LC-MS/MS-Based Proteomics Approach for the Identification of Candidate Serum Biomarkers in Patients with Narcolepsy Type 1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
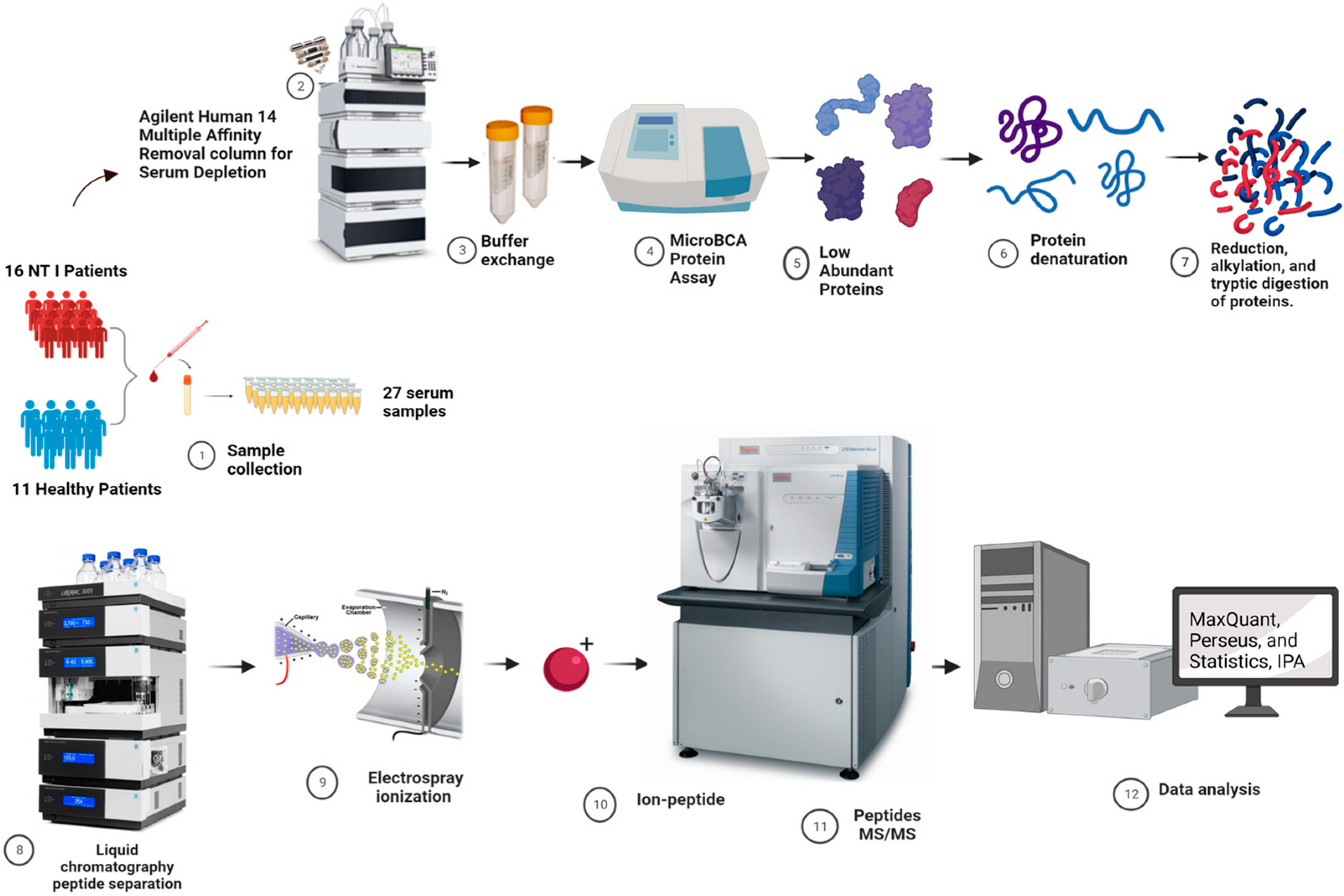
2.2. Study Participants
2.3. Depletion of High-Abundance Proteins in Serum Samples
2.4. Tryptic Digestion
2.5. Identification of Potential Biomarkers Using Untargeted LC-MS/MS Proteomics Analysis
2.6. Untargeted LC-MS/MS Data Analysis
2.7. Ingenuity Pathway Analysis (IPA) and Gene Ontology for System Biology
2.8. Subcellular Localization
2.9. Targeted Proteomics (LC-PRM-MS) Strategy
3. Results
3.1. Unsupervised PCA for Comparative Proteomics Analysis of NT1 and Control Samples
3.2. Heatmap of DEPs
3.3. Sex and Age-Based Comparison and Volcano Plot
3.4. Gene Ontology for the Untargeted Proteomics Result
3.5. PRM Validates DEPs
3.6. GO, KEGG and Subcellular Localization of the Three Quantitatively Validated DEPs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABC | ammonium bicarbonate |
| ACN | acetonitrile |
| NT1 | narcolepsy type 1 |
| FDR | false discovery rate |
| DTT | dithiothreitol |
| HLA | human leukocyte antigen |
| FA | formic acid |
| IAA | iodoacetamide |
| IPA | ingenuity pathway analysis |
| DEPs | differentially expressed proteins |
| LFQP | label-free quantitation proteomics |
| PCA | principal component analysis |
| HPLC | high-performance liquid chromatography |
| KEGG | Kyoto encylopedia of genes and genomes |
References
- Kornum, B.R.; Knudsen, S.; Ollila, H.M.; Pizza, F.; Jennum, P.J.; Dauvilliers, Y.; Overeem, S. Narcolepsy. Nat. Rev. Dis. Prim. 2017, 3, 16100. [Google Scholar] [CrossRef]
- Bassetti, C.L.A.; Adamantidis, A.; Burdakov, D.; Han, F.; Gay, S.; Kallweit, U.; Khatami, R.; Koning, F.; Kornum, B.R.; Lammers, G.J.; et al. Narcolepsy—Clinical spectrum, aetiopathophysiology, diagnosis and treatment. Nat. Rev. Neurol. 2019, 15, 519–539. [Google Scholar] [CrossRef] [PubMed]
- Tsujino, N.; Sakurai, T. Orexin/hypocretin: A neuropeptide at the interface of sleep, energy homeostasis, and reward system. Pharmacol. Rev. 2009, 61, 162–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammadi, S.; Dolatshahi, M.; Zare-Shahabadi, A.; Rahmani, F. Untangling narcolepsy and diabetes: Pathomechanisms with eyes on therapeutic options. Brain Res. 2019, 1718, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Yoss, R.E.; Daly, D.D. Criteria for the diagnosis of the narcoleptic syndrome. Proc. Staff Meet. Mayo Clin. 1957, 32, 320–328. [Google Scholar]
- Sturzenegger, C.; Bassetti, C.L. The clinical spectrum of narcolepsy with cataplexy: A reappraisal. J. Sleep Res. 2004, 13, 395–406. [Google Scholar] [CrossRef]
