Pediatric Glioma: An Update of Diagnosis, Biology, and Treatment
Abstract
Simple Summary
Abstract
1. Introduction
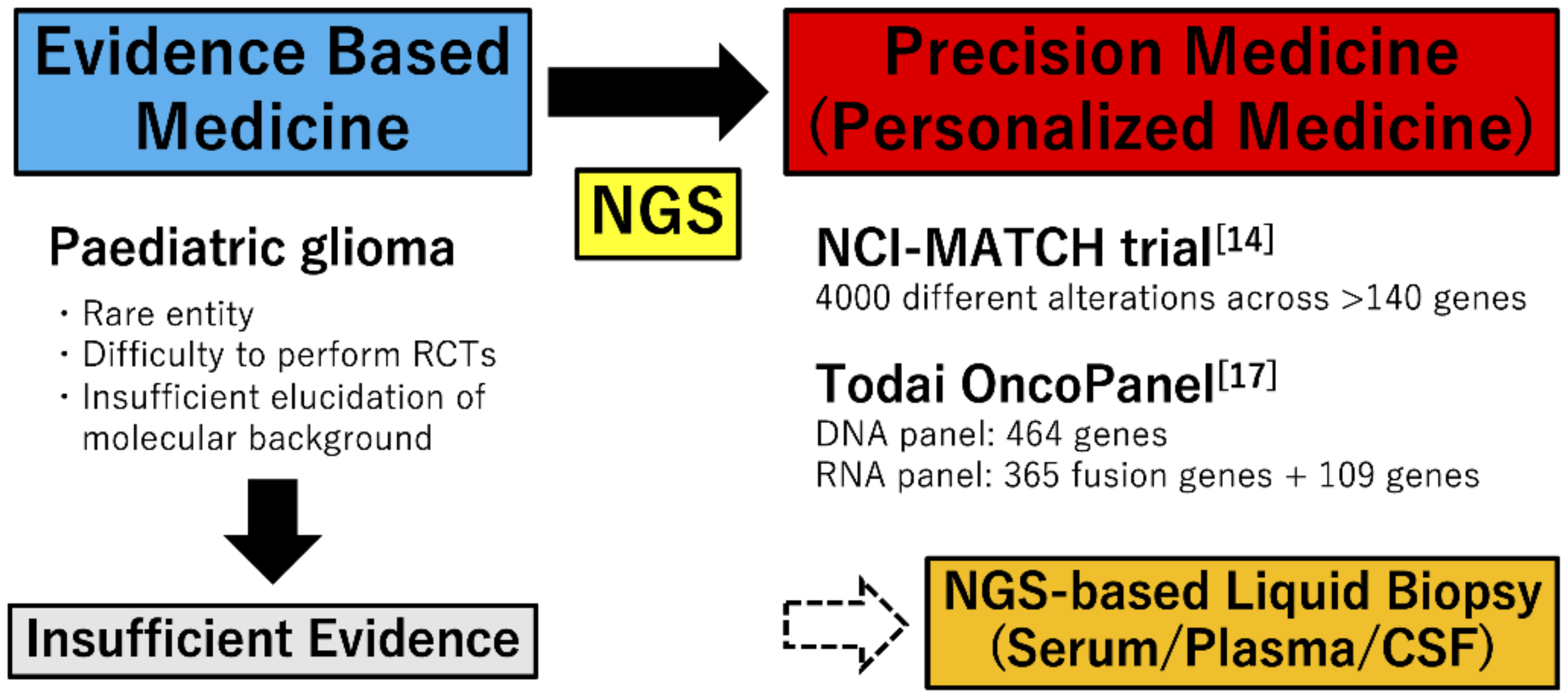
2. Diagnostic Approach for Pediatric Brain Tumors
2.1. Integrated Diagnosis with Histological and Genetical Classification
2.2. DNA Methylation Profiling
2.3. Molecular Approaches for Precision Therapy
3. Histopathologic Subtypes of Pediatric CNS Tumors
4. Low Grade Gliomas
4.1. Molecular Landscape in pLGG
4.1.1. RAS/Mitogen-Activated Protein Kinase (MAPK) Pathway
4.1.2. Non-RAS/MAPK Pathway
4.2. Integrated Diagnosis of Pediatric Diffuse Gliomas in cIMPACT-NOW Update 4
- Diffuse glioma, MYB-altered;
- Diffuse glioma, MYBL1-altered;
- Diffuse glioma, FGFR1 TKD-duplicated;
- Diffuse glioma, FGFR1-mutant;
- Diffuse glioma, BRAF V600E-mutant;
- Diffuse glioma, other MAPK pathway alteration.
4.3. Targeted Therapy for pLGG
5. High Grade Gliomas
5.1. Molecular Landscape in pHGG (Table 1)
5.2. Targeted Therapy for pHGG
6. Infantile Gliomas
- Group 1, hemispheric receptor tyrosine kinase-driven gliomas, including ALK, ROS1, NTRK, and MET fusions, which are enriched for high-grade glioma, and have an intermediate clinical outcome.
- Group 2, hemispheric RAS/MAPK-driven gliomas, which demonstrate excellent long-term survival with minimal post-surgery clinical intervention.
- Group 3, midline RAS/MAPK-driven gliomas, which are enriched for LGG, such as PA, with BRAF alternations, and have a poor outcome.
6.1. Fusion Genes in Infantile Gliomas (Table 1)
6.2. Treatment for Each Group in Infantile Gliomas
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ward, E.; DeSantis, C.; Robbins, A.; Kohler, B.; Jemal, A. Childhood and Adolescent Cancer Statistics, 2014. C.A. Cancer J. Clin. 2014, 64, 83–103. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, G.T.; Liu, Q.; Yasui, Y.; Huang, S.; Ness, K.K.; Leisenring, W.; Hudson, M.M.; Donaldson, S.S.; King, A.A.; Stovall, M.; et al. Long-Term Outcomes Among Adult Survivors of Childhood Central Nervous System Malignancies in the Childhood Cancer Survivor Study. J. Natl. Cancer Inst. 2009, 101, 946–958. [Google Scholar] [CrossRef] [PubMed]
- Vinchon, M.; Baroncini, M.; Leblond, P.; Delestret, I. Morbidity and Tumor-Related Mortality Among Adult Survivors of Pediatric Brain Tumors: A Review. Childs Nerv. Syst. 2011, 27, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Glod, J.; Rahme, G.J.; Kaur, H.; Raabe, E.; Hwang, E.I.; Israel, M.A. Pediatric Brain Tumors: Current Knowledge and Therapeutic Opportunities. J. Pediatr. Hematol. Oncol. 2016, 38, 249–260. [Google Scholar] [CrossRef]
- Lassaletta, A.; Zapotocky, M.; Bouffet, E.; Hawkins, C.; Tabori, U. An Integrative Molecular and Genomic Analysis of Pediatric Hemispheric Low-Grade Gliomas: An Update. Childs Nerv. Syst. 2016, 32, 1789–1797. [Google Scholar] [CrossRef]
- Park, S.H.; Won, J.; Kim, S.I.; Lee, Y.; Park, C.K.; Kim, S.K. Molecular Testing of Brain Tumor. J. Pathol. Transl. Med. 2017, 51, 205–223. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Louis, D.N.; Wesseling, P.; Aldape, K.; Brat, D.J.; Capper, D.; Cree, I.A.; Eberhart, C.; Figarella-Branger, D.; Fouladi, M.; Fuller, G.N.; et al. cIMPACT-NOW update 6: New Entity and Diagnostic Principle Recommendations of the cIMPACT-Utrecht Meeting on Future CNS Tumor Classification and Grading. Brain Pathol. 2020, 30, 844–856. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Burger, P.; Ellison, D.W.; Reifenberger, G.; von Deimling, A.; Aldape, K.; Brat, D.; Collins, V.P.; Eberhart, C.; et al. International Society of Neuropathology—Haarlem Consensus Guidelines for Nervous System Tumor Classification and Grading. Brain Pathol. 2014, 24, 429–435. [Google Scholar] [CrossRef]
- Louis, D.N.; Wesseling, P.; Brandner, S.; Brat, D.J.; Ellison, D.W.; Giangaspero, F.; Hattab, E.M.; Hawkins, C.; Judge, M.J.; Kleinschmidt-DeMasters, B.; et al. Data Sets for the Reporting of Tumors of the Central Nervous System. Arch. Pathol. Lab. Med. 2019, 144, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA Methylation-Based Classification of Central Nervous System Tumours. Nature 2018, 255, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Liu, A.P.Y.; Orr, B.A.; Northcott, P.A.; Robinson, G.W. Advances in the Classification of Pediatric Brain Tumors Through DNA Methylation Profiling: From Research Tool to Frontline Diagnostic. Cancer 2018, 124, 4168–4180. [Google Scholar] [CrossRef]
- Mack, S.C.; Northcott, P.A. Genomic Analysis of Childhood Brain Tumors: Methods for Genome-Wide Discovery and Precision Medicine Become Mainstream. J. Clin. Oncol. 2017, 35, 2346–2354. [Google Scholar] [CrossRef] [PubMed]
- Seibel, N.L.; Janeway, K.; Allen, C.E.; Chi, S.N.; Cho, Y.J.; Glade Bender, J.L.; Kim, A.; Laetsch, T.W.; Irwin, M.S.; Takebe, N.; et al. Pediatric Oncology Enters an Era of Precision Medicine. Curr. Probl. Cancer 2017, 41, 194–200. [Google Scholar] [CrossRef]
- Fujioka, Y.; Hata, N.; Akagi, Y.; Kuga, D.; Hatae, R.; Sangatsuda, Y.; Michiwaki, Y.; Amemiya, T.; Takigawa, K.; Funakoshi, Y.; et al. Molecular diagnosis of diffuse glioma using a chip-based digital PCR system to analyze IDH, TERT, and H3 mutations in the cerebrospinal fluid. J. Neurooncol. 2021. [Google Scholar] [CrossRef]
- Miller, A.M.; Shah, R.H.; Pentsova, E.I.; Pourmaleki, M.; Briggs, S.; Distefano, N.; Zheng, Y.; Skakodub, A.; Mehta, S.A.; Campos, C.; et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature 2019, 565, 654–658. [Google Scholar] [CrossRef] [PubMed]
