1 | −8.9 | −7.1 | −7.7 |
| ZINC70669788 | NAT | −11.4 | −12.8 | −7.5 | −8.4 | −8.7 | −10.1 | −7.9 | −8.7 | −7.7 | −7.6 | −7.6 | −6.5 | |
| ZINC70669789 | NAT | −11.5 | −12.7 | −7.3 | −8.3 | −8.8 | −9.8 | −8.1 | −9 | −7.9 | −7.8 | −7.8 | −6.3 | |
| ZINC34257514 | NAT | −12.6 | −12.6 | −6.7 | −8.8 | −7.7 | −9.8 | −6.9 | −8.7 | −6.8 | −7.6 | −6.7 | −7.6 | |
| ZINC04817561 | NAT | −10.3 | −12.4 | −10.1 | −8.3 | −8.6 | −12.4 | −7.2 | −9.2 | −7.1 | −7.9 | −7.2 | −7 | |
| ZINC67912808 | NAT | −12.3 | −12.5 | −8.2 | −8.8 | −8.4 | −9.1 | −7.6 | −9.5 | −7.6 | −8.1 | −7.1 | −6.8 | |
| ZINC70686498 | NAT | −12.2 | −12.2 | −7.8 | −8.2 | −8 | −9.9 | −7.4 | −8.1 | −7.3 | −9.6 | −6.6 | −7.1 | |
| ZINC04015296 | NAT | −11.7 | −11.6 | −8.4 | −9.8 | −9 | −11.7 | −9.3 | −11.4 | −7.5 | −9.3 | −7.4 | −9.8 | |
| ZINC08594547 | LRG | −11.6 | −11.7 | −7.7 | −8.5 | −8.1 | −11.4 | −7.6 | −8.6 | −7.1 | −8.2 | −6.5 | −8.3 | |
| 12 | ZINC00970517 | SM | −9.4 | −9.4 | −9 | −8.2 | −7 | −9 | −6.7 | −7.7 | −6.3 | −7.5 | −5.9 | −7.3 |
| 12 | ZINC01033612 | SM | −9.3 | −9.5 | −8.7 | −7.7 | −7.3 | −9.4 | −7.4 | −7.8 | −7 | −8.1 | −7.4 | −7.5 |
| 12 | ZINC08647964 | SM | −9.7 | −9.8 | −9.4 | −7.9 | −7.4 | −10.4 | −6.9 | −8.4 | −6.8 | −8.7 | −6.2 | −7.5 |
| 12 | ZINC12369546 | SM | −9.5 | −10 | −9.1 | −8.3 | −7.5 | −9.8 | −7.9 | −8.3 | −6.7 | −7.8 | −6.8 | −8.5 |
| 12 | ZINC19924906 | SM | −11.1 | −11 | −7.2 | −8.2 | −7.1 | −9.2 | −6.9 | −8.3 | −6.7 | −7.5 | −6.8 | −7.5 |
| 12 | ZINC03120327 | SM | −9.6 | −9.7 | −10.1 | −7.9 | −7.1 | −10.1 | −6.7 | −8 | −7.1 | −8 | −6.9 | −8.5 |
| 12 | ZINC19835160 | SM | −10.3 | −9.5 | −6.9 | −8.7 | −7.7 | −9.7 | −6.6 | −8.4 | −6.7 | −8 | −5.8 | −6.9 |
| 12 | ZINC19835187 | SM | −10.7 | −10.7 | −9.2 | −8.8 | −8.5 | −10.3 | −8.1 | −8.7 | −7.5 | −8.4 | −6.9 | −8.8 |
| 12 | ZINC19924939 | SM | −10.4 | −10.4 | −9 | −8.9 | −7.2 | −8.1 | −7.6 | −7.5 | −7.1 | −8 | −6.4 | −7.9 |
| 12 | ZINC59608258 | SM | −9.4 | −10.1 | −7.8 | −7.9 | −7.2 | −8.9 | −6.8 | −7.7 | −6.1 | −7.6 | −6.3 | −6.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rivera-Quiroga, R.E.; Cardona, N.; Padilla, L.; Rivera, W.; Rocha-Roa, C.; Diaz De Rienzo, M.A.; Morales, S.M.; Martinez, M.C. In Silico Selection and In Vitro Evaluation of New Molecules That Inhibit the Adhesion of Streptococcus mutans through Antigen I/II. Int. J. Mol. Sci. 2021, 22, 377. https://doi.org/10.3390/ijms22010377
Rivera-Quiroga RE, Cardona N, Padilla L, Rivera W, Rocha-Roa C, Diaz De Rienzo MA, Morales SM, Martinez MC. In Silico Selection and In Vitro Evaluation of New Molecules That Inhibit the Adhesion of Streptococcus mutans through Antigen I/II. International Journal of Molecular Sciences. 2021; 22(1):377. https://doi.org/10.3390/ijms22010377
Chicago/Turabian StyleRivera-Quiroga, Raúl E., Néstor Cardona, Leonardo Padilla, Wbeimar Rivera, Cristian Rocha-Roa, Mayri A. Diaz De Rienzo, Sandra M. Morales, and María C. Martinez. 2021. "In Silico Selection and In Vitro Evaluation of New Molecules That Inhibit the Adhesion of Streptococcus mutans through Antigen I/II" International Journal of Molecular Sciences 22, no. 1: 377. https://doi.org/10.3390/ijms22010377