- Han, F.; Lin, L.; Schormair, B.; Pizza, F.; Plazzi, G.; Ollila, H.M.; Nevsimalova, S.; Jennum, P.; Knudsen, S.; Winkelmann, J.; et al. HLA DQB1*06:02 negative narcolepsy with hypocretin/orexin deficiency. Sleep 2014, 37, 1601–1608. [Google Scholar] [CrossRef]
- Sun, X.; Sun, H.; Han, X.; Chen, P.C.; Jiao, Y.; Wu, Z.; Zhang, X.; Wang, Z.; Niu, M.; Yu, K.; et al. Deep Single-Cell-Type Proteome Profiling of Mouse Brain by Nonsurgical AAV-Mediated Proximity Labeling. Anal. Chem. 2022, 94, 5325–5334. [Google Scholar] [CrossRef]
- Kulasingam, V.; Diamandis, E.P. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat. Clin. Pract. Oncol. 2008, 5, 588–599. [Google Scholar] [CrossRef]
- Srinivas, P.R.; Verma, M.; Zhao, Y.; Srivastava, S. Proteomics for cancer biomarker discovery. Clin. Chem. 2002, 48, 1160–1169. [Google Scholar]
- Zhong, L.; Zhu, L.; Cai, Z.W. Mass Spectrometry-based Proteomics and Glycoproteomics in COVID-19 Biomarkers Identification: A Mini-review. J. Anal. Test 2021, 5, 298–313. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.; Zhao, J.; Peng, W.; Banazadeh, A.; Williamson, S.D.; Goli, M.; Huang, Y.; Mechref, Y. Advances in mass spectrometry-based glycoproteomics. Electrophoresis 2018, 39, 3104–3122. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhu, G.; Yan, X.; Dovichi, N.J. High sensitivity capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry for the rapid analysis of complex proteomes. Curr. Opin. Chem. Biol. 2013, 17, 795–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pejchinovski, M.; Hrnjez, D.; Ramirez-Torres, A.; Bitsika, V.; Mermelekas, G.; Vlahou, A.; Zürbig, P.; Mischak, H.; Metzger, J.; Koeck, T. Capillary zone electrophoresis on-line coupled to mass spectrometry: A perspective application for clinical proteomics. Proteom. Clin. Appl. 2015, 9, 453–468. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Jemal, M. High-throughput quantitative bioanalysis by LC/MS/MS. Biomed Chromatogr. 2000, 14, 422–429. [Google Scholar] [CrossRef]
- Gallien, S.; Duriez, E.; Demeure, K.; Domon, B. Selectivity of LC-MS/MS analysis: Implication for proteomics experiments. J. Proteom. 2013, 81, 148–158. [Google Scholar] [CrossRef]
- Lindström, M.; Schinkelshoek, M.; Tienari, P.J.; Kukkonen, J.P.; Renkonen, R.; Fronczek, R.; Lammers, G.J.; Itkonen, O. Orexin-A measurement in narcolepsy: A stability study and a comparison of LC-MS/MS and immunoassays. Clin. Biochem. 2021, 90, 34–39. [Google Scholar] [CrossRef]
- Azzam, S.; Schlatzer, D.; Nethery, D.; Saleh, D.; Li, X.; Akladious, A.; Chance, M.R.; Strohl, K.P. Proteomic profiling of the hypothalamus in two mouse models of narcolepsy. Proteomics 2017, 17, 1600478. [Google Scholar] [CrossRef]