- Kohsaka, S.; Tatsuno, K.; Ueno, T.; Nagano, M.; Shinozaki-Ushiku, A.; Ushiku, T.; Takai, D.; Ikegami, M.; Kobayashi, H.; Kage, H.; et al. Comprehensive Assay for the Molecular Profiling of Cancer by Target Enrichment from Formalin-Fixed Paraffin-Embedded Specimens. Cancer Sci. 2019, 110, 1464–1479. [Google Scholar] [CrossRef]
- About Us—Accelerate Platform. Available online: https://www.accelerate-platform.org/about-us/ (accessed on 5 February 2021).
- Ostrom, Q.T.; Gittleman, H.; Farah, P.; Ondracek, A.; Chen, Y.; Wolinsky, Y. CBTRUS statistical report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2006–2010. Neuro Oncol. 2013, 15 (Suppl. 2), ii1–ii56. [Google Scholar] [CrossRef]
- Sturm, D.; Pfister, S.M.; Jones, D.T.W. Pediatric Gliomas: Current Concepts on Diagnosis, Biology, and Clinical Management. J. Clin. Oncol. 2017, 35, 2370–2377. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; De Blank, P.M.; Kruchko, C.; Petersen, C.M.; Liao, P.; Finlay, J.L.; Stearns, D.S.; Wolff, J.E.; Wolinsky, Y.; Letterio, J.J.; et al. Alex’s Lemonade Stand Foundation Infant and Childhood Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2007–2011. Neuro. Oncol. 2015, 16 (Suppl. 10), x1–x36. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Gittleman, H.; Truitt, G.; Boscia, A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro. Oncol. 2018, 20 (Suppl. 4), iv1–iv86. [Google Scholar] [CrossRef]
- Ryall, S.; Tabori, U.; Hawkins, C. Pediatric Low-Grade Glioma in the Era of Molecular Diagnostics. Acta Neuropathol. Commun. 2020, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Wisoff, J.H.; Sanford, R.A.; Heier, L.A.; Sposto, R.; Burger, P.C.; Yates, A.J.; Holmes, E.J.; Kun, L.E. Primary Neurosurgery for Pediatric Low-Grade Gliomas: A Prospective Multi-Institutional Study from the Children’s Oncology Group. Neurosurgery 2011, 68, 1548–1555. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, G.T.; Conklin, H.M.; Huang, S.; Srivastava, D.; Sanford, R.; Ellison, D.W.; Merchant, T.E.; Hudson, M.M.; Hoehn, M.E.; Robison, L.L. Survival and Long-Term Health and Cognitive Outcomes After Low-Grade Glioma. Neuro Oncol. 2011, 13, 223–234. [Google Scholar] [CrossRef]
- Erkal, H.S.; Serin, M.; Çakmak, A. Management of Optic Pathway and Chiasmatic-Hypothalamic Gliomas in Children with Radiation Therapy. Radiother. Oncol. 1997, 45, 11–15. [Google Scholar] [CrossRef]
- Merchant, T.E.; Conklin, H.M.; Wu, S.; Lustig, R.H.; Xiong, X. Late Effects of Conformal Radiation Therapy for Pediatric Patients with Low-Grade Glioma: Prospective Evaluation of Cognitive, Endocrine, and Hearing Deficits. J. Clin. Oncol. 2009, 27, 3691–3697. [Google Scholar] [CrossRef]
- Krishnatry, R.; Zhukova, N.; Guerreiro Stucklin, A.S.; Pole, J.D.; Mistry, M.; Fried, I.; Ramaswamy, V.; Bartels, U.; Huang, A.; Laperriere, N.; et al. Clinical and Treatment Factors Determining Long-Term Outcomes for Adult Survivors of Childhood Low-Grade Glioma: A Population-Based Study. Cancer 2016, 122, 1261–1269. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Gittleman, H.; Xu, J.; Kromer, C.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2009–2013. Neuro. Oncol. 2016, 18 (Suppl. 5), v1–v75. [Google Scholar] [CrossRef]
- Collins, V.P.; Jones, D.T.W.; Giannini, C. Pilocytic Astrocytoma: Pathology, Molecular Mechanisms and Markers. Acta Neuropathol. 2015, 129, 775–788. [Google Scholar] [CrossRef]
- Jones, D.T.W.; Gronych, J.; Lichter, P.; Witt, O.; Pfister, S.M. MAPK Pathway Activation in Pilocytic Astrocytoma. Cell. Mol. Life Sci. 2012, 69, 1799–1811. [Google Scholar] [CrossRef]
- Northcott, P.A.; Pfister, S.M.; Jones, D.T.W. Next-Generation (Epi)Genetic Drivers of Childhood Brain Tumours and the Outlook for Targeted Therapies. Lancet Oncol. 2015, 16, e293–e302. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, G.; Miller, C.P.; Tatevossian, R.G.; Dalton, J.D.; Tang, B.; Orisme, W.; Punchihewa, C.; Parker, M.; Qaddoumi, I.; et al. Whole-Genome Sequencing Identifies Genetic Alterations in Pediatric Low-Grade Gliomas. Nat. Genet. 2013, 45, 602–612. [Google Scholar]
- Lassaletta, A.; Zapotocky, M.; Mistry, M.; Ramaswamy, V.; Honnorat, M.; Krishnatry, R.; Guerreiro Stucklin, A.; Zhukova, N.; Arnoldo, A.; Ryall, S.; et al. Therapeutic and Prognostic Implications of BRAF V600E in Pediatric Low-Grade Gliomas. J. Clin. Oncol. 2017, 35, 2934–2941. [Google Scholar] [CrossRef]
- Hatae, R.; Hata, N.; Suzuki, S.O.; Yoshimoto, K.; Kuga, D.; Murata, H.; Akagi, Y.; Sangatsuda, Y.; Iwaki, T.; Mizoguchi, M.; et al. A Comprehensive Analysis Identifies BRAF Hotspot Mutations Associated with Gliomas with Peculiar Epithelial Morphology. Neuropathology 2017, 37, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Goetz, R.; Mohammadi, M. Exploring Mechanisms of FGF Signalling Through the Lens of Structural Biology. Nat. Rev. Mol. Cell Biol. 2013, 14, 166–180. [Google Scholar] [CrossRef]
- Dai, S.; Zhou, Z.; Chen, Z.; Xu, G.; Chen, Y. Fibroblast Growth Factor Receptors (FGFRs): Structures and Small Molecule Inhibitors. Cells 2019, 8, 614. [Google Scholar] [CrossRef] [PubMed]
- Lehtinen, B.; Raita, A.; Kesseli, J.; Annala, M.; Nordfors, K.; Yli-Harja, O.; Zhang, W.; Visakorpi, T.; Nykter, M.; Haapasalo, H.; et al. Clinical Association Analysis of Ependymomas and Pilocytic Astrocytomas Reveals Elevated FGFR3 and FGFR1 Expression in Aggressive Ependymomas. BMC Cancer 2017, 17, 310. [Google Scholar] [CrossRef]
- Holzhauser, S.; Lukoseviciute, M.; Andonova, T.; Ursu, R.G.; Dalianis, T.; Wickström, M.; Kostopoulou, O.N. Targeting Fibroblast Growth Factor Receptor (FGFR) and Phosphoinositide 3-Kinase (PI3K) Signaling Pathways in Medulloblastoma Cell Lines. Anticancer Res. 2020, 40, 53–66. [Google Scholar] [CrossRef]
