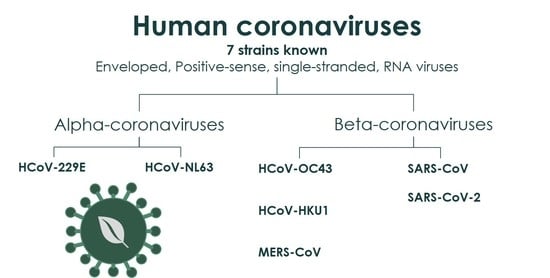
Natural and Nature-Derived Products Targeting Human Coronaviruses
Abstract
:1. Introduction
2. SARS-CoV-2 and SARS-CoV: Structural Aspects and Therapeutic Targeting
3. NPs with Anti-HCoV Potential
3.1. “Common Cold” HCoVs
3.2. MERS-CoV
3.3. SARS-CoV
3.4. SARS-CoV-2
3.4.1. Virtual Screening Approaches
3.4.2. Actual Screening of NPs for Anti-SARS-CoV-2 Activity
3.4.3. Host Interactions and Future Prospects
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maier, H.J.; Bickerton, E.; Britton, P. Coronaviruses: Methods and Protocols; Humana Press: Heidelberg, Germany, 2015; pp. 1–282. ISBN 9781493924387. [Google Scholar]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar]
- Memish, Z.A.; Perlman, S.; Van Kerkhove, M.D.; Zumla, A. Middle East respiratory syndrome. Lancet 2020, 395, 1063–1077. [Google Scholar] [CrossRef]
- Cheng, V.C.C.; Lau, S.K.P.; Woo, P.C.Y.; Yuen, K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007, 20, 660–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef]
- Laha, S.; Chakraborty, J.; Das, S.; Manna, S.K.; Biswas, S.; Chatterjee, R. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect. Genet. Evol. 2020, 85, 104445. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [Green Version]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Matsuyama, S.; Nagata, N.; Shirato, K.; Kawase, M.; Takeda, M.; Taguchi, F. Efficient Activation of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein by the Transmembrane Protease TMPRSS2. J. Virol. 2010, 84, 12658–12664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirato, K.; Kawase, M.; Matsuyama, S. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology 2018, 517, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Simmons, G.; Gosalia, D.N.; Rennekamp, A.J.; Reeves, J.D.; Diamond, S.L.; Bates, P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 2005, 102, 11876–11881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawase, M.; Shirato, K.; van der Hoek, L.; Taguchi, F.; Matsuyama, S. Simultaneous Treatment of Human Bronchial Epithelial Cells with Serine and Cysteine Protease Inhibitors Prevents Severe Acute Respiratory Syndrome Coronavirus Entry. J. Virol. 2012, 86, 6537–6545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, S.K.; Li, W.; Moore, M.J.; Choe, H.; Farzan, M. A 193-Amino Acid Fragment of the SARS Coronavirus S Protein Efficiently Binds Angiotensin-converting Enzyme 2. J. Biol. Chem. 2004, 279, 3197–3201. [Google Scholar] [CrossRef] [Green Version]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.; Decroly, E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020, 94, 00127-20. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Chen, P.; Wang, J.; Feng, J.; Zhou, H.; Li, X.; Zhong, W.; Hao, P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020, 63, 457–460. [Google Scholar] [CrossRef] [Green Version]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, W.; Mao, C.; Luan, X.; Shen, D.-D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M.; et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020, 368, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Ahn, D.-G.; Choi, J.-K.; Taylor, D.R.; Oh, J.-W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012, 157, 2095–2104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanner, J.A.; Watt, R.M.; Chai, Y.-B.; Lu, L.-Y.; Lin, M.C.; Peiris, J.S.M.; Poon, L.L.M.; Kung, H.-F.; Huang, J.-D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 2003, 278, 39578–39582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Yang, M.; Ding, Y.; Liu, Y.; Lou, Z.; Zhou, Z.; Sun, L.; Mo, L.; Ye, S.; Pang, H.; et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. USA 2003, 100, 13190–13195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratia, K.; Saikatendu, K.S.; Santarsiero, B.D.; Barreto, N.; Baker, S.C.; Stevens, R.C.; Mesecar, A.D. Severe acute respiratory syndrome coronavirus papain-like-protease: Structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA 2006, 103, 5717–5722. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-emergency-use-authorization-potential-covid-19-treatment (accessed on 15 January 2021).
- Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/214787Orig1s000lbl.pdf (accessed on 15 January 2021).
- Xue, X.; Yu, H.; Yang, H.; Xue, F.; Wu, Z.; Shen, W.; Li, J.; Zhou, Z.; Ding, Y.; Zhao, Q.; et al. Structures of Two Coronavirus Main Proteases: Implications for Substrate Binding and Antiviral Drug Design. J. Virol. 2008, 82, 2515–2527. [Google Scholar] [CrossRef] [Green Version]
- Báez-Santos, Y.M.; John, S.E.S.; Mesecar, A.D. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015, 115, 21–38. [Google Scholar] [CrossRef]
- Tong, T.R. Therapies for coronaviruses. Part 2: Inhibitors of intracellular life cycle. Expert Opin. Ther. Pat. 2009, 19, 415–431. [Google Scholar] [CrossRef]
- Mielech, A.M.; Chen, Y.; Mesecar, A.D.; Baker, S.C. Nidovirus papain-like proteases: Multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 2014, 194, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Xiong, B.; Gui, C.-S.; Xu, X.-Y.; Luo, C.; Chen, J.; Luo, H.-B.; Chen, L.-L.; Li, G.-W.; Sun, T.; Yu, C.-Y.; et al. A 3D model of SARS_CoV 3CL proteinase and its inhibitors design by virtual screening. Acta Pharm. Sin. 2003, 24, 497–504+619. [Google Scholar]
- Pillaiyar, T.; Manickam, M.; Namasivayam, V.; Hayashi, Y.; Jung, S.H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: Peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016, 59, 6595–6628. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-Y.; Jan, J.-T.; Ma, S.-H.; Kuo, C.-J.; Juan, H.-F.; Cheng, Y.-S.E.; Hsu, H.-H.; Huang, H.-C.; Wu, D.; Brik, A.; et al. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. USA 2004, 101, 10012–10017. [Google Scholar] [CrossRef] [Green Version]
- Kao, R.Y.; Tsui, W.H.W.; Lee, T.S.W.; Tanner, J.A.; Watt, R.M.; Huang, J.-D.; Hu, L.; Chen, G.; Chen, Z.; Zhang, L.; et al. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem. Biol. 2004, 11, 1293–1299. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.T.; Sarkar, C.; El-Kersh, D.M.; Jamaddar, S.; Uddin, S.J.; Shilpi, J.A.; Mubarak, M.S. Natural products and their derivatives against coronavirus: A review of the non-clinical and pre-clinical data. Phyther. Res. 2020, 34, 2471–2492. [Google Scholar] [CrossRef]
- Yang, Y.; Islam, M.S.; Wang, J.; Li, Y.; Chen, X. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): A review and perspective. Int. J. Biol. Sci. 2020, 16, 1708–1717. [Google Scholar] [CrossRef]