- Cartwright, R.D. Alcohol and NREM parasomnias: Evidence versus opinions in the international classification of sleep disorders, 3rd edition. J. Clin. Sleep Med. 2014, 10, 1039–1040. [Google Scholar] [CrossRef] [Green Version]
- Mondello, S.; Kobeissy, F.; Mechref, Y.; Zhao, J.; Talih, F.R.; Cosentino, F.; Antelmi, E.; Moresco, M.; Plazzi, G.; Ferri, R. Novel biomarker signatures for idiopathic REM sleep behavior disorder: A proteomic and system biology approach. Neurology 2018, 91, e1710–e1715. [Google Scholar] [CrossRef] [PubMed]
- Mondello, S.; Kobeissy, F.H.; Mechref, Y.; Zhao, J.; El Hayek, S.; Zibara, K.; Moresco, M.; Plazzi, G.; Cosentino, F.I.I.; Ferri, R. Searching for Novel Candidate Biomarkers of RLS in Blood by Proteomic Analysis. Nat. Sci. Sleep 2021, 13, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.L.; Song, J.Y.; Sun, Z.G. Quantitative label-free proteomic analysis of human follicle fluid to identify novel candidate protein biomarker for endometriosis-associated infertility. J. Proteom. 2022, 266, 104680. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic. Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rauniyar, N. Parallel Reaction Monitoring: A Targeted Experiment Performed Using High Resolution and High Mass Accuracy Mass Spectrometry. Int. J. Mol. Sci. 2015, 16, 28566–28581. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez Reyes, C.D.; Huang, Y.; Atashi, M.; Zhang, J.; Zhu, J.; Liu, S.; Parikh, N.D.; Singal, A.G.; Dai, J.; Lubman, D.M.; et al. PRM-MS Quantitative Analysis of Isomeric N-Glycopeptides Derived from Human Serum Haptoglobin of Patients with Cirrhosis and Hepatocellular Carcinoma. Metabolites 2021, 11, 563. [Google Scholar] [CrossRef]
- Tsai, T.H.; Song, E.; Zhu, R.; Di Poto, C.; Wang, M.; Luo, Y.; Varghese, R.S.; Tadesse, M.G.; Ziada, D.H.; Desai, C.S.; et al. LC-MS/MS-based serum proteomics for identification of candidate biomarkers for hepatocellular carcinoma. Proteomics 2015, 15, 2369–2381. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Valverde, A. Insights into the area under the receiver operating characteristic curve (AUC) as a discrimination measure in species distribution modelling. Glob. Ecol. Biogeogr. 2012, 21, 498–507. [Google Scholar] [CrossRef]
- Straat, M.E.; Schinkelshoek, M.S.; Fronczek, R.; Lammers, G.J.; Rensen, P.C.N.; Boon, M.R. Role of Brown Adipose Tissue in Adiposity Associated With Narcolepsy Type 1. Front Endocrinol. 2020, 11, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barateau, L.; Liblau, R.; Peyron, C.; Dauvilliers, Y. Narcolepsy Type 1 as an Autoimmune Disorder: Evidence, and Implications for Pharmacological Treatment. CNS Drugs 2017, 31, 821–834. [Google Scholar] [CrossRef] [PubMed]