- Jones, D.T.W.; Hutter, B.; Jäger, N.; Korshunov, A.; Kool, M.; Warnatz, H.J.; Zichner, T.; Lambert, S.R.; Ryzhova, M.; Quang, D.A.K.; et al. Recurrent Somatic Alterations of FGFR1 and NTRK2 in Pilocytic Astrocytoma. Nat. Genet. 2013, 45, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Qaddoumi, I.; Orisme, W.; Wen, J.; Santiago, T.; Gupta, K.; Dalton, J.D.; Tang, B.; Haupfear, K.; Punchihewa, C.; Easton, J.; et al. Genetic Alterations in Uncommon Low-Grade Neuroepithelial Tumors: BRAF, FGFR1, and MYB Mutations Occur at High Frequency and Align with Morphology. Acta Neuropathol. 2016, 131, 833–845. [Google Scholar] [CrossRef]
- Friedman, J.M. Epidemiology of Neurofibromatosis type 1. Am. J. Med. Genet. Semin. Med. Genet. 1999, 89, 1–6. [Google Scholar] [CrossRef]
- Rasmussen, S.A.; Friedman, J.M. NF1 Gene and Neurofibromatosis 1. Am. J. Epidemiol. 2000, 151, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, G.; Lafforgue, M.P.; Lion-François, L.; Kemlin, I.; Rodriguez, D.; Castelnau, P.; Meyer, P.; Rivier, F.; Barbarot, S.; Chaix, Y.; et al. Systematic MRI in NF1 Children Under Six Years of Age for the Diagnosis of Optic Pathway Gliomas. Study and Outcome of a French Cohort. Eur. J. Paediatr. Neurol. 2016, 20, 275–281. [Google Scholar] [CrossRef]
- Uusitalo, E.; Rantanen, M.; Kallionpää, R.A.; Pöyhönen, M.; Leppävirta, J.; Ylä-Outinen, H.; Riccardi, V.M.; Pukkala, E.; Pitkäniemi, J.; Peltonen, S.; et al. Distinctive Cancer Associations in Patients with Neurofibromatosis type 1. J. Clin. Oncol. 2016, 34, 1978–1986. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.J.; Loguidice, M.; Gutmann, D.H.; Listernick, R.; Ferner, R.E.; Ullrich, N.J.; Packer, R.J.; Tabori, U.; Hoffman, R.O.; Ardern-Holmes, S.L.; et al. Visual Outcomes in Children with Neurofibromatosis type 1-Associated Optic Pathway Glioma Following Chemotherapy: A Multicenter Retrospective Analysis. Neuro. Oncol. 2012, 14, 790–797. [Google Scholar] [CrossRef]
- Pattabiraman, D.R.; Gonda, T.J. Role and Potential for Therapeutic Targeting of MYB in Leukemia. Leukemia 2013, 27, 269–277. [Google Scholar] [CrossRef]
- Zhou, Y.; Ness, S.A. Myb Proteins: Angels and Demons in Normal and Transformed Cells. Front. Biosci. 2011, 16, 1109–1131. [Google Scholar] [CrossRef] [PubMed]
- Bandopadhayay, P.; Ramkissoon, L.A.; Jain, P.; Bergthold, G.; Wala, J.; Zeid, R.; Schumacher, S.E.; Urbanski, L.; O’Rourke, R.; Gibson, W.J.; et al. MYB-QKI Rearrangements in Angiocentric Glioma Drive Tumorigenicity Through a Tripartite Mechanism. Nat. Genet. 2016, 48, 273–282. [Google Scholar] [CrossRef]
- Ramkissoon, L.A.; Horowitz, P.M.; Craig, J.M.; Ramkissoon, S.H.; Rich, B.E.; Schumacher, S.E.; McKenna, A.; Lawrence, M.S.; Bergthold, G.; Brastianos, P.K.; et al. Genomic Analysis of Diffuse Pediatric Low-Grade Gliomas Identifies Recurrent Oncogenic Truncating Rearrangements in the Transcription Factor MYBL1. Proc. Natl. Acad. Sci. USA 2013, 110, 8188–8193. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.; Harreld, J.H.; Tinkle, C.L.; Moreira, D.C.; Li, X.; Acharya, S.; Qaddoumi, I.; Ellison, D.W. A Single-Center Study of the Clinicopathologic Correlates of Gliomas with a MYB or MYBL1 Alteration. Acta Neuropathol. 2019, 138, 1091–1092. [Google Scholar] [CrossRef]
- Appay, R.; Dehais, C.; Maurage, C.A.; Alentorn, A.; Carpentier, C.; Colin, C.; Ducray, F.; Escande, F.; Idbaih, A.; Kamoun, A.; et al. CDKN2A Homozygous Deletion Is a Strong Adverse Prognosis Factor in Diffuse Malignant IDH-Mutant Gliomas. Neuro Oncol. 2019, 21, 1519–1528. [Google Scholar] [CrossRef]
- Reis, G.F.; Pekmezci, M.; Hansen, H.M.; Rice, T.; Marshall, R.E.; Molinaro, A.M.; Phillips, J.J.; Vogel, H.; Wiencke, J.K.; Wrensch, M.R.; et al. CDKN2A Loss Is Associated with Shortened Overall Survival in Lower-Grade (World Health Organization Grades II–III) Astrocytomas. J. Neuropathol. Exp. Neurol. 2015, 74, 442–452. [Google Scholar] [CrossRef] [PubMed]
- Brat, D.J.; Aldape, K.; Colman, H.; Figrarella-Branger, D.; Fuller, G.N.; Giannini, C.; Holland, E.C.; Jenkins, R.B.; Kleinschmidt-DeMasters, B.; Komori, T.; et al. cIMPACT-NOW update 5: Recommended Grading Criteria and Terminologies for IDH-Mutant Astrocytomas. Acta Neuropathol. 2020, 139, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Rudra, S.; Campian, J.L.; Dahiya, S.; Dunn, G.P.; Johanns, T.; Goldstein, M.; Kim, A.H.; Huang, J. Prognostic Impact of CDKN2A/B Deletion, Tert Mutation, and EGFR Amplification on Histological and Molecular IDH-Wildtype Glioblastoma. Neuro. Oncol. Adv. 2020, 2, vdaa126. [Google Scholar] [CrossRef]
- Mistry, M.; Zhukova, N.; Merico, D.; Rakopoulos, P.; Krishnatry, R.; Shago, M.; Stavropoulos, J.; Alon, N.; Pole, J.D.; Ray, P.N.; et al. BRAF Mutation and CDKN2A Deletion Define a Clinically Distinct Subgroup of Childhood Secondary High-Grade Glioma. J. Clin. Oncol. 2015, 33, 1015–1022. [Google Scholar] [CrossRef]
- Ellison, D.W.; Hawkins, C.; Jones, D.T.W.; Onar-Thomas, A.; Pfister, S.M.; Reifenberger, G.; Louis, D.N. cIMPACT-NOW update 4: Diffuse Gliomas Characterized by MYB, MYBL1, or FGFR1 Alterations or BRAF V600E Mutation. Acta Neuropathol. 2019, 137, 683–687. [Google Scholar] [CrossRef] [PubMed]
- Hofer, S.; Berthod, G.; Riklin, C.; Rushing, E.; Feilchenfeldt, J. BRAF V600E Mutation: A Treatable Driver Mutation in Pleomorphic Xanthoastrocytoma (PXA). Acta Oncol. 2016, 55, 122–123. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Q.; Ruland, S.; Leboeuf, N.R.; Wen, P.Y.; Santagata, S. Successful Treatment of a Progressive BRAF V600E-Mutated Anaplastic Pleomorphic Xanthoastrocytoma with Vemurafenib Monotherapy. J. Clin. Oncol. 2016, 34, e87–e89. [Google Scholar] [CrossRef]