- Al-Hatamleh, M.A.I.; Hatmal, M.M.; Sattar, K.; Ahmad, S.; Mustafa, M.Z.; Bittencourt, M.D.C.; Mohamud, R. Antiviral and Immunomodulatory Effects of Phytochemicals from Honey against COVID-19: Potential mechanisms of action and future Directions. Molecules 2020, 25, 5017. [Google Scholar] [CrossRef]
- Boozari, M.; Hosseinzadeh, H. Natural products for COVID-19 prevention and treatment regarding to previous coronavirus infections and novel studies. Phyther. Res. 2020. [Google Scholar] [CrossRef]
- Liu, Y.; Liang, C.; Xin, L.; Ren, X.; Tian, L.; Ju, X.; Li, H.; Wang, Y.; Zhao, Q.; Liu, H.; et al. The development of Coronavirus 3C-Like protease (3CLpro) inhibitors from 2010 to 2020. Eur. J. Med. Chem. 2020, 206, 112711. [Google Scholar] [CrossRef]
- Tiwari, V.; Beer, J.C.; Sankaranarayanan, N.V.; Swanson-Mungerson, M.; Desai, U.R. Discovering small-molecule therapeutics against SARS-CoV-2. Drug Discov. Today 2020, 25, 1535–1544. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Huang, J. Current Findings Regarding Natural Components with Potential Anti-2019-nCoV Activity. Front. Cell Dev. Biol. 2020, 8, 589. [Google Scholar] [CrossRef] [PubMed]
- da Silva Antonio, A.; Wiedemann, L.S.M.; Veiga-Junior, V.F. Natural products’ role against COVID-19. RSC Adv. 2020, 10, 23379–23393. [Google Scholar] [CrossRef]
- Khalifa, S.A.M.; Yosri, N.; El-Mallah, M.F.; Ghonaim, R.; Guo, Z.; Musharraf, S.G.; Du, M.; Khatib, A.; Xiao, J.; Saeed, A.; et al. Screening for natural and derived bio-active compounds in preclinical and clinical studies: One of the frontlines of fighting the coronaviruses pandemic. Phytomedicine 2020, 153311. [Google Scholar] [CrossRef]
- Verma, S.; Twilley, D.; Esmear, T.; Oosthuizen, C.B.; Reid, A.M.; Nel, M.; Lall, N. Anti-SARS-CoV Natural Products with the Potential to Inhibit SARS-CoV-2 (COVID-19). Front. Pharmacol. 2020, 11, 561334. [Google Scholar] [CrossRef]
- Adhikari, B.; Marasini, B.P.; Rayamajhee, B.; Bhattarai, B.R.; Lamichhane, G.; Khadayat, K.; Adhikari, A.; Khanal, S.; Parajuli, N. Potential roles of medicinal plants for the treatment of viral diseases focusing on COVID-19: A review. Phyther. Res. 2020. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, L. Turning the Tide: Natural Products and Natural-Product-Inspired Chemicals as Potential Counters to SARS-CoV-2 Infection. Front. Pharmacol. 2020, 11, 1013. [Google Scholar] [CrossRef]
- Hamre, D.; Procknow, J.J. A New Virus Isolated froiii the Human Respiratory Tract. Exp. Biol. Med. 1966, 121, 190–193. [Google Scholar] [CrossRef]
- McIntosh, K.; Dees, J.H.; Becker, W.B.; Kapikian, A.Z.; Chanock, R.M. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. USA 1967, 57, 933–940. [Google Scholar] [CrossRef] [Green Version]
- McIntosh, K.; Becker, W.B.; Chanock, R.M. Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc. Natl. Acad. Sci. USA 1967, 58, 2268–2273. [Google Scholar] [CrossRef] [Green Version]
- Van Der Hoek, L. Human coronaviruses: What do they cause? Antivir. Ther. 2007, 12, 651–658. [Google Scholar] [PubMed]
- Gaunt, E.R.; Hardie, A.; Claas, E.C.J.; Simmonds, P.; Templeton, K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010, 48, 2940–2947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poppe, M.; Wittig, S.; Jurida, L.; Bartkuhn, M.; Wilhelm, J.; Müller, H.; Beuerlein, K.; Karl, N.; Bhuju, S.; Ziebuhr, J.; et al. The NF-κB-dependent and -independent transcriptome and chromatin landscapes of human coronavirus 229E-infected cells. PLoS Pathog. 2017, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeager, C.L.; Ashmun, R.A.; Williams, R.K.; Cardellichio, C.B.; Shapiro, L.H.; Look, A.T.; Holmes, K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 1992, 357, 420–422. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Tomlinson, A.C.; Wong, A.H.; Zhou, D.; Desforges, M.; Talbot, P.J.; Benlekbir, S.; Rubinstein, J.L.; Rini, J.M. The human coronavirus HCoV-229E S-protein structure and receptor binding. Elife 2019, 8. [Google Scholar] [CrossRef]
- Liu, C.; Feng, Y.; Gao, F.; Zhang, Q.; Wang, M. Characterization of HCoV-229E fusion core: Implications for structure basis of coronavirus membrane fusion. Biochem. Biophys. Res. Commun. 2006, 345, 1108–1115. [Google Scholar] [CrossRef]
- Zhang, W.; Zheng, Q.; Yan, M.; Chen, X.; Yang, H.; Zhou, W.; Rao, Z. Structural characterization of the HCoV-229E fusion core. Biochem. Biophys. Res. Commun. 2018, 497, 705–712. [Google Scholar] [CrossRef]
- Yan, L.; Meng, B.; Xiang, J.; Wilson, I.A.; Yang, B. Crystal structure of the post-fusion core of the Human coronavirus 229E spike protein at 1.86 Å resolution. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 841–851. [Google Scholar] [CrossRef] [Green Version]
- Bertram, S.; Dijkman, R.; Habjan, M.; Heurich, A.; Gierer, S.; Glowacka, I.; Welsch, K.; Winkler, M.; Schneider, H.; Hofmann-Winkler, H.; et al. TMPRSS2 Activates the Human Coronavirus 229E for Cathepsin-Independent Host Cell Entry and Is Expressed in Viral Target Cells in the Respiratory Epithelium. J. Virol. 2013, 87, 6150–6160. [Google Scholar] [CrossRef] [Green Version]
- Kawase, M.; Shirato, K.; Matsuyama, S.; Taguchi, F. Protease-Mediated Entry via the Endosome of Human Coronavirus 229E. J. Virol. 2009, 83, 712–721. [Google Scholar] [CrossRef] [Green Version]
- Bonnin, A.; Danneels, A.; Dubuisson, J.; Goffard, A.; Belouzard, S. HCoV-229E spike protein fusion activation by trypsin-like serine proteases is mediated by proteolytic processing in the S2′ region. J. Gen. Virol. 2018, 99, 908–912. [Google Scholar] [CrossRef] [PubMed]
- van der Hoek, L.; Pyrc, K.; Berkhout, B. Human coronavirus NL63, a new respiratory virus. FEMS Microbiol. Rev. 2006, 30, 760–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Hoek, L.; Pyrc, K.; Jebbink, M.F.; Vermeulen-Oost, W.; Berkhout, R.J.M.; Wolthers, K.C.; Wertheim-van Dillen, P.M.E.; Kaandorp, J.; Spaargaren, J.; Berkhout, B. Identification of a new human coronavirus. Nat. Med. 2004, 10, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, H.; Pyrc, K.; Van Der Hoek, L.; Geier, M.; Berkhout, B.; Pöhlmann, S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA 2005, 102, 7988–7993. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.; Li, W.; Peng, G.; Li, F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. USA 2009, 106, 19970–19974. [Google Scholar] [CrossRef] [Green Version]
- Michaelis, M.; Doerr, H.W.; Cinatl, J. Investigation of the influence of EPs® 7630, a herbal drug preparation from Pelargonium sidoides, on replication of a broad panel of respiratory viruses. Phytomedicine 2011, 18, 384–386. [Google Scholar] [CrossRef]
- Kedik, S.A.; Yartsev, E.I.; Stanishevskaya, I.E. Antiviral activity of dried extract of Stevia. Pharm. Chem. J. 2009, 43, 198–199. [Google Scholar] [CrossRef]