- Yüceer, H.; Gezen Ak, D.; Şenel, G.B.; Dursun, E.; Yılmaz, V.; Karadeniz, D.; Tüzün, E.; Küçükali, C. Classical complement pathway factor alterations in narcolepsy. Acta Neuropsychiatr. 2022, 34, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Rose, N.R.; Bona, C. Defining criteria for autoimmune diseases (Witebsky’s postulates revisited). Immunol. Today 1993, 14, 426–430. [Google Scholar] [CrossRef]
- Latorre, D.; Kallweit, U.; Armentani, E.; Foglierini, M.; Mele, F.; Cassotta, A.; Jovic, S.; Jarrossay, D.; Mathis, J.; Zellini, F.; et al. T cells in patients with narcolepsy target self-antigens of hypocretin neurons. Nature 2018, 562, 63–68. [Google Scholar] [CrossRef]
- Giannoccaro, M.P.; Liguori, R.; Plazzi, G.; Pizza, F. Reviewing the Clinical Implications of Treating Narcolepsy as an Autoimmune Disorder. Nat. Sci. Sleep 2021, 13, 557–577. [Google Scholar] [CrossRef]
- de Jesus, J.R.; de Campos, B.K.; Galazzi, R.M.; Martinez, J.L.; Arruda, M.A. Bipolar disorder: Recent advances and future trends in bioanalytical developments for biomarker discovery. Anal. Bioanal. Chem. 2015, 407, 661–667. [Google Scholar] [CrossRef]
- Polaskova, V.; Kapur, A.; Khan, A.; Molloy, M.P.; Baker, M.S. High-abundance protein depletion: Comparison of methods for human plasma biomarker discovery. Electrophoresis 2010, 31, 471–482. [Google Scholar] [CrossRef]
- Araújo, J.E.; Lodeiro, C.; Capelo, J.L.; Rodríguez-González, B.; dos Santos, A.A.; Santos, H.M.; Fernández-Lodeiro, J. Novel nanocomposites based on a strawberry-like gold-coated magnetite (Fe@Au) for protein separation in multiple myeloma serum samples. Nano Res. 2015, 8, 1189–1198. [Google Scholar] [CrossRef]
- Wang, J.; Tong, L.; Lin, G.; Wang, H.; Zhang, L.; Yang, X. Immunological and clinicopathological characteristics of C1RL in 2120 glioma patients. BMC Cancer 2020, 20, 931. [Google Scholar] [CrossRef]
- Fan, S.; Wu, N.; Chang, S.; Chen, L.; Sun, X. The immune regulation of BCL3 in glioblastoma with mutated IDH1. Aging Albany N. Y. 2022, 14, 3856–3873. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Feng, T.; Cao, W.; Yu, H.; Zhang, Z. Identification of Key Genes in Nasopharyngeal Carcinoma Based on Bioinformatics Analysis. Comput. Intell. Neurosci. 2022, 2022, 9022700. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhou, X.; Yang, L.; Zhou, H.; Meng, M.; Wu, H.; Liu, Z.; Zhang, L.; Li, C. Glioma stem cell signature predicts the prognosis and the response to tumor treating fields treatment. CNS Neurosci. 2022, 28, 2148–2162. [Google Scholar] [CrossRef] [PubMed]
- Mogavero, M.P.; Silvani, A.; DelRosso, L.M.; Salemi, M.; Ferri, R. Focus on the Complex Interconnection between Cancer, Narcolepsy and Other Neurodegenerative Diseases: A Possible Case of Orexin-Dependent Inverse Comorbidity. Cancers 2021, 13, 2612. [Google Scholar] [CrossRef]