- Usubalieva, A.; Pierson, C.R.; Kavran, C.A.; Huntoon, K.; Kryvenko, O.N.; Mayer, T.G.; Zhao, W.; Rock, J.; Ammirati, M.; Puduvalli, V.K.; et al. Primary Meningeal Pleomorphic Xanthoastrocytoma with Anaplastic Features: A Report of 2 Cases, One with BRAFV600E Mutation and Clinical Response to the BRAF Inhibitor Dabrafenib. J. Neuropathol. Exp. Neurol. 2015, 74, 960–969. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brown, N.F.; Carter, T.; Kitchen, N.; Mulholland, P. Dabrafenib and Trametinib in BRAFV600E Mutated Glioma. CNS Oncol. 2017, 6, 291–296. [Google Scholar] [CrossRef]
- Hargrave, D.R.; Bouffet, E.; Tabori, U.; Broniscer, A.; Cohen, K.J.; Hansford, J.R.; Geoerger, B.; Hingorani, P.; Dunkel, I.J.; Russo, M.W.; et al. Efficacy and Safety of Dabrafenib in Pediatric Patients with BRAF V600 Mutation–Positive Relapsed or Refractory Low-Grade Glioma: Results from a phase I/IIa Study. Clin. Cancer Res. 2019, 25, 7303–7311. [Google Scholar] [CrossRef]
- Solit, D.B.; Rosen, N. Resistance to BRAF Inhibition in Melanomas. N. Engl. J. Med. 2011, 364, 772–774. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Karaszewska, B.; Schachter, J.; Rutkowski, P.; Mackiewicz, A.; Stroiakovski, D.; Lichinitser, M.; Dummer, R.; Grange, F.; Mortier, L.; et al. Improved Overall Survival in Melanoma with Combined Dabrafenib and Trametinib. N. Engl. J. Med. 2015, 72, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Stroyakovskiy, D.; Gogas, H.; Levchenko, E.; De Braud, F.; Larkin, J.; Garbe, C.; Jouary, T.; Hauschild, A.; Grob, J.J.; et al. Dabrafenib and Trametinib Versus Dabrafenib and Placebo for Val600 BRAF-Mutant Melanoma: A Multicentre, Double-Blind, phase 3 Randomised Controlled Trial. Lancet 2015, 386, 444–451. [Google Scholar] [CrossRef]
- Brastianos, P.K.; Shankar, G.M.; Gill, C.M.; Taylor-Weiner, A.; Nayyar, N.; Panka, D.J.; Sullivan, R.J.; Frederick, D.T.; Abedalthagafi, M.; Jones, P.S.; et al. Dramatic Response of BRAF V600E Mutant Papillary Craniopharyngioma to Targeted Therapy. J. Natl. Cancer Inst. 2016, 108, djv310. [Google Scholar] [CrossRef]
- Migliorini, D.; Aguiar, D.; Vargas, M.I.; Lobrinus, A.; Dietrich, P.Y. BRAF/MEK Double Blockade in Refractory Anaplastic Pleomorphic Xanthoastrocytoma. Neurology 2017, 88, 1291–1293. [Google Scholar] [CrossRef]
- Fangusaro, J.; Onar-Thomas, A.; Young Poussaint, T.; Wu, S.; Ligon, A.H.; Lindeman, N.; Banerjee, A.; Packer, R.J.; Kilburn, L.B.; Goldman, S.; et al. Selumetinib in Paediatric Patients with BRAF-Aberrant or Neurofibromatosis type 1-Associated Recurrent, Refractory, or Progressive Low-Grade Glioma: A Multicentre, phase 2 Trial. Lancet Oncol. 2019, 20, 1011–1022. [Google Scholar] [CrossRef]
- Wang, H.; Long-Boyle, J.; Winger, B.A.; Nicolaides, T.; Mueller, S.; Prados, M.; Ivaturi, V. Population Pharmacokinetics of Vemurafenib in Children with Recurrent/Refractory BRAF Gene V600E-Mutant Astrocytomas. J. Clin. Pharmacol. 2020, 60, 1209–1219. [Google Scholar] [CrossRef]
- Nicolaides, T.; Nazemi, K.J.; Crawford, J.; Kilburn, L.; Minturn, J.; Gajjar, A.; Gauvain, K.; Leary, S.; Dhall, G.; Aboian, M.; et al. Phase I Study of Vemurafenib in Children with Recurrent or Progressive BRAFV600E Mutant Brain Tumors: Pacific Pediatric Neuro-Oncology Consortium Study (PNOC-002). Oncotarget 2020, 11, 1942–1952. [Google Scholar] [CrossRef]
- Gavine, P.R.; Mooney, L.; Kilgour, E.; Thomas, A.P.; Al-Kadhimi, K.; Beck, S.; Rooney, C.; Coleman, T.; Baker, D.; Mellor, M.J.; et al. AZD4547: An Orally Bioavailable, Potent, and Selective Inhibitor of the Fibroblast Growth Factor Receptor Tyrosine Kinase Family. Cancer Res. 2012, 72, 2045–2056. [Google Scholar] [CrossRef] [PubMed]
- Chae, Y.K.; Hong, F.; Vaklavas, C.; Cheng, H.H.; Hammerman, P.; Mitchell, E.P.; Zwiebel, J.A.; Ivy, S.P.; Gray, R.J.; Li, S.; et al. Phase II Study of AZD4547 in Patients with Tumors Harboring Aberrations in the FGFR Pathway: Results from the NCI-MATCH Trial (EAY131) Subprotocol W. J. Clin. Oncol. 2020, 38, 2407–2417. [Google Scholar] [CrossRef] [PubMed]
- Wetmore, C.; Daryani, V.M.; Billups, C.A.; Boyett, J.M.; Leary, S.; Tanos, R.; Goldsmith, K.C.; Stewart, C.F.; Blaney, S.M.; Gajjar, A. Phase II Evaluation of Sunitinib in the Treatment of Recurrent or Refractory High-Grade Glioma or Ependymoma in Children: A Children’s Oncology Group Study ACNS1021. Cancer Med. 2016, 5, 1416–1424. [Google Scholar] [CrossRef]
- Becher, O.J.; Gilheeney, S.W.; Khakoo, Y.; Lyden, D.C.; Haque, S.; De Braganca, K.C.; Kolesar, J.M.; Huse, J.T.; Modak, S.; Wexler, L.H.; et al. A Phase I Study of Perifosine with Temsirolimus for Recurrent Pediatric Solid Tumors. Pediatr. Blood Cancer 2017, 64. [Google Scholar] [CrossRef] [PubMed]
- Broniscer, A.; Jia, S.; Mandrell, B.; Hamideh, D.; Huang, J.; Onar-Thomas, A.; Gajjar, A.; Raimondi, S.C.; Tatevossian, R.G.; Stewart, C.F. Phase 1 Trial, Pharmacokinetics, and Pharmacodynamics of Dasatinib Combined with Crizotinib in Children with Recurrent or Progressive High-Grade and Diffuse Intrinsic Pontine Glioma. Pediatr. Blood Cancer 2018, 65, e27035. [Google Scholar] [CrossRef] [PubMed]
- Chi, A.S.; Tarapore, R.S.; Hall, M.D.; Shonka, N.; Gardner, S.; Umemura, Y.; Sumrall, A.; Khatib, Z.; Mueller, S.; Kline, C.; et al. Pediatric and Adult H3 K27M-Mutant Diffuse Midline Glioma Treated with the Selective DRD2 Antagonist ONC201. J. Neuro Oncol. 2019, 145, 97–105. [Google Scholar] [CrossRef]