- Tsai, Y.-C.; Lee, C.-L.; Yen, H.-R.; Chang, Y.-S.; Lin, Y.-P.; Huang, S.-H.; Lin, C.-W. Antiviral action of tryptanthrin isolated from strobilanthes cusia leaf against human coronavirus nl63. Biomolecules 2020, 10, 366. [Google Scholar] [CrossRef] [Green Version]
- Cheng, P.-W.; Ng, L.-T.; Chiang, L.-C.; Lin, C.-C. Antiviral effects of saikosaponins on human coronavirus 229E in vitro. Clin. Exp. Pharmacol. Physiol. 2006, 33, 612–616. [Google Scholar] [CrossRef]
- Chang, F.-R.; Yen, C.-T.; Ei-Shazly, M.; Lin, W.-H.; Yen, M.-H.; Lin, K.-H.; Wu, Y.-C. Anti-human coronavirus (anti-HCoV) triterpenoids from the leaves of Euphorbia neriifolia. Nat. Prod. Commun. 2012, 7, 1415–1417. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.-C.; Wang, L.-T.; Khalil, A.T.; Chiang, L.C.; Cheng, P.-W. Bioactive pyranoxanthones from the roots of Calophyllum blancoi. Chem. Pharm. Bull. 2005, 53, 244–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, C.; Schulte, F.W.; Lange-Grünweller, K.; Obermann, W.; Madhugiri, R.; Pleschka, S.; Ziebuhr, J.; Hartmann, R.K.; Grünweller, A. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antivir. Res. 2018, 150, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, H.; Iwami, M.; Kino, T.; Goto, T.; Okuhara, M. FR-900520 and FR-900523, novel immunosuppressants isolated from a Streptomyces. I. Taxonomy of the producing strain. J. Antibiot. (Tokyo) 1988, 41, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Carbajo-Lozoya, J.; Müller, M.A.; Kallies, S.; Thiel, V.; Drosten, C.; Von Brunn, A. Replication of human coronaviruses SARS-CoV, HCoV-NL63 and HCoV-229E is inhibited by the drug FK506. Virus Res. 2012, 165, 112–117. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Chu, C.-M.; Chan, K.-H.; Tsoi, H.-W.; Huang, Y.; Wong, B.H.L.; Poon, R.W.S.; Cai, J.J.; Luk, W.-k.; et al. Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia. J. Virol. 2005, 79, 884–895. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Q.; Langereis, M.A.; Van Vliet, A.L.W.; Huizinga, E.G.; De Groot, R.J. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 9065–9069. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Dong, W.; Milewska, A.; Golda, A.; Qi, Y.; Zhu, Q.K.; Marasco, W.A.; Baric, R.S.; Sims, A.C.; Pyrc, K.; et al. Human Coronavirus HKU1 Spike Protein Uses O-Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme. J. Virol. 2015, 89, 7202–7213. [Google Scholar] [CrossRef] [Green Version]
- Hulswit, R.J.G.; Lang, Y.; Bakkers, M.J.G.; Li, W.; Li, Z.; Schouten, A.; Ophorst, B.; Van Kuppeveld, F.J.M.; Boons, G.-J.; Bosch, B.-J.; et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. USA 2019, 116, 2681–2690. [Google Scholar] [CrossRef] [Green Version]
- Tortorici, M.A.; Walls, A.C.; Lang, Y.; Wang, C.; Li, Z.; Koerhuis, D.; Boons, G.-J.; Bosch, B.-J.; Rey, F.A.; de Groot, R.J.; et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019, 26, 481–489. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.E.; Min, J.S.; Jang, M.S.; Lee, J.Y.; Shin, Y.S.; Park, C.M.; Song, J.H.; Kim, H.R.; Kim, S.; Jin, Y.-H.; et al. Natural bis-benzylisoquinoline alkaloids-tetrandrine, fangchinoline, and cepharanthine, inhibit human coronavirus oc43 infection of mrc-5 human lung cells. Biomolecules 2019, 9, 696. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.; Niu, J.; Wang, C.; Huang, B.; Wang, W.; Zhu, N.; Deng, Y.; Wang, H.; Ye, F.; Cen, S.; et al. High-Throughput Screening and Identification of Potent Broad-Spectrum Inhibitors of Coronaviruses. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, B.Y.; Su, B.-N.; Chai, H.; Mi, Q.; Kardono, L.B.S.; Afriastini, J.J.; Riswan, S.; Santarsiero, B.D.; Mesecar, A.D.; Wild, R.; et al. Silvestrol and Episilvestrol, Potential Anticancer Rocaglate Derivatives from Aglaia silvestris. J. Org. Chem. 2004, 69, 3350–3358. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-Y.; Yuk, H.J.; Ryu, H.W.; Lim, S.H.; Kim, K.S.; Park, K.H.; Ryu, Y.B.; Lee, W.S. Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J. Enzyme Inhib. Med. Chem. 2017, 32, 504–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.Y.; Kim, Y., II; Park, S.J.; Kim, L.K.; Choi, Y.K.; Kim, S.-H. Safe, high-throughput screening of natural compounds of MERS-CoV entry inhibitors using a pseudovirus expressing MERS-CoV spike protein. Int. J. Antimicrob. Agents 2018, 52, 730–732. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.-C.; Shyur, L.-F.; Jan, J.-T.; Liang, P.-H.; Kuo, C.-J.; Arulselvan, P.; Wu, J.-B.; Kuo, S.C.; Yang, N.-S. Traditional chinese medicine herbal extracts of cibotium barometz, gentiana scabra, dioscorea batatas, cassia tora, and taxillus chinensis inhibit sars-cov replication. J. Tradit. Complement. Med. 2011, 1, 41–50. [Google Scholar] [CrossRef] [Green Version]
- Li, S.-Y.; Chen, C.; Zhang, H.-Q.; Guo, H.-Y.; Wang, H.; Wang, L.; Zhang, X.; Hua, S.-N.; Yu, J.; Xiao, P.-G. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antivir. Res. 2005, 67, 18–23. [Google Scholar] [CrossRef]
- Chen, C.-J.; Michaelis, M.; Hsu, H.-K.; Tsai, C.-C.; Yang, K.D.; Wu, Y.-C.; Cinatl, J.; Doerr, H.W. Toona sinensis Roem tender leaf extract inhibits SARS coronavirus replication. J. Ethnopharmacol. 2008, 120, 108–111. [Google Scholar] [CrossRef]
- Zhuang, M.; Jiang, H.; Suzuki, Y.; Li, X.; Xiao, P.; Tanaka, T.; Ling, H.; Yang, B.; Saitoh, H.; Zhang, L.; et al. Procyanidins and butanol extract of Cinnamomi Cortex inhibit SARS-CoV infection. Antivir. Res. 2009, 82, 73–81. [Google Scholar] [CrossRef]
- Keyaerts, E.; Li, S.; Vijgen, L.; Rysman, E.; Verbeeck, J.; Van Ranst, M.; Maes, P. Antiviral Activity of Chloroquine against Human Coronavirus OC43 Infection in Newborn Mice. Antimicrob. Agents Chemother. 2009, 53, 3416–3421. [Google Scholar] [CrossRef] [Green Version]
- Luo, W.; Su, X.; Gong, S.; Qin, Y.; Liu, W.; Li, J.; Yu, H.; Xu, Q. Anti-SARS coronavirus 3C-like protease effects of Rheum palmatum L. extracts. Biosci. Trends 2009, 3, 124–126. [Google Scholar]
- Kumar, V.; Jung, Y.-S.; Liang, P.-H. Anti-SARS coronavirus agents: A patent review (2008 – present). Expert Opin. Ther. Pat. 2013, 23, 1337–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, T.-Y.; Wu, S.-L.; Chen, J.-C.; Li, C.-C.; Hsiang, C.-Y. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antivir. Res. 2007, 74, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Wang, K.; Yu, W.; Sun, B.; Schwarz, W. Emodin inhibits current through SARS-associated coronavirus 3a protein. Antivir. Res. 2011, 90, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-W.; Tsai, F.-J.; Tsai, C.-H.; Lai, C.-C.; Wan, L.; Ho, T.-Y.; Hsieh, C.-C.; Chao, P.-D.L. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antivir. Res. 2005, 68, 36–42. [Google Scholar] [CrossRef]