- Gadjeva, M.; Kugadas, A.; Petenkova, A.; Geddes-McAlister, J.; Mansour, M.K.; Sykes, D. Neutrophil maturation and their response to infectious pathogens are regulated by microbiota. J. Immunol. 2019, 202, 127-22. [Google Scholar] [CrossRef]
- Tsujino, N.; Sakurai, T. Role of orexin in modulating arousal, feeding, and motivation. Front. Behav. Neurosci. 2013, 7, 28. [Google Scholar] [CrossRef] [Green Version]
- Tsuneki, H.; Murata, S.; Anzawa, Y.; Soeda, Y.; Tokai, E.; Wada, T.; Kimura, I.; Yanagisawa, M.; Sakurai, T.; Sasaoka, T. Age-related insulin resistance in hypothalamus and peripheral tissues of orexin knockout mice. Diabetologia 2008, 51, 657–667. [Google Scholar] [CrossRef] [Green Version]
- Bonvalet, M.; Ollila, H.M.; Ambati, A.; Mignot, E. Autoimmunity in narcolepsy. Curr. Opin. Pulm. Med. 2017, 23, 522–529. [Google Scholar] [CrossRef]
- Berczi, I.; Stephano, A.Q. Chapter 8—Vasopressin, the Acute Phase Response and Healing. In Insights to Neuroimmune Biology, 2nd ed.; Berczi, I., Ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 185–199. [Google Scholar] [CrossRef]
- Zhang, W.; Tettamanti, G.; Bassal, T.; Heryanto, C.; Eleftherianos, I.; Mohamed, A. Regulators and signalling in insect antimicrobial innate immunity: Functional molecules and cellular pathways. Cell Signal 2021, 83, 110003. [Google Scholar] [CrossRef]
- Chang, J.R.; Ghafouri, M.; Mukerjee, R.; Bagashev, A.; Chabrashvili, T.; Sawaya, B.E. Role of p53 in neurodegenerative diseases. Neuro-Degener. Dis. 2012, 9, 68–80. [Google Scholar] [CrossRef] [Green Version]
- Conigliaro, P.; Triggianese, P.; Ballanti, E.; Perricone, C.; Perricone, R.; Chimenti, M.S. Complement, infection, and autoimmunity. Curr. Opin. Rheumatol. 2019, 31, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Mogavero, M.P.; DelRosso, L.M.; Bruni, O.; Salemi, M.; Salsone, M.; Novellino, F.; Zucconi, M.; Ferini Strambi, L.; Ferri, R. Genetics and Epigenetics of Rare Hypersomnia. A Possible Keystone for their Diagnosis and Treatment within the Framework of Precision Medicine. Trends Genet. TIG 2023, in press. [Google Scholar]



| Group Information | Narcolepsy Type 1 | Controls |
|---|---|---|
| Sample number | 16 | 11 |
| Sex | 11 males, 5 females | 7 males, 4 females |
| Age range (years) | 19–71 | 28–73 |
| HLA DQB1*0602 allele | 15 samples | 2 Samples |
| Cerebrospinal Fluid Orexin, pg/mL | 0–119.6 (Undetectable in 6 patients, not available in 3 patients) | Not available |
| Body Mass Index | 20.2–28.6 | Not available |
| Protein Accession | Gene Name | p-Value | Fold Change | ROC/AUC |
|---|---|---|---|---|
| Downregulated Proteins | Combined AUC = 0.95 | |||
| O14786 | NRP1 | 0.0001 | 0.83 | 0.80 |
| P00450 | CP | 0.0002 | 0.82 | 0.80 |
| P00734 | F2 | 0.0008 | 0.86 | 0.81 |
| P00736 | C1R | 0.001 | 0.88 | 0.81 |