- Bondy, M.L.; Scheurer, M.E.; Malmer, B.; Barnholtz-Sloan, J.S.; Davis, F.G.; Il’yasova, D.; Kruchko, C.; McCarthy, B.J.; Rajaraman, P.; Schwartzbaum, J.A.; et al. Brain Tumor Epidemiology: Consensus from the Brain Tumor Epidemiology Consortium. Cancer 2008, 113, 1953–1968. [Google Scholar] [CrossRef]
- McCrea, H.J.; Bander, E.D.; Venn, R.A.; Reiner, A.S.; Iorgulescu, J.B.; Puchi, L.A.; Schaefer, P.M.; Cederquist, G.; Greenfield, J.P. Sex, Age, Anatomic Location, and Extent of Resection Influence Outcomes in Children with High-Grade Glioma. Neurosurgery 2015, 77, 443–453. [Google Scholar] [CrossRef]
- Cohen, K.J.; Pollack, I.F.; Zhou, T.; Buxton, A.; Holmes, E.J.; Burger, P.C.; Brat, D.J.; Rosenblum, M.K.; Hamilton, R.L.; Lavey, R.S.; et al. Temozolomide in the Treatment of High-Grade Gliomas in Children: A Report from the Children’s Oncology Group. Neuro Oncol. 2011, 13, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Buczkowicz, P.; Bartels, U.; Bouffet, E.; Becher, O.; Hawkins, C. Histopathological Spectrum of Paediatric Diffuse Intrinsic Pontine Glioma: Diagnostic and Therapeutic Implications. Acta Neuropathol. 2014, 128, 573–581. [Google Scholar] [CrossRef]
- Okuda, T.; Hata, N.; Suzuki, S.O.; Yoshimoto, K.; Arimura, K.; Amemiya, T.; Akagi, Y.; Kuga, D.; Oba, U.; Koga, Y.; et al. Pediatric Ganglioglioma with an H3 K27M Mutation Arising from the Cervical Spinal Cord. Neuropathology 2018. [Google Scholar] [CrossRef] [PubMed]
- Mackay, A.; Burford, A.; Carvalho, D.; Izquierdo, E.; Fazal-Salom, J.; Taylor, K.R.; Bjerke, L.; Clarke, M.; Vinci, M.; Nandhabalan, M.; et al. Integrated Molecular Meta-Analysis of 1,000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell 2017, 32, 520–537.e5. [Google Scholar] [CrossRef]
- D’Angelo, F.; Ceccarelli, M.; Tala; Garofano, L.; Zhang, J.; Frattini, V.; Caruso, F.P.; Lewis, G.; Alfaro, K.D.; Bauchet, L.; et al. The molecular landscape of glioma in patients with Neurofibromatosis 1. Nat. Med. 2019, 25, 176–187. [Google Scholar]
- Korshunov, A.; Capper, D.; Reuss, D.; Schrimpf, D.; Ryzhova, M.; Hovestadt, V.; Sturm, D.; Meyer, J.; Jones, C.; Zheludkova, O.; et al. Histologically Distinct Neuroepithelial Tumors with Histone 3 G34 Mutation Are Molecularly Similar and Comprise a Single Nosologic Entity. Acta Neuropathol. 2016, 131, 137–146. [Google Scholar] [CrossRef]
- Schwartzentruber, J.; Korshunov, A.; Liu, X.Y.; Jones, D.T.W.; Pfaff, E.; Jacob, K.; Sturm, D.; Fontebasso, A.M.; Quang, D.A.; Tönjes, M.; et al. Driver Mutations in Histone H3.3 and Chromatin Remodelling Genes in Paediatric Glioblastoma. Nature 2012, 482, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Broniscer, A.; McEachron, T.A.; Lu, C.; Paugh, B.S.; Becksfort, J.; Qu, C.; Ding, L.; Huether, R.; Parker, M.; et al. Somatic Histone H3 Alterations in Pediatric Diffuse Intrinsic Pontine Gliomas and Non-Brainstem Glioblastomas. Nat. Genet. 2012, 44, 251–253. [Google Scholar] [PubMed]
- Khuong-Quang, D.A.; Buczkowicz, P.; Rakopoulos, P.; Liu, X.Y.; Fontebasso, A.M.; Bouffet, E.; Bartels, U.; Albrecht, S.; Schwartzentruber, J.; Letourneau, L.; et al. K27M Mutation in Histone H3.3 Defines Clinically and Biologically Distinct Subgroups of Pediatric Diffuse Intrinsic Pontine Gliomas. Acta Neuropathol. 2012, 124, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Sturm, D.; Witt, H.; Hovestadt, V.; Khuong-Quang, D.A.; Jones, D.W.; Konermann, C.; Pfaff, E.; Tönjes, M.; Sill, M.; Bender, S.; et al. Hotspot Mutations in H3F3A and IDH1 Define Distinct Epigenetic and Biological Subgroups of Glioblastoma. Cancer Cell 2012, 22, 425–437. [Google Scholar] [CrossRef]
- Wu, G.; Diaz, A.K.; Paugh, B.S.; Rankin, S.L.; Ju, B.; Li, Y.; Zhu, X.; Qu, C.; Chen, X.; Zhang, J.; et al. The Genomic Landscape of Diffuse Intrinsic Pontine Glioma and Pediatric Non-Brainstem High-Grade Glioma. Nat. Genet. 2014, 46, 444–450. [Google Scholar] [PubMed]
- Bender, S.; Tang, Y.; Lindroth, A.M.; Hovestadt, V.; Jones, D.W.; Kool, M.; Zapatka, M.; Northcott, P.; Sturm, D.; Wang, W.; et al. Reduced H3K27me3 and DNA Hypomethylation Are Major Drivers of Gene Expression in K27M Mutant Pediatric High-Grade Gliomas. Cancer Cell 2013, 24, 660–672. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.M.; Fang, D.; Gan, H.; Hashizume, R.; Yu, C.; Schroeder, M.; Gupta, N.; Mueller, S.; James, C.D.; Jenkins, R.; et al. The Histone H3.3K27M Mutation in Pediatric Glioma Reprograms H3K27 Methylation and Gene Expression. Genes Dev. 2013, 27, 985–990. [Google Scholar] [CrossRef]
- Stafford, J.M.; Lee, C.H.; Voigt, P.; Descostes, N.; Saldaña-Meyer, R.; Yu, J.R.; Leroy, G.; Oksuz, O.; Chapman, J.R.; Suarez, F.; et al. Multiple Modes of PRC2 Inhibition Elicit Global Chromatin Alterations in H3K27M Pediatric Glioma. Sci. Adv. 2018, 4, eaau5935. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Sweha, S.R.; Pratt, D.; Tamrazi, B.; Panwalkar, P.; Banda, A.; Bayliss, J.; Hawes, D.; Yang, F.; Lee, H.J.; et al. Integrated Metabolic and Epigenomic Reprogramming by H3K27M Mutations in Diffuse Intrinsic Pontine Gliomas. Cancer Cell 2020, 38, 334–349.e9. [Google Scholar] [CrossRef]