- Yi, L.; Li, Z.; Yuan, K.; Qu, X.; Chen, J.; Wang, G.; Zhang, H.; Luo, H.; Zhu, L.; Jiang, P.; et al. Small Molecules Blocking the Entry of Severe Acute Respiratory Syndrome Coronavirus into Host Cells. J. Virol. 2004, 78, 11334–11339. [Google Scholar] [CrossRef] [Green Version]
- De Wilde, A.; Raj, V.; Oudshoorn, D.; Bestebroer, T.; Van Nieuwkoop, S.; Limpens, R.W.A.L.; Posthuma, C.; Van Der Meer, Y.; Bárcena, M.; Haagmans, B.; et al. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J. Gen. Virol. 2013, 94, 1749–1760. [Google Scholar] [CrossRef]
- Yang, C.-W.; Lee, Y.-Z.; Kang, I.-J.; Barnard, D.L.; Jan, J.-T.; Lin, D.; Huang, C.-W.; Yeh, T.-K.; Chao, Y.-S.; Lee, S.-J. Identification of phenanthroindolizines and phenanthroquinolizidines as novel potent anti-coronaviral agents for porcine enteropathogenic coronavirus transmissible gastroenteritis virus and human severe acute respiratory syndrome coronavirus. Antivir. Res. 2010, 88, 160–168. [Google Scholar] [CrossRef]
- Hoever, G.; Baltina, L.; Michaelis, M.; Kondratenko, R.; Tolstikov, G.A.; Doerr, H.W.; Cinatl, J. Antiviral Activity of Glycyrrhizic Acid Derivatives against SARS−Coronavirus. J. Med. Chem. 2005, 48, 1256–1259. [Google Scholar] [CrossRef]
- Wen, C.-C.; Kuo, Y.-H.; Jan, J.-T.; Liang, P.-H.; Wang, S.-Y.; Liu, H.-G.; Lee, C.-K.; Chang, S.-T.; Kuo, C.-J.; Lee, S.-S.; et al. Specific Plant Terpenoids and Lignoids Possess Potent Antiviral Activities against Severe Acute Respiratory Syndrome Coronavirus. J. Med. Chem. 2007, 50, 4087–4095. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Eo, E.-Y.; Park, H.; Kim, Y.-C.; Park, S.; Shin, H.; Kim, K. Medicinal herbal extracts of Sophorae radix, Acanthopanacis cortex, Sanguisorbae radix and Torilis fructus inhibit coronavirus replication in vitro. Antivir. Ther. 2010, 15, 697–709. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.-Y.; Tian, X.-Y.; Fang, W.-S. Bioactive coumarins from Boenninghausenia sessilicarpa. J. Asian Nat. Prod. Res. 2007, 9, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chan, K.; Jiang, Y.; Kao, R.; Lu, H.; Fan, K.; Cheng, V.; Tsui, W.; Hung, I.; Lee, T. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. 2004, 31, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.B.; Jeong, H.J.; Kim, J.H.; Kim, Y.M.; Park, J.-Y.; Kim, D.; Naguyen, T.T.H.; Park, S.-J.; Chang, J.S.; Park, K.H. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorg. Med. Chem. 2010, 18, 7940–7947. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, J.; Luo, C.; Liu, H.; Xu, W.; Chen, G.; Liew, O.W.; Zhu, W.; Puah, C.; Shen, X.; et al. Binding interaction of quercetin-3-β-galactoside and its synthetic derivatives with SARS-CoV 3CLpro: Structure–activity relationship studies reveal salient pharmacophore features. Bioorg. Med. Chem. 2006, 14, 8295–8306. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.B.; Park, S.-J.; Kim, Y.M.; Lee, J.-Y.; Seo, W.D.; Chang, J.S.; Park, K.H.; Rho, M.-C.; Lee, W.S. SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii. Bioorg. Med. Chem. Lett. 2010, 20, 1873–1876. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zuckerman, D.M.; Brantley, S.; Sharpe, M.; Childress, K.; Hoiczyk, E.; Pendleton, A.R. Sambucus nigra extracts inhibit infectious bronchitis virus at an early point during replication. BMC Veter Res. 2014, 10, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.-R.; Wang, Y.-C.; Lin, Y.W.; Chou, S.-Y.; Chen, S.-F.; Liu, L.T.; Wu, Y.-T.; Kuo, C.-J.; Chen, T.S.-S.; Juang, S.-H. Synthesis and evaluation of isatin derivatives as effective SARS coronavirus 3CL protease inhibitors. Bioorg. Med. Chem. Lett. 2005, 15, 3058–3062. [Google Scholar] [CrossRef]
- Liu, W.; Zhu, H.-M.; Niu, G.-J.; Shi, E.-Z.; Chen, J.; Sun, B.; Chen, W.; Zhou, H.; Yang, C. Synthesis, modification and docking studies of 5-sulfonyl isatin derivatives as SARS-CoV 3C-like protease inhibitors. Bioorg. Med. Chem. 2014, 22, 292–302. [Google Scholar] [CrossRef]
- Schmidtke, M.; Meier, C.; Schacke, M.; Helbig, B.; Makarov, V.; Rabenau, H.F.; Cinatl, J.; Wutzler, P. Antiviral activity of phenolic polymers and cycloSal-pronucleotides against a SARS-associated coronavirus. Chemother. J. 2005, 14, 16–21. [Google Scholar]
- Park, J.-Y.; Kim, J.H.; Kim, Y.M.; Jeong, H.J.; Kim, D.W.; Park, K.H.; Kwon, H.-J.; Park, S.-J.; Lee, W.S.; Ryu, Y.B. Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorg. Med. Chem. 2012, 20, 5928–5935. [Google Scholar] [CrossRef]
- Park, J.-Y.; Kim, J.H.; Kwon, J.M.; Kwon, H.-J.; Jeong, H.J.; Kim, Y.M.; Kim, D.; Lee, W.S.; Ryu, Y.B. Dieckol, a SARS-CoV 3CLpro inhibitor, isolated from the edible brown algae Ecklonia cava. Bioorg. Med. Chem. 2013, 21, 3730–3737. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-Y.; Ko, J.-A.; Kim, D.W.; Kim, Y.M.; Kwon, H.-J.; Jeong, H.J.; Kim, C.Y.; Park, K.H.; Lee, W.S.; Ryu, Y.B. Chalcones isolated fromAngelica keiskeiinhibit cysteine proteases of SARS-CoV. J. Enzym. Inhib. Med. Chem. 2016, 31, 23–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.-Y.; Jeong, H.J.; Kim, J.H.; Kim, Y.M.; Park, S.-J.; Kim, D.; Park, K.H.; Lee, W.S.; Ryu, Y.B. Diarylheptanoids from Alnus japonica Inhibit Papain-Like Protease of Severe Acute Respiratory Syndrome Coronavirus. Biol. Pharm. Bull. 2012, 35, 2036–2042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.W.; Seo, K.H.; Curtis-Long, M.J.; Oh, K.Y.; Oh, J.-W.; Cho, J.K.; Lee, K.H.; Park, K.H. Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J. Enzym. Inhib. Med. Chem. 2013, 29, 59–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.H.; Kim, D.W.; Curtis-Long, M.J.; Yuk, H.J.; Wang, Y.; Zhuang, N.; Lee, K.H.; Jeon, K.S.; Park, K.H. Papain-Like Protease (PLpro) Inhibitory Effects of Cinnamic Amides from Tribulus terrestris Fruits. Biol. Pharm. Bull. 2014, 37, 1021–1028. [Google Scholar] [CrossRef] [Green Version]
- Cho, J.K.; Curtis-Long, M.J.; Lee, K.H.; Kim, D.W.; Ryu, H.W.; Yuk, H.J.; Park, K.H. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorg. Med. Chem. 2013, 21, 3051–3057. [Google Scholar] [CrossRef]
- Yu, M.-S.; Lee, J.; Lee, J.M.; Kim, Y.; Chin, Y.-W.; Jee, J.-G.; Keum, Y.-S.; Jeong, Y.-J. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 2012, 22, 4049–4054. [Google Scholar] [CrossRef]
- Keyaerts, E.; Vijgen, L.; Maes, P.; Neyts, J.; Van Ranst, M. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Commun. 2004, 323, 264–268. [Google Scholar] [CrossRef]