| P00742 | F10 | 0.002 | 0.80 | 1.0 |
| P00748 | F12 | 0.002 | 0.68 | 0.91 |
| P00751 | CFB | 0.003 | 0.83 | 0.74 |
| P01031 | C5 | 0.005 | 0.87 | 0.77 |
| P02749 | APOH | 0.006 | 0.77 | 0.76 |
| P02760 | AMBP | 0.007 | 0.89 | 0.76 |
| P02774 | GC | 0.008 | 0.76 | 0.87 |
| P04217 | A1BG | 0.009 | 0.86 | 0.87 |
| P06681 | C2 | 0.009 | 0.82 | 0.81 |
| P07225 | PROS | 0.009 | 0.79 | 0.81 |
| P08185 | CBG | 0.009 | 0.44 | 0.85 |
| P09871 | C1S | 0.009 | 0.84 | 0.85 |
| P12111 | COL6A3 | 0.010 | 0.54 | 0.77 |
| P14151 | SELL | 0.010 | 0.75 | 0.74 |
| P17936 | IGFBP3 | 0.009 | 0.74 | 0.78 |
| P19823 | ITIH2 | 0.01 | 0.88 | 0.79 |
| P19827 | ITIH1 | 0.02 | 0.88 | 0.82 |
| P25311 | AZGP1 | 0.02 | 0.77 | 0.85 |
| P33908 | MAN1A1 | 0.02 | 0.61 | 0.76 |
| P35858 | IGFALS (ALS) | 0.03 | 0.75 | 0.76 |
| P48740 | MASP1 | 0.03 | 0.78 | 0.80 |
| P54108 | CRIS3 | 0.03 | 0.82 | 0.73 |
| Q03591 | CFHR1 | 0.04 | 0.64 | 0.74 |
| Q14624 | ITIH4 | 0.04 | 0.89 | 0.73 |
| Q15485 | FCN2 | 0.04 | 0.78 | 0.73 |
| Q6EMK4 | VASN | 0.04 | 0.81 | 0.84 |
| Q96IY4 | CPB2 | 0.04 | 0.85 | 0.77 |
| Q9BXR6 | CFHR5 | 0.04 | 0.79 | 0.73 |
| Upregulated Proteins | Combined AUC = 0.76 | |||
| Q9NZP8 | C1RL | 0.04 | 1.08 | 0.45 |
| P14543 | NID1 | 0.03 | 2.27 | 0.60 |
| P02751 | FN1 | 0.006 | 1.61 | 0.69 |
| Q9UHG3 | PCYOX | 0.0001 | 1.49 | 0.68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanni, A.; Goli, M.; Zhao, J.; Wang, J.; Barsa, C.; El Hayek, S.; Talih, F.; Lanuzza, B.; Kobeissy, F.; Plazzi, G.; et al. LC-MS/MS-Based Proteomics Approach for the Identification of Candidate Serum Biomarkers in Patients with Narcolepsy Type 1. Biomolecules 2023, 13, 420. https://doi.org/10.3390/biom13030420
Sanni A, Goli M, Zhao J, Wang J, Barsa C, El Hayek S, Talih F, Lanuzza B, Kobeissy F, Plazzi G, et al. LC-MS/MS-Based Proteomics Approach for the Identification of Candidate Serum Biomarkers in Patients with Narcolepsy Type 1. Biomolecules. 2023; 13(3):420. https://doi.org/10.3390/biom13030420
Chicago/Turabian StyleSanni, Akeem, Mona Goli, Jingfu Zhao, Junyao Wang, Chloe Barsa, Samer El Hayek, Farid Talih, Bartolo Lanuzza, Firas Kobeissy, Giuseppe Plazzi, and et al. 2023. "LC-MS/MS-Based Proteomics Approach for the Identification of Candidate Serum Biomarkers in Patients with Narcolepsy Type 1" Biomolecules 13, no. 3: 420. https://doi.org/10.3390/biom13030420
APA StyleSanni, A., Goli, M., Zhao, J., Wang, J., Barsa, C., El Hayek, S., Talih, F., Lanuzza, B., Kobeissy, F., Plazzi, G., Moresco, M., Mondello, S., Ferri, R., & Mechref, Y. (2023). LC-MS/MS-Based Proteomics Approach for the Identification of Candidate Serum Biomarkers in Patients with Narcolepsy Type 1. Biomolecules, 13(3), 420. https://doi.org/10.3390/biom13030420