- Amary, F.; Berisha, F.; Ye, H.; Gupta, M.; Gutteridge, A.; Baumhoer, D.; Gibbons, R.; Tirabosco, R.; O’Donnell, P.; Flanagan, A. H3F3A (Histone 3.3) G34W Immunohistochemistry: A Reliable Marker Defining Benign and Malignant Giant Cell Tumor of Bone. Am. J. Surg. Pathol. 2017, 41, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Neumann, J.E.; Dorostkar, M.M.; Korshunov, A.; Mawrin, C.; Koch, A.; Giese, A.; Schüller, U. Distinct Histomorphology in Molecular Subgroups of Glioblastomas in Young Patients. J. Neuropathol. Exp. Neurol. 2016, 75, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, K.; Hatae, R.; Sangatsuda, Y.; Suzuki, S.O.; Hata, N.; Akagi, Y.; Kuga, D.; Hideki, M.; Yamashita, K.; Togao, O.; et al. Prevalence and Clinicopathological Features of H3.3 G34-Mutant High-Grade Gliomas: A Retrospective Study of 411 Consecutive Glioma Cases in a Single Institution. Brain Tumor Pathol. 2017, 34, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Sangatsuda, Y.; Miura, F.; Araki, H.; Mizoguchi, M.; Hata, N.; Kuga, D.; Hatae, R.; Akagi, Y.; Amemiya, T.; Fujioka, Y.; et al. Base-Resolution Methylomes of Gliomas Bearing Histone H3.3 Mutations Reveal a G34 Mutant-Specific Signature Shared with Bone Tumors. Sci. Rep. 2020, 10, 16162. [Google Scholar] [CrossRef] [PubMed]
- Lewis, P.W.; Müller, M.M.; Koletsky, M.S.; Cordero, F.; Lin, S.; Banaszynski, L.A.; Garcia, B.A.; Muir, T.W.; Becher, O.J.; Allis, C.D. Inhibition of PRC2 Activity by a Gain-of-Function H3 Mutation Found in Pediatric Glioblastoma. Science 2013, 340, 857–861. [Google Scholar] [CrossRef]
- Mohammad, F.; Weissmann, S.; Leblanc, B.; Pandey, D.P.; Højfeldt, J.W.; Comet, I.; Zheng, C.; Johansen, J.V.; Rapin, N.; Porse, B.T.; et al. EZH2 Is a Potential Therapeutic Target for H3K27M-Mutant Pediatric Gliomas. Nat. Med. 2017, 23, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Pfaff, E.; El Damaty, A.; Balasubramanian, G.P.; Blattner-Johnson, M.; Worst, B.C.; Stark, S.; Witt, H.; Pajtler, K.W.; van Tilburg, C.M.; Witt, R.; et al. Brainstem Biopsy in Pediatric Diffuse Intrinsic Pontine Glioma in the Era of Precision Medicine: The INFORM Study Experience. Eur. J. Cancer 2019, 114, 27–35. [Google Scholar] [CrossRef]
- Mueller, S.; Jain, P.; Liang, W.S.; Kilburn, L.; Kline, C.; Gupta, N.; Panditharatna, E.; Magge, S.N.; Zhang, B.; Zhu, Y.; et al. A Pilot Precision Medicine Trial for Children with Diffuse Intrinsic Pontine Glioma—PNOC003: A Report from the Pacific Pediatric Neuro-Oncology Consortium. Int. J. Cancer 2019, 145, 1889–1901. [Google Scholar] [CrossRef]
- Li, J.; Zhu, S.; Kozono, D.; Ng, K.; Futalan, D.; Shen, Y.; Akers, J.C.; Steed, T.; Kushwaha, D.; Schlabach, M.; et al. Genome-Wide shRNA Screen Revealed Integrated Mitogenic Signaling Between Dopamine Receptor D2 (DRD2) and Epidermal Growth Factor Receptor (EGFR) in Glioblastoma. Oncotarget 2014, 5, 882–893. [Google Scholar] [CrossRef]
- Hall, M.D.; Odia, Y.; Allen, J.E.; Tarapore, R.; Khatib, Z.; Niazi, T.N.; Daghistani, D.; Schalop, L.; Chi, A.S.; Oster, W.; et al. First Clinical Experience with DRD2/3 Antagonist ONC201 in H3 K27M–Mutant Pediatric Diffuse Intrinsic Pontine Glioma: A Case Report. J. Neurosurg. Pediatr. 2019. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Ribas, A.; Schachter, J.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.M.; Lotem, M.; et al. Pembrolizumab Versus Ipilimumab in Advanced Melanoma (KEYNOTE-006): Post-hoc 5-Year Results from an Open-Label, Multicentre, Randomised, Controlled, phase 3 Study. Lancet Oncol. 2019, 20, 1239–1251. [Google Scholar] [CrossRef]
- Wolchok, J.D.; Chiarion-Sileni, V.; Gonzalez, R.; Rutkowski, P.; Grob, J.-J.; Cowey, C.L.; Lao, C.D.; Wagstaff, J.; Schadendorf, D.; Ferrucci, P.F.; et al. Overall Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2017, 377, 1345–1356. [Google Scholar] [CrossRef]
- Omuro, A.; Vlahovic, G.; Lim, M.; Sahebjam, S.; Baehring, J.; Cloughesy, T.; Voloschin, A.; Ramkissoon, S.H.; Ligon, K.L.; Latek, R.; et al. Nivolumab with or Without Ipilimumab in Patients with Recurrent Glioblastoma: Results from Exploratory Phase i Cohorts of CheckMate. Neuro Oncol. 2018, 20, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Filley, A.C.; Henriquez, M.; Dey, M. Recurrent Glioma Clinical Trial, CheckMate-143: The Game Is Not Over Yet. Oncotarget 2017, 8, 91779–91794. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, M.; Hata, N.; Suzuki, S.O.; Fujioka, Y.; Murata, H.; Amano, T.; Nakamizo, A.; Yoshimoto, K.; Iwaki, T.; Sasaki, T. Pediatric Glioblastoma with Oligodendroglioma Component: Aggressive Clinical Phenotype with Distinct Molecular Characteristics. Neuropathology 2013, 33, 652–657. [Google Scholar] [CrossRef]
- Szybka, M.; Bartkowiak, J.; Zakrzewski, K.; Polis, L.; Liberski, P.P.; Kordek, R. Microsatellite Instability and Expression of DNA Mismatch Repair Genes in Malignant Astrocytic Tumors from Adult and Pediatric Patients. Clin. Neuropathol. 2003, 22, 180–186. [Google Scholar] [PubMed]
- Duffner, P.K.; Krischer, J.P.; Burger, P.C.; Cohen, M.E.; Backstrom, J.W.; Horowitz, M.E.; Sanford, R.; Friedman, H.; Kun, L. Treatment of Infants with Malignant Gliomas: The Pediatric Oncology Group Experience. J. Neurooncol. 1996, 28, 245–256. [Google Scholar] [CrossRef]
- Ater, J.L.; Zhou, T.; Holmes, E.; Mazewski, C.M.; Booth, T.N.; Freyer, D.R.; Lazarus, K.H.; Packer, R.J.; Prados, M.; Sposto, R.; et al. Randomized Study of Two Chemotherapy Regimens for Treatment of Low-Grade Glioma in Young Children: A Report from the Children’s Oncology Group. J. Clin. Oncol. 2012, 30, 2641–2647. [Google Scholar] [CrossRef] [PubMed]
- Mirow, C.; Pietsch, T.; Berkefeld, S.; Kwiecien, R.; Warmuth-Metz, M.; Falkenstein, F.; Diehl, B.; von Hornstein, S.; Gnekow, A.K. Children <1 year show an inferior outcome when treated according to the traditional LGG treatment strategy: A report from the german multicenter trial HIT-LGG 1996–2003 for children with low grade glioma (LGG). Pediatr. Blood Cancer 2014, 61, 457–463. [Google Scholar]