- Barnard, D.L.; Day, C.W.; Bailey, K.; Heiner, M.; Montgomery, R.; Lauridsen, L.; Chan, P.K.; Sidwell, R.W. Evaluation of Immunomodulators, Interferons and Knownin VitroSARS-CoV Inhibitors for Inhibition of SARS-Cov Replication in BALB/c Mice. Antivir. Chem. Chemother. 2006, 17, 275–284. [Google Scholar] [CrossRef] [Green Version]
- Mehra, M.R.; Desai, S.S.; Ruschitzka, F.; Patel, A.N. RETRACTED: Hydroxychloroquine or chloroquine with or without a macrolide for treatment of COVID-19: A multinational registry analysis. Lancet 2020. [Google Scholar] [CrossRef]
- Jo, S.; Kim, S.; Shin, D.H.; Kim, M.-S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzym. Inhib. Med. Chem. 2020, 35, 145–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cinatl, J.; Morgenstern, B.; Bauer, G.; Chandra, P.; Rabenau, H.; Doerr, H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet 2003, 361, 2045–2046. [Google Scholar] [CrossRef] [Green Version]
- Pyrc, K.; Bosch, B.J.; Berkhout, B.; Jebbink, M.F.; Dijkman, R.; Rottier, P.; Van Der Hoek, L. Inhibition of Human Coronavirus NL63 Infection at Early Stages of the Replication Cycle. Antimicrob. Agents Chemother. 2006, 50, 2000–2008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huan, C.-C.; Wang, H.-X.; Sheng, X.-X.; Wang, R.; Wang, X.; Mao, X. Glycyrrhizin inhibits porcine epidemic diarrhea virus infection and attenuates the proinflammatory responses by inhibition of high mobility group box-1 protein. Arch. Virol. 2017, 162, 1467–1476. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, M.; Geiler, J.; Naczk, P.; Sithisarn, P.; Leutz, A.; Doerr, H.W.; Cinatl, J. Glycyrrhizin Exerts Antioxidative Effects in H5N1 Influenza A Virus-Infected Cells and Inhibits Virus Replication and Pro-Inflammatory Gene Expression. PLoS ONE 2011, 6, e19705. [Google Scholar] [CrossRef] [Green Version]
- DeDiego, M.L.; Nieto-Torres, J.L.; Regla-Nava, J.A.; Jimenez-Guardeño, J.M.; Fernandez-Delgado, R.; Fett, C.; Castaño-Rodriguez, C.; Perlman, S.; Enjuanes, L. Inhibition of NF-B-Mediated Inflammation in Severe Acute Respiratory Syndrome Coronavirus-Infected Mice Increases Survival. J. Virol. 2014, 88, 913–924. [Google Scholar] [CrossRef] [Green Version]
- Khare, P.; Sahu, U.; Pandey, S.C.; Samant, M. Current approaches for target-specific drug discovery using natural com-pounds against SARS-CoV-2 infection. Virus Res. 2020, 290, 198169. [Google Scholar] [CrossRef]
- Naik, B.; Gupta, N.; Ojha, R.; Singh, S.; Prajapati, V.K.; Prusty, D. High throughput virtual screening reveals SARS-CoV-2 multi-target binding natural compounds to lead instant therapy for COVID-19 treatment. Int. J. Biol. Macromol. 2020, 160, 1–17. [Google Scholar] [CrossRef]
- Chávez-Hernández, A.L.; Sánchez-Cruz, N.; Medina-Franco, J.L. Fragment Library of Natural Products and Compound Databases for Drug Discovery. Biomolecules 2020, 10, 1518. [Google Scholar] [CrossRef]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020, 10, 766–788. [Google Scholar] [CrossRef]
- Rakib, A.; Paul, A.; Chy, N.U.; Sami, S.A.; Baral, S.K.; Majumder, M.; Tareq, A.M.; Amin, M.N.; Shahriar, A.; Uddin, Z.; et al. Biochemical and Computational Approach of Selected Phytocompounds from Tinospora crispa in the Management of COVID-19. Molecules 2020, 25, 3936. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.A.A.; Abdeljawaad, K.A.A.; Abdelrahman, A.H.M.; Hegazy, M.E.F. Natural-like products as potential SARS-CoV-2 Mpro inhibitors: In-silico drug discovery. J. Biomol. Struct. Dyn. 2020, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gyebi, G.A.; Ogunro, O.B.; Adegunloye, A.P.; Ogunyemi, O.M.; Afolabi, S.O. Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CLpro): An in silico screening of alkaloids and terpenoids from African medicinal plants. J. Biomol. Struct. Dyn. 2020, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Sarmah, S.; Lyndem, S.; Roy, A.S. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J. Biomol. Struct. Dyn. 2020, 1–11. [Google Scholar] [CrossRef]
- Quimque, M.T.J.; Notarte, K.I.R.; Fernandez, R.A.T.; Mendoza, M.A.O.; Liman, R.A.D.; Lim, J.A.K.; Pilapil, L.A.E.; Ong, J.K.H.; Pastrana, A.M.; Khan, A.; et al. Virtual screening-driven drug discovery of SARS-CoV2 enzyme inhibitors targeting viral attachment, replication, post-translational modification and host immunity evasion infection mechanisms. J. Biomol. Struct. Dyn. 2020, 1–18. [Google Scholar] [CrossRef]
- Badavath, V.N.; Kumar, A.; Samanta, P.K.; Maji, S.; Das, A.; Blum, G.; Jha, A.; Sen, A. Determination of potential inhibitors based on isatin derivatives against SARS-CoV-2 main protease (mpro): A molecular docking, molecular dynamics and structure-activity relationship studies. J. Biomol. Struct. Dyn. 2020, 1–19. [Google Scholar] [CrossRef]
- Narkhede, R.R.; Pise, A.V.; Cheke, R.S.; Shinde, S.D. Recognition of Natural Products as Potential Inhibitors of COVID-19 Main Protease (Mpro): In-Silico Evidences. Nat. Prod. Bioprospecting 2020, 10, 297–306. [Google Scholar] [CrossRef]
- Abdelrheem, D.A.; Ahmed, S.A.; El-Mageed, H.R.A.; Mohamed, H.S.; Rahman, A.A.; Elsayed, K.N.M.; Ahmed, S.A. The inhibitory effect of some natural bioactive compounds against SARS-CoV-2 main protease: Insights from molecular docking analysis and molecular dynamic simulation. J. Environ. Sci. Heal. Part A 2020, 55, 1373–1386. [Google Scholar] [CrossRef]
- Tripathi, N.; Goel, B.; Bhardwaj, N.; Sahu, B.; Kumar, H.; Jain, S.K. Virtual screening and molecular simulation study of natural products database for lead identification of novel coronavirus main protease inhibitors. J. Biomol. Struct. Dyn. 2020, 1–13. [Google Scholar] [CrossRef]
- Borquaye, L.S.; Gasu, E.N.; Ampomah, G.B.; Kyei, L.K.; Amarh, M.A.; Mensah, C.N.; Nartey, D.; Commodore, M.; Adomako, A.K.; Acheampong, P.; et al. Alkaloids from Cryptolepis sanguinolenta as Potential Inhibitors of SARS-CoV-2 Viral Proteins: An In Silico Study. BioMed Res. Int. 2020, 2020, 1–14. [Google Scholar] [CrossRef]
- Qamar, M.T.U.; Alqahtani, S.M.; Alamri, M.A.; Chen, L.-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020, 10, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, G.C.S.; Maia, M.D.S.; De Menezes, R.P.B.; Cavalcanti, A.B.S.; De Sousa, N.F.; De Moura, É.P.; Monteiro, A.F.M.; Scotti, L.; Scotti, M.T. Ligand and Structure-based Virtual Screening of Lamiaceae Diterpenes with Potential Activity against a Novel Coronavirus (2019-nCoV). Curr. Top. Med. Chem. 2020, 20, 2126–2145. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, I.; Muangchoo, K.; Muhammad, A.; Ajingi, Y.S.; Muhammad, I.; Umar, I.D.; Muhammad, A.B. A Computational Study to Identify Potential Inhibitors of SARS-CoV-2 Main Protease (Mpro) from Eucalyptus Active Compounds. Computation 2020, 8, 79. [Google Scholar] [CrossRef]
- Ebada, S.S.; Al-Jawabri, N.A.; Youssef, F.S.; El-Kashef, D.H.; Knedel, T.-O.; Albohy, A.; Korinek, M.; Hwang, T.-L.; Chen, B.-H.; Lin, G.-H.; et al. Anti-inflammatory, antiallergic and COVID-19 protease inhibitory activities of phytochemicals from the Jordanian hawksbeard: Identification, structure–activity relationships, molecular modeling and impact on its folk medicinal uses. RSC Adv. 2020, 10, 38128–38141. [Google Scholar] [CrossRef]
- Joshi, R.S.; Jagdale, S.S.; Bansode, S.B.; Shankar, S.S.; Tellis, M.B.; Pandya, V.K.; Chugh, A.; Giri, A.P.; Kulkarni, M.J. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J. Biomol. Struct. Dyn. 2020, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Lakshmi, S.A.; Rajamohamed, B.S.; Priya, A.; Pandian, S.K. Ethnomedicines of Indian origin for combating COVID-19 infection by hampering the viral replication: Using structure-based drug discovery approach. J. Biomol. Struct. Dyn. 2020, 1–16. [Google Scholar] [CrossRef]
- Fakhar, Z.; Faramarzi, B.; Pacifico, S.; Faramarzi, S. Anthocyanin derivatives as potent inhibitors of SARS-CoV-2 main protease: An in-silico perspective of therapeutic targets against COVID-19 pandemic. J. Biomol. Struct. Dyn. 2020, 1–13. [Google Scholar] [CrossRef]
- Khalifa, I.; Nawaz, A.; Sobhy, R.; Althwab, S.A.; Barakat, H. Polyacylated anthocyanins constructively network with catalytic dyad residues of 3CLpro of 2019-nCoV than monomeric anthocyanins: A structural-relationship activity study with 10 anthocyanins using in-silico approaches. J. Mol. Graph. Model. 2020, 100, 107690. [Google Scholar] [CrossRef]
- Iheagwam, F.N.; Rotimi, S.O. Computer-Aided Analysis of Multiple SARS-CoV-2 Therapeutic Targets: Identification of Potent Molecules from African Medicinal Plants. Scientifica 2020, 2020, 1–25. [Google Scholar] [CrossRef]
- Gahlawat, A.; Kumar, N.; Kumar, R.; Sandhu, H.; Singh, I.P.; Singh, S.; Sjöstedt, A.; Garg, P. Structure-Based Virtual Screening to Discover Potential Lead Molecules for the SARS-CoV-2 Main Protease. J. Chem. Inf. Model. 2020, 60, 5781–5793. [Google Scholar] [CrossRef]
- Santos-Filho, O. Identification of Potential Inhibitors of Severe Acute Respiratory Syndrome-Related Coronavirus 2 (SARS-CoV-2) Main Protease from Non-Natural and Natural Sources: A Molecular Docking Study. J. Braz. Chem. Soc. 2020, 31, 2638–2643. [Google Scholar] [CrossRef]
- Sampangi-Ramaiah, M.H.; Vishwakarma, R.; Shaanker, R.U. Molecular docking analysis of selected natural products from plants for inhibition of SARS-CoV-2 main protease. Curr. Sci. 2020, 118, 1087–1092. [Google Scholar] [CrossRef]
- Owis, A.I.; El-Hawary, M.S.; El Amir, D.; Aly, O.M.; Abdelmohsen, U.R.; Kamel, M.S. Molecular docking reveals the potential of Salvadora persica flavonoids to inhibit COVID-19 virus main protease. RSC Adv. 2020, 10, 19570–19575. [Google Scholar] [CrossRef]
- Karankumar, B.; Faheem; Sekhar, K.V.G.C.; Ojha, R.; Prajapati, V.K.; Pai, A.; Murugesan, S. Pharmacophore based virtual screening, molecular docking, molecular dynamics and MM-GBSA approach for identification of prospective SARS-CoV-2 inhibitor from natural product databases. J. Biomol. Struct. Dyn. 2020, 1–24. [Google Scholar] [CrossRef]
- Majumder, R.; Mandal, M. Screening of plant-based natural compounds as a potential COVID-19 main protease inhibitor: An in silico docking and molecular dynamics simulation approach. J. Biomol. Struct. Dyn. 2020, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Zia, K.; Ashraf, S.; Uddin, R.; Ul-Haq, Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J. Biomol. Struct. Dyn. 2020, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, T.; Sharma, P.; Mathpal, S.; Pundir, H.; Bhatt, V.; Chandra, S. In silico screening of natural compounds against COVID-19 by targeting Mpro and ACE2 using molecular docking. Eur. Rev. Med. Pharmacol. Sci 2020, 24, 4529–4536. [Google Scholar]
- El-Mordy, F.M.A.; El-Hamouly, M.M.; Ibrahim, M.T.; El-Rheem, G.A.; Aly, O.M.; El-Kader, A.M.A.; Youssif, K.A.; Abdelmohsen, U.R. Inhibition of SARS-CoV-2 main protease by phenolic compounds from Manilkara hexandra (Roxb.) Dubard assisted by metabolite profiling and in silico virtual screening. RSC Adv. 2020, 10, 32148–32155. [Google Scholar] [CrossRef]
- Mazzini, S.; Musso, L.; Dallavalle, S.; Artali, R. Putative SARS-CoV-2 Mpro Inhibitors from an In-House Library of Natural and Nature-Inspired Products: A Virtual Screening and Molecular Docking Study. Molecules 2020, 25, 3745. [Google Scholar] [CrossRef]
- Mitra, K.; Ghanta, P.; Acharya, S.; Chakrapani, G.; Ramaiah, B.; Doble, M. Dual inhibitors of SARS-CoV-2 proteases: Pharmacophore and molecular dynamics based drug repositioning and phytochemical leads. J. Biomol. Struct. Dyn. 2020, 1–14. [Google Scholar] [CrossRef]
- Singh, R.; Gautam, A.; Chandel, S.; Ghosh, A.; Dey, D.; Roy, S.; Ravichandiran, V.; Ghosh, D. Protease Inhibitory Effect of Natural Polyphenolic Compounds on SARS-CoV-2: An In Silico Study. Molecules 2020, 25, 4604. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.A.A.; Abdelrahman, A.H.; Hussien, T.A.; Badr, E.A.; Mohamed, T.A.; El-Seedi, H.R.; Pare, P.W.; Efferth, T.; Hegazy, M.-E.F. In silico drug discovery of major metabolites from spices as SARS-CoV-2 main protease inhibitors. Comput. Biol. Med. 2020, 126, 104046. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.N.; De Sá, É.R.A.; Bezerra, R.D.S.; Souza, J.L.; Lima, F.D.C.A. Constituents of buriti oil (Mauritia flexuosa L.) like inhibitors of the SARS-Coronavirus main peptidase: An investigation by docking and molecular dynamics. J. Biomol. Struct. Dyn. 2020, 1–8. [Google Scholar] [CrossRef]
- Yang, J.; Chiang, J.-H.; Tsai, S.; Hsu, Y.-M.; Bau, D.-T.; Lee, K.-H.; Tsai, F.-J. In Silico De Novo Curcuminoid Derivatives from the Compound Library of Natural Products Research Laboratories Inhibit COVID-19 3CLpro Activity. Nat. Prod. Commun. 2020, 15. [Google Scholar] [CrossRef]
- Caruso, F.; Rossi, M.; Pedersen, J.Z.; Incerpi, S. Computational studies reveal mechanism by which quinone derivatives can inhibit SARS-CoV-2. Study of embelin and two therapeutic compounds of interest, methyl prednisolone and dexamethasone. J. Infect. Public Heal. 2020, 13, 1868–1877. [Google Scholar] [CrossRef]
- Didemnins Inhibit COVID-19 Main Protease (Mpro). Biointerface Res. Appl. Chem. 2020, 11, 8204–8209. [CrossRef]
- Basu, A.; Sarkar, A.; Maulik, U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Hasan, A.; Paray, B.A.; Hussain, A.; Qadir, F.A.; Attar, F.; Aziz, F.M.; Sharifi, M.; Derakhshankhah, H.; Rasti, B.; Mehrabi, M.; et al. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J. Biomol. Struct. Dyn. 2020, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Rahman, N.; Basharat, Z.; Yousuf, M.; Castaldo, G.; Rastrelli, L.; Khan, H. Virtual Screening of Natural Products against Type II Transmembrane Serine Protease (TMPRSS2), the Priming Agent of Coronavirus 2 (SARS-CoV-2). Molecules 2020, 25, 2271. [Google Scholar] [CrossRef]