- Gnekow, A.K.; Walker, D.A.; Kandels, D.; Picton, S.; Perilongo, G.; Grill, J.; Stokland, T.; Sandstrom, P.E.; Warmuth-Metz, M.; Pietsch, T.; et al. A European Randomised Controlled Trial of the Addition of Etoposide to Standard Vincristine and Carboplatin Induction as Part of an 18-Month Treatment Programme for Childhood (≤16 Years) Low Grade Glioma—A Final Report. Eur. J. Cancer 2017, 81, 206–225. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro Stucklin, A.S.; Ryall, S.; Fukuoka, K.; Zapotocky, M.; Lassaletta, A.; Li, C.; Bridge, T.; Kim, B.; Arnoldo, A.; Kowalski, P.E.; et al. Alterations in ALK/ROS1/NTRK/MET Drive a Group of Infantile Hemispheric Gliomas. Nat. Commun. 2019, 10, 4343. [Google Scholar] [CrossRef]
- Vaishnavi, A.; Le, A.T.; Doebele, R.C. TRKing down an Old Oncogene in a New Era of Targeted Therapy. Cancer Discov. 2015, 5, 25–34. [Google Scholar] [CrossRef]
- Amatu, A.; Sartore-Bianchi, A.; Siena, S. NTRK Gene Fusions as Novel Targets of Cancer Therapy Across Multiple Tumour Types. ESMO Open 2016, 1, e000023. [Google Scholar] [CrossRef]
- Chao, M.V. Neurotrophins and Their Receptors: A Convergence Point for Many Signalling Pathways. Nat. Rev. Neurosci. 2003, 4, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Khotskaya, Y.B.; Holla, V.R.; Farago, A.F.; Mills Shaw, K.R.; Meric-Bernstam, F.; Hong, D.S. Targeting TRK Family Proteins in Cancer. Pharmacol. Ther. 2017, 173, 58–66. [Google Scholar] [CrossRef]
- Nguyen, N.; Lee, S.B.; Lee, Y.S.; Lee, K.H.; Ahn, J.Y. Neuroprotection by NGF and BDNF Against Neurotoxin-Exerted Apoptotic Death in Neural Stem Cells Are Mediated Through TRK Receptors, Activating PI3-Kinase and MAPK Pathways. Neurochem. Res. 2009, 34, 942–951. [Google Scholar] [CrossRef]
- Frattini, V.; Trifonov, V.; Chan, J.M.; Castano, A.; Lia, M.; Abate, F.; Keir, S.T.; Ji, A.X.; Zoppoli, P.; Niola, F.; et al. The Integrated Landscape of Driver Genomic Alterations in Glioblastoma. Nat. Genet. 2013, 45, 1141–1149. [Google Scholar] [CrossRef]
- Kurozumi, K.; Nakano, Y.; Ishida, J.; Tanaka, T.; Doi, M.; Hirato, J.; Yoshida, A.; Washio, K.; Shimada, A.; Kohno, T.; et al. High-Grade Glioneuronal Tumor with an ARHGEF2-NTRK1 Fusion Gene. Brain Tumor Pathol. 2019, 36, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, F.; Duplantier, M.M.; Trempat, P.; Hieblot, C.; Lamant, L.; Espinos, E.; Racaud-Sultan, C.; Allouche, M.; Campo, E.; Delsol, G.; et al. Differential Effects of X-ALK Fusion Proteins on Proliferation, Transformation, and Invasion Properties of NIH3T3 Cells. Oncogene 2004, 23, 6071–6082. [Google Scholar] [CrossRef]
- Aghajan, Y.; Levy, M.L.; Malicki, D.M.; Crawford, J.R. Novel PPP1CB-ALK Fusion Protein in a High-Grade Glioma of Infancy. BMJ Case Rep. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.; Levy, M.L.; Malicki, D.M.; Crawford, J.R. Unusual High-Grade and Low-Grade Glioma in an Infant with PPP1CB-ALK Gene Fusion. BMJ Case Rep. 2019, 12. [Google Scholar] [CrossRef]
- Olsen, T.K.; Panagopoulos, I.; Meling, T.R.; Micci, F.; Gorunova, L.; Thorsen, J.; Due-Tønnessen, B.; Scheie, D.; Lund-Iversen, M.; Krossnes, B.; et al. Fusion Genes with ALK as Recurrent Partner in Ependymoma-Like Gliomas: A New Brain Tumor Entity? Neuro. Oncol. 2015, 17, 1365–1373. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Koga, Y.; Ono, H.; Asai, H.; Ono, K.; Hatae, R.; Hata, N.; Mizoguchi, M.; Yamamoto, H.; Suzuki, O.S.; et al. High-grade glioma with a novel fusion gene of VCL-ALK. Neuro. Oncol. 2020, 22, iii348. [Google Scholar] [CrossRef]
- Coccé, M.C.; Mardin, B.R.; Bens, S.; Stütz, A.M.; Lubieniecki, F.; Vater, I.; Korbel, J.O.; Siebert, R.; Alonso, C.N.; Gallego, M.S. Identification of ZCCHC8 as Fusion Partner of ROS1 in a Case of Congenital Glioblastoma multiforme with a t(6;12) (q21;q24.3). Genes Chromosom. Cancer 2016, 55, 677–687. [Google Scholar] [CrossRef]
- Davare, M.A.; Henderson, J.J.; Agarwal, A.; Wagner, J.P.; Iyer, S.R.; Shah, N.; Woltjer, R.; Somwar, R.; Gilheeney, S.W.; DeCarvalo, A.; et al. Rare but Recurrent ROS1 Fusions Resulting from chromosome 6q22 Microdeletions Are Targetable Oncogenes in Glioma. Clin. Cancer Res. 2018, 24, 6471–6482. [Google Scholar] [CrossRef]
- Nakano, Y.; Tomiyama, A.; Kohno, T.; Yoshida, A.; Yamasaki, K.; Ozawa, T.; Fukuoka, K.; Fukushima, H.; Inoue, T.; Hara, J.; et al. Identification of a Novel KLC1-ROS1 Fusion in a Case of Pediatric Low-Grade Localized Glioma. Brain Tumor Pathol. 2019, 36, 14–19. [Google Scholar] [CrossRef]
- Drilon, A.; Somwar, R.; Wagner, J.P.; Vellore, N.A.; Eide, C.A.; Zabriskie, M.S.; Arcila, M.E.; Hechtman, J.F.; Wang, L.; Smith, R.S.; et al. A Novel Crizotinib-Resistant Solvent-Front Mutation Responsive to Cabozantinib Therapy in a Patient with ROS1-Rearranged Lung Cancer. Clin. Cancer Res. 2016, 22, 2351–2358. [Google Scholar] [CrossRef]
- Shaw, A.T.; Ou, S.-H.I.; Bang, Y.-J.; Camidge, D.R.; Solomon, B.J.; Salgia, R.; Riely, G.J.; Varella-Garcia, M.; Shapiro, G.I.; Costa, D.; et al. Crizotinib in ROS1-Rearranged Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2014, 371, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Siena, S.; Ou, S.H.I.; Patel, M.; Ahn, M.J.; Lee, J.; Bauer, T.M.; Farago, A.F.; Wheler, J.J.; Liu, S.V.; et al. Safety and Antitumor Activity of the Multitargeted Pan-TRK, ROS1, and ALK Inhibitor Entrectinib: Combined Results from Two Phase I Trials (ALKA-372-001 and STARTRK-1). Cancer Discov. 2017, 7, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Lassaletta, A.; Guerreiro Stucklin, A.; Ramaswamy, V.; Zapotocky, M.; McKeown, T.; Hawkins, C.; Bouffet, E.; Tabori, U. Profound Clinical and Radiological Response to BRAF Inhibition in a 2-Month-Old Diencephalic Child with Hypothalamic/Chiasmatic Glioma. Pediatr. Blood Cancer 2016, 63, 2038–2041. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Jakacki, R.I.; Onar-Thomas, A.; Wu, S.; Nicolaides, T.; Young Poussaint, T.; Fangusaro, J.; Phillips, J.; Perry, A.; Turner, D.; et al. A phase i Trial of the MEK Inhibitor Selumetinib (AZD6244) in Pediatric Patients with Recurrent or Refractory Low-Grade Glioma: A Pediatric Brain Tumor Consortium (PBTC) Study. Neuro. Oncol. 2017, 19, 1135–1144. [Google Scholar] [CrossRef] [PubMed]
| Molecular Alteration | Function | Tumor Type | Potential Biomarker |
|---|---|---|---|
| pLGG (RAS/MAPK pathway) | |||
| KIAA1549-BRAF fusion | Activation of BRAF kinase domain Deregulation of the RAS/MAPK pathway | PA | Diagnostic marker Poor prognostic marker |
| BRAF V600E mutation | PA/PXA/GG/DA | ||
| FGFR1 | Upregulation of the RAS/MAPK pathway | CNS tumors | NA |
| NF-1 | Negative regulator of RAS | PA/DA | NA |
| pLGG (non-RAS/MAPK pathway) | |||
| MYB and MYBL1 | Control of proliferation and differentiation of hematopoietic and other progenitor cells | DA | NA |
| CDKN2A homozygous deletion | Non-coding of the gene for tumor suppressors, protein p14ARF and p16INK4A | PXA | Poor prognostic marker |
| pHGG | |||
| H3K27M mutation | Decrease levels of lysine 27 methylation | GBM/DIPG | Diagnostic marker Poor prognostic marker |
| H3G34R/V mutation | Changes the distribution of lysine 36 methylation | ||
| Infantile glioma | |||
| NTRK fusions | Upregulation of the RAS/MAPK and PI3K/AKT/mTOR pathways | Hemispheric HGG | Intermediate prognostic marker |
| ALK fusions | |||
| ROS1 fusions | |||
| Author | Year | ClinicalTrials.gov ID | Phase | Patients | Disease | Molecular Target | Treatment | Outcome |
|---|---|---|---|---|---|---|---|---|
| LGG | ||||||||
| Fangusaro et al. [68] | 2019 | NCT01089101 | II | 3–21 y n = 38 | Recurrent/refractory LGG | MEK | Selumetinib | Positive antitumor activity Well-tolerated |
| Hargrave et al. [62] | 2019 | NCT01677741 | I/II | 2–18 y n = 32 | Recurrent/refractory LGG | BRAF V600E mutant | Dabrafenib | Positive antitumor activity Well-tolerated |
| Nicolaides et al. [70] | 2020 | NCT01748149 | I | 3–17 y n = 19 | Recurrent/refractory gliomas | BRAF V600E mutant | Vemurafenib | Positive antitumor activity Well-tolerated |
| HGG | ||||||||
| Wetmore et al. [73] | 2016 | NCT01462695 | II | 18 m–22 y n = 30 | HGG or ependymoma | VEGFR PDGFR KIT | Sunitinib | No antitumor activity Well-tolerated |
| Becher et al. [74] | 2017 | NCT01049841 | I | 4–24 y n = 23 | Recurrent/refractory brain tumor | AKT mTOR | Perifosine Temsirolimus | Well-tolerated |
| Broniscer et al. [75] | 2018 | NCT01644773 | I | 2–21 y n = 25 | Recurrent/progressive HGG or DIPG | PDGFRA c-Met | Dasatinib Crizotinib | Minimal antitumor activity Poorly tolerated |
| Chi et al. [76] | 2019 | NCT03134131 | II | 3–42 y n = 18 | H3K27M mutant diffuse midline glioma/DIPG | DRD2/3 | ONC201 | Positive antitumor activity |
| ClinicalTrials.gov ID | Phase | Patients | Disease | Molecular Target | Treatment | Status |
|---|---|---|---|---|---|---|
| RAS/MAPK pathway targeted therapy | ||||||
| NCT01734512 | II | 3–21 y | Recurrent/progressive LGG | mTOR | Everolimus | Active, not recruiting |
| NCT01748149 | I | Up to 25 y | Recurrent/refractory glioma | BRAF V600E mutant | Vemurafenib | Active, not recruiting |
| NCT02684058 | II | 12 m–17 y | LGG or relapsed/refractory HGG | BRAF V600E mutant MEK | Dabrafenib Trametinib | Recruiting |
| NCT03363217 | I/II | 1 m–25 y | NF-1, Recurrent/refractory LGG | MAPK/ERK pathway BRAF fusion | Trametinib | Recruiting |
| NCT04485559 | I | 1–25 y | Recurrent grade 2 glioma | MAPK/ERK pathway mTOR | Trametinib Everolimus | Recruiting |
| NCT03429803 | I | 1–25 y | Recurrent/progressive LGG | BRAF fusion | TAK-580 | Recruiting |
| NCT02285439 | I/II | 1–18 y | NF-1, Recurrent/refractory LGG | MEK | MEK162 | Recruiting |
| NCT03696355 | I | 2–21 y | DIPC or other diffuse midline H3K27M mutant gliomas | PI3K/Akt/mTOR | GDC-0084 | Active, not recruiting |
| NCT02650401 | I/II | Up to 18 y | CNS tumor | NTRK or ROS1 fusion | Entrectinib | Recruiting |
| NCT04655404 | I | Up to 21 y | Newly diagnosed HGG | NTRK fusion | Larotrectinib | Not yet recruiting |
| Other targeted therapy | ||||||
| NCT03749187 | I | 13–25 y | Gliomas, IDH1/2 mutant | PARP | BGB-290 + TMZ | Recruiting |
| NCT03416530 | I | 2–18 y | Newly diagnosed DIPG Recurrent/refractory H3K27M gliomas | DRD2 | ONC201 | Recruiting |
| NCT01922076 | I | 37 m–21 y | Newly diagnosed DIPG | Tyrosine kinase WEE1 | Adavosertib | Active, not recruiting |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Funakoshi, Y.; Hata, N.; Kuga, D.; Hatae, R.; Sangatsuda, Y.; Fujioka, Y.; Takigawa, K.; Mizoguchi, M. Pediatric Glioma: An Update of Diagnosis, Biology, and Treatment. Cancers 2021, 13, 758. https://doi.org/10.3390/cancers13040758
Funakoshi Y, Hata N, Kuga D, Hatae R, Sangatsuda Y, Fujioka Y, Takigawa K, Mizoguchi M. Pediatric Glioma: An Update of Diagnosis, Biology, and Treatment. Cancers. 2021; 13(4):758. https://doi.org/10.3390/cancers13040758
Chicago/Turabian StyleFunakoshi, Yusuke, Nobuhiro Hata, Daisuke Kuga, Ryusuke Hatae, Yuhei Sangatsuda, Yutaka Fujioka, Kosuke Takigawa, and Masahiro Mizoguchi. 2021. "Pediatric Glioma: An Update of Diagnosis, Biology, and Treatment" Cancers 13, no. 4: 758. https://doi.org/10.3390/cancers13040758
APA StyleFunakoshi, Y., Hata, N., Kuga, D., Hatae, R., Sangatsuda, Y., Fujioka, Y., Takigawa, K., & Mizoguchi, M. (2021). Pediatric Glioma: An Update of Diagnosis, Biology, and Treatment. Cancers, 13(4), 758. https://doi.org/10.3390/cancers13040758