- Vivek-Ananth, R.; Rana, A.; Rajan, N.; Biswal, H.S.; Samal, A. In Silico Identification of Potential Natural Product Inhibitors of Human Proteases Key to SARS-CoV-2 Infection. Molecules 2020, 25, 3822. [Google Scholar] [CrossRef]
- Chikhale, R.V.; Gupta, V.K.; E Eldesoky, G.; Wabaidur, S.M.; A Patil, S.; Islam, A. Identification of potential anti-TMPRSS2 natural products through homology modelling, virtual screening and molecular dynamics simulation studies. J. Biomol. Struct. Dyn. 2020, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, M.; Schleich, S.; Prade, I.; Degen, S.; Roell, D.; Schubert, U.; Tanner, T.; Claessens, F.; Matusch, R.; Baniahmad, A. The natural compound atraric acid is an antagonist of the human androgen receptor inhibiting cellular invasiveness and prostate cancer cell growth. J. Cell. Mol. Med. 2008, 13, 2210–2223. [Google Scholar] [CrossRef] [PubMed]
- Meimetis, L.G.; Williams, D.E.; Mawji, N.R.; Banuelos, C.A.; Lal, A.A.; Park, J.J.; Tien, A.H.; Fernandez, J.G.; De Voogd, N.J.; Sadar, M.D.; et al. Niphatenones, Glycerol Ethers from the Sponge Niphates digitalis Block Androgen Receptor Transcriptional Activity in Prostate Cancer Cells: Structure Elucidation, Synthesis, and Biological Activity. J. Med. Chem. 2012, 55, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.C.; Zhang, J.; Dev, A.; Wing, A.; Bjeldanes, L.F.; Firestone, G.L. Indole-3-carbinol inhibition of androgen receptor expression and downregulation of androgen responsiveness in human prostate cancer cells. Carcinogenesis 2005, 26, 1896–1904. [Google Scholar] [CrossRef] [Green Version]
- Qiao, Y.; Wang, X.-M.; Mannan, R.; Pitchiaya, S.; Zhang, Y.; Wotring, J.W.; Xiao, L.; Robinson, D.R.; Wu, Y.-M.; Tien, J.C.-Y.; et al. Targeting transcriptional regulation of SARS-CoV-2 entry factors ACE2 and TMPRSS2. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Zhang, Z.-R.; Zhang, Y.-N.; Li, X.-D.; Zhang, H.-Q.; Xiao, S.-Q.; Deng, F.; Yuan, Z.-M.; Ye, H.-Q.; Zhang, B. A cell-based large-scale screening of natural compounds for inhibitors of SARS-CoV-2. Signal Transduct. Target. Ther. 2020, 5. [Google Scholar] [CrossRef]
- Kanjanasirirat, P.; Suksatu, A.; Manopwisedjaroen, S.; Munyoo, B.; Tuchinda, P.; Jearawuttanakul, K.; Seemakhan, S.; Charoensutthivarakul, S.; Wongtrakoongate, P.; Rangkasenee, N.; et al. High-Content Screening of Thai Medicinal Plants Reveals Boesenbergia rotunda Extract and its Component Panduratin A as Anti-SARS-CoV-2 Agents. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Su, H.X.; Yao, S.; Zhao, W.F.; Li, M.J.; Liu, J.; Shang, W.J.; Xie, H.; Ke, C.Q.; Hu, H.C.; Gao, M.N.; et al. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020, 41, 1167–1177. [Google Scholar] [CrossRef]
- Shi, T.-H.; Huang, Y.-L.; Chen, C.-C.; Pi, W.-C.; Hsu, Y.-L.; Lo, L.-C.; Chen, W.-Y.; Fu, S.-L.; Lin, C.-H. Andrographolide and its fluorescent derivative inhibit the main proteases of 2019-nCoV and SARS-CoV through covalent linkage. Biochem. Biophys. Res. Commun. 2020, 533, 467–473. [Google Scholar] [CrossRef]
- Coelho, C.; Gallo, G.; Campos, C.B.; Hardy, L.; Würtele, M. Biochemical screening for SARS-CoV-2 main protease inhibitors. PLoS ONE 2020, 15, e0240079. [Google Scholar] [CrossRef]
- Abian, O.; Ortega-Alarcon, D.; Jimenez-Alesanco, A.; Ceballos-Laita, L.; Vega, S.; Reyburn, H.T.; Rizzuti, B.; Velázquez-Campoy, A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int. J. Biol. Macromol. 2020, 164, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Shanavas, A. Natural derivatives with dual binding potential against SARS-CoV-2 main protease and human ACE2 possess low oral bioavailability: A brief computational analysis. J. Biomol. Struct. Dyn. 2020, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Efferth, T.; Romero, M.R.; Wolf, D.G.; Stamminger, T.; Marin, J.J.G.; Marschall, M. The Antiviral Activities of Artemisinin and Artesunate. Clin. Infect. Dis. 2008, 47, 804–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baglivo, M.; Baronio, M.; Natalini, G.; Beccari, T.; Chiurazzi, P.; Fulcheri, E.; Petralia, P.P.; Michelini, S.; Fiorentini, G.; Miggiano, G.A.D.; et al. Natural small molecules as inhibitors of coronavirus lipid-dependent attachment to host cells: A possible strategy for reducing SARS-COV-2 infectivity? Acta Biomed. 2020, 91, 161–164. [Google Scholar]
- Bailly, C.; Vergoten, G. Glycyrrhizin: An alternative drug for the treatment of COVID-19 infection and the associated res-piratory syndrome? Pharmacol. Ther. 2020, 214, 107618. [Google Scholar] [CrossRef]
- Wang, S.; Li, W.; Hui, H.; Tiwari, S.K.; Zhang, Q.; Croker, B.A.; Rawlings, S.; Smith, D.; Carlin, A.F.; Rana, T.M. Cholesterol 25-Hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 2020, 39. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Han, J.W.; Ahn, S.H.; Park, S.H.; Wang, S.Y.; Bae, G.U.; Seo, D.W.; Kwon, H.K.; Hong, S.; Lee, H.Y.; Lee, Y.W.; et al. Apicidin, a histone deacetylase inhibitor, inhibits proliferation of tumor cells via induction of p21WAF1/Cip1 and gelsolin. Cancer Res. 2000, 60, 6068–6074. [Google Scholar]
- Kindrachuk, J.; Ork, B.; Hart, B.J.; Mazur, S.; Holbrook, M.R.; Frieman, M.B.; Traynor, D.; Johnson, R.F.; Dyall, J.; Kuhn, J.H.; et al. Antiviral Potential of ERK/MAPK and PI3K/AKT/mTOR Signaling Modulation for Middle East Respiratory Syndrome Coronavirus Infection as Identified by Temporal Kinome Analysis. Antimicrob. Agents Chemother. 2015, 59, 1088–1099. [Google Scholar] [CrossRef] [Green Version]
- Coleman, C.M.; Sisk, J.M.; Mingo, R.M.; Nelson, E.A.; White, J.M.; Frieman, M.B. Abelson Kinase Inhibitors Are Potent Inhibitors of Severe Acute Respiratory Syndrome Coronavirus and Middle East Respiratory Syndrome Coronavirus Fusion. J. Virol. 2016, 90, 8924–8933. [Google Scholar] [CrossRef] [Green Version]
- Emadi, A.; Chua, J.V.; Talwani, R.; Bentzen, S.M.; Baddley, J.W. Safety and Efficacy of Imatinib for Hospitalized Adults with COVID-19: A structured summary of a study protocol for a randomised controlled trial. Trials 2020, 21, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Mendenhall, M.; Deininger, M.W. Imatinib is not a potent anti-SARS-CoV-2 drug. Leukemia 2020, 34, 3085–3087. [Google Scholar] [CrossRef] [PubMed]
- Pairo-Castineira, E.; Clohisey, S.; Klaric, L.; Bretherick, A.D.; Rawlik, K.; Pasko, D.; Walker, S.; Parkinson, N.; Fourman, M.H.; Russell, C.D.; et al. Genetic mechanisms of critical illness in Covid-19. Nat. Cell Biol. 2020, 1. [Google Scholar] [CrossRef]
- Sigurdsson, S.; Nordmark, G.; Göring, H.H.H.; Lindroos, K.; Wiman, A.-C.; Sturfelt, G.; Jönsen, A.; Rantapää-Dahlqvist, S.; Möller, B.; Kere, J.; et al. Polymorphisms in the Tyrosine Kinase 2 and Interferon Regulatory Factor 5 Genes Are Associated with Systemic Lupus Erythematosus. Am. J. Hum. Genet. 2005, 76, 528–537. [Google Scholar] [CrossRef] [Green Version]
- Dowty, M.E.; Lin, T.H.; Jesson, M.I.; Hegen, M.; Martin, D.A.; Katkade, V.; Menon, S.; Telliez, J. Janus kinase inhibitors for the treatment of rheumatoid arthritis demonstrate similar profiles of in vitro cytokine receptor inhibition. Pharmacol. Res. Perspect. 2019, 7, e00537. [Google Scholar] [CrossRef] [Green Version]
- The Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene) Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat. Genet. 2009, 41, 824–828. [CrossRef]
- Forman, S.B.; Pariser, D.M.; Poulin, Y.; Vincent, M.S.; A Gilbert, S.; Kieras, E.M.; Qiu, R.; Yu, D.; Papacharalambous, J.; Tehlirian, C.; et al. TYK2/JAK1 Inhibitor PF-06700841 in Patients with Plaque Psoriasis: Phase IIa, Randomized, Double-blind, Placebo-controlled Trial. J. Investig. Dermatol 2020, 140, 2359–2370.e5. [Google Scholar] [CrossRef]
- Page, K.M.; Suarez-Farinas, M.; Suprun, M.; Zhang, W.; Garcet, S.; Fuentes-Duculan, J.; Li, X.; Scaramozza, M.; Kieras, E.; Banfield, C.; et al. Molecular and Cellular Responses to the TYK2/JAK1 Inhibitor PF-06700841 Reveal Reduction of Skin In-flammation in Plaque Psoriasis. J. Investig. Dermatol. 2020, 140, 1546–1555.e4. [Google Scholar] [CrossRef]
- Gerstenberger, B.S.; Ambler, C.M.; Arnold, E.P.; Banker, M.-E.; Brown, M.F.; Clark, J.D.; Dermenci, A.; Dowty, M.E.; Fensome, A.; Fish, S.; et al. Discovery of Tyrosine Kinase 2 (TYK2) Inhibitor (PF-06826647) for the Treatment of Autoimmune Diseases. J. Med. Chem. 2020, 63, 13561–13577. [Google Scholar] [CrossRef]
- Liu, M.; Xiao, C.; Sun, M.; Tan, M.; Hu, L.; Yu, Q. Parthenolide Inhibits STAT3 Signaling by Covalently Targeting Janus Kinases. Molecules 2018, 23, 1478. [Google Scholar] [CrossRef] [Green Version]



| Structure | Name | Source | Target | Assay | Ref. | |
|---|---|---|---|---|---|---|
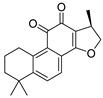 | Cryptotanshinone | Salvia miltiorrhiza Lamiaceae | SARS-CoV | Enzyme inhibition (IC50) | PLpro 0.8 μΜ, 3CLpro 226.7 μΜ | [113] |
| SARS-CoV-2 | CPE inhibition (EC50) | 5.024 μΜ | [179] | |||
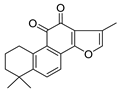 | Tanshinone IIA | Salvia miltiorrhiza Lamiaceae | SARS-CoV | Enzyme inhibition (IC50) | PLpro 1.6 μΜ, 3CLpro 89.1 μΜ | [113] |
| SARS-CoV-2 | CPE inhibition (EC50) | <11 μΜ | [179] | |||
 | Isobavachalcone | Psoralea sp. Fabaceae | SARS-CoV | Enzyme inhibition (IC50) | PLpro 7.3 μΜ | [117] |
| SARS-CoV-2 | CPE inhibition (EC50) | <11 μΜ | [179] | |||
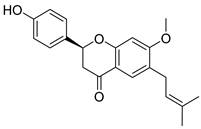 | Bavachin | Psoralea sp. Fabaceae | SARS-CoV | Enzyme inhibition (IC50) | PLpro 38.4 μΜ | [117] |
| SARS-CoV-2 | CPE inhibition (EC50) | <11 μΜ | [179] | |||
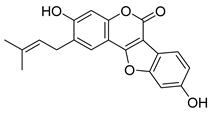 | Psoralidin | Psoralea sp. Fabaceae | SARS-CoV | Enzyme inhibition (IC50) | PLpro 4.2 μΜ | [117] |
| SARS-CoV-2 | CPE inhibition (EC50) | <11 μΜ | [179] | |||
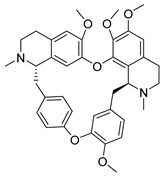 | Tetrandrine | Stephania tetrandra Menispermaceae | HCoV-NL63 | CPE inhibition (EC50) | 2.05 μΜ | [84] |
| HCoV-OC43 | 0.29 μΜ/0.33 μΜ | [83,84] | ||||
| MERS-CoV | 12.68 μΜ | [84] | ||||
| SARS-CoV-2 | CPE inhibition (EC50) | <11 μΜ | [179] | |||
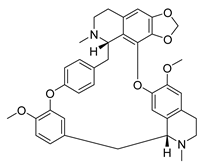 | Cepharanthine | Stephania tetrandra Menispermaceae | HCoV-OC43 | CPE inhibition (EC50) | 0.83 μΜ | [83] |
| SARS-CoV-2 | CPE inhibition (EC50) | <11 μΜ | [179] | |||
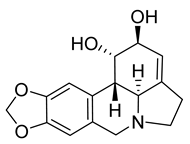 | Lycorine | Lycoris sp. Amaryllidaceae | HCoV-NL63 | CPE inhibition (EC50) | 0.47 μΜ | [84] |
| HCoV-OC43 | 0.15 μΜ | |||||
| MERS-CoV | 1.63 μΜ | |||||
| SARS-CoV | 169.8 μΜ | [89] | ||||
| SARS-CoV-2 | CPE inhibition (EC50) | <11 μΜ | [179] | |||
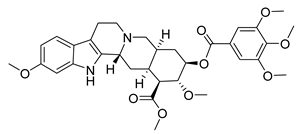 | Reserpine | Rauvolfia serpentina Apocynaceae | SARS-CoV | CPE inhibition (EC50) | 3.4 μΜ | [37] |
| SARS-CoV-2 | CPE inhibition (EC50) | <11 μΜ | [179] | |||
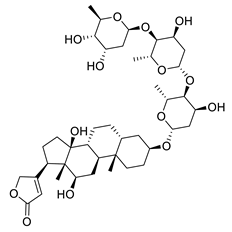 | Digoxin | Digitalis sp. Plantaginaceae | SARS-CoV-2 | CPE inhibition (EC50) | 0.1541 μΜ | [179] |
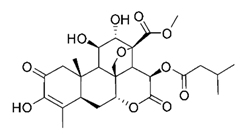 | Bruceine A | Brucea javanica Simaroubaceae | 0.011 μΜ | |||
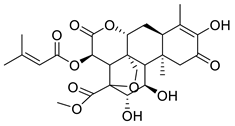 | Brusatol | Brucea javanica Simaroubaceae | 0.0492 μΜ | |||
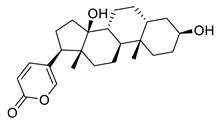 | Bufalin | Toad venom Bufonidae | 0.018 μΜ | |||
 | Bufotalin | Toad venom Bufonidae | 0.0259 μΜ | |||
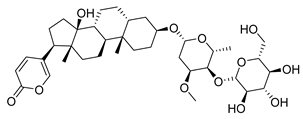 | Toad venom Bufonidae | 0.0657 μΜ | ||||
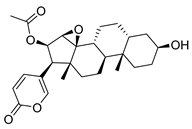 | Cinobufagin | Toad venom Bufonidae | 0.018 μΜ | |||
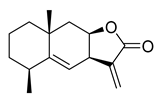 | Alantolactone | Inula helenium Asteraceae | 1.724 μΜ | |||
 | Isoalantolactone | Inula helenium Asteraceae | 1.483 μΜ | |||
 | Dehydrocostus lactone | Saussurea costus Asteraceae | 2.322 μΜ | |||
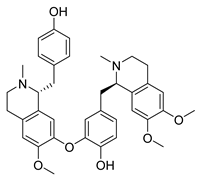 | Liensinine | Nelumbo nucifera Nelumbonaceae | 2.537 μΜ | |||
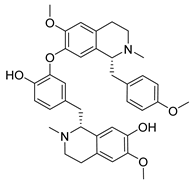 | Isoliensinine | Nelumbo nucifera Nelumbonaceae | 1.615 μΜ | |||
 | Momordinic | Bassia scoparia Amaranthaceae | 3.529 μΜ | |||
 | Oridonin | Isodon sp. Lamiaceae | 1.462 μΜ | |||
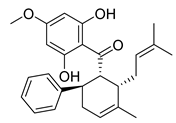 | Panduratin A | Boesenbergia rotunda Zingiberaceae | SARS-CoV-2 | CPE inhibition (EC50) | 0.81 μΜ | [180] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vougogiannopoulou, K.; Corona, A.; Tramontano, E.; Alexis, M.N.; Skaltsounis, A.-L. Natural and Nature-Derived Products Targeting Human Coronaviruses. Molecules 2021, 26, 448. https://doi.org/10.3390/molecules26020448
Vougogiannopoulou K, Corona A, Tramontano E, Alexis MN, Skaltsounis A-L. Natural and Nature-Derived Products Targeting Human Coronaviruses. Molecules. 2021; 26(2):448. https://doi.org/10.3390/molecules26020448
Chicago/Turabian StyleVougogiannopoulou, Konstantina, Angela Corona, Enzo Tramontano, Michael N. Alexis, and Alexios-Leandros Skaltsounis. 2021. "Natural and Nature-Derived Products Targeting Human Coronaviruses" Molecules 26, no. 2: 448. https://doi.org/10.3390/molecules26020448
APA StyleVougogiannopoulou, K., Corona, A., Tramontano, E., Alexis, M. N., & Skaltsounis, A.-L. (2021). Natural and Nature-Derived Products Targeting Human Coronaviruses. Molecules, 26(2), 448. https://doi.org/10.3390/molecules26020448